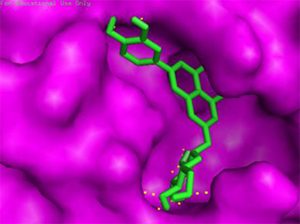
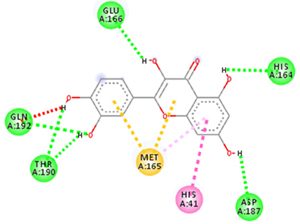
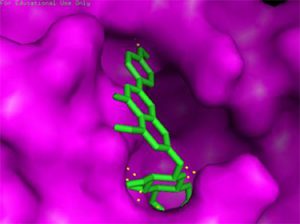
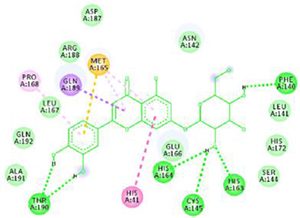
Coronavirus Disease (COVID-19) is a new disease that emerged in Wuhan, China which spreads through close contact of people, often by small droplets produced during coughing or sneezing. Detail mechanism by which it spreads between people are under investigation. The World Health Organization (WHO) declared this disease as a pandemic after the severity of the disease increased. Many scientific reports gathered have suggested many drugs that could be potential candidates for the treatment. Although, clinical effectiveness has not been fully evaluated. In this review, we have aggregated the data from few research articles, official news websites and few review papers regarding its phylogenetic relation, genomic constitution, transmission, replication and in-silico analysis done by researchers for few potent drugs that are currently used to cure COVID-19. SARS-CoV-2 belongs to Betacoronavirus genus with Genome structure consists 14 Open Reading Frames (ORFs) that encode 27 proteins. Coronavirus replicates into the host cells having unique mechanisms like ribosome frame-shifting and synthesis of genomic and sub genomic RNAs. In-silico methods have the advantage that they can make fast predictions for a large set of compounds in a high-throughput mode and also make their prediction based on the structure of a compound even before it has been synthesized. In-silico softwares have been used to find or to improve a novel bioactive compound, which may exhibit a strong affinity to a particular target in the drug development process.
COVID-19, SARS-CoV-2, SARS-CoV, MERS-CoV, Pandemic, WHO
An outbreak of coronavirus disease was reported in China in December, 2019. A novel strain of coronavirus was identified as a causative agent of the disease. As the species was identified as a new species, it was named as 2019 novel coronavirus (2019-nCoV, where n stands for novel). Since the virus is showing similar genetic structure and symptoms as SARS-CoV which was responsible for the SARS (Severe Acute Respiratory Syndrome) epidemic 2003, the virus was also named as SARS-CoV-2 by Chinese Center of Disease Control and Prevention (CCDC)1. The rapid spread of the COVID-19, Coronavirus Disease-2019 has caused worldwide fear in almost 213 countries. This is the third zoonotic outbreak of last two decades following SARS-CoV (Severe Acute Respiratory Syndrome-CoV, 2003) and MERS-CoV(Middle East Respiratory Syndrome-CoV, 2012) that spread through human to human transmission by physical contact1. Depending upon the severity of the global spread of the disease, The World Health Organization officially declared COVID-19 as a pandemic and Public Health Emergency of International Concern on 30thJanuary 20202. Reproductive number (R0) represents the transmissibility of the infectious disease, as virus transmits to new people it reproduces itself. If R0 is >1 then infection is likely to be increased and if R0 is <1 then the infection is likely to decline within a short period. R0 of SARS-CoV-2 is ranging from 2 to 2.5 which is higher than SARS-CoV (1.7 to 1.9) and MERS-CoV (<1), due to this novel coronavirus can easily spread among people(Petrosillo, Viceconte, Ergonul, Ippolito, & Petersen et al., 2020). Coronavirus is the genus of enveloped, positive sense, ssRNA Coronaviridae family which has spike proteins on the outer surface. These spike proteins play an important role in transmission of the virus into the host cells4. SARS-CoV and SARS-CoV-2 shares same human receptor that is Angiotensin Converting Enzyme 2 (ACE2), an ectoenzyme anchored on the plasma membrane of the cell of several tissues of lower respiratory tract, heart, kidney, gastrointestinal tract. Spike proteins of the virus bind to the protein receptors on the plasma membrane, which facilitates insertion of viral RNA into the cytoplasm of the host cell. Inoculation of 2019-nCoV onto the surface layers of the airway highly replicates type I and type II pneumocytes which down regulates ACE2 receptors and causes acute lung injury and ARDS (Acute Respiratory Distress Syndrome)3,5. No potent drugs are available for this novel coronavirus but, there are some drugs which can be tried for instance. The main viral protease MPro has been recently used as a target for drug designing against SARS infection due to its main role in processing the polyproteins which are necessary for the viral reproduction6. Several dockings have been done for various drugs which are discussed in this review. There are no vaccines or antiviral treatments available till date. People with infection are being analyzed and kept in surveillance. Here we tried to concise the information about SARS-CoV-2 and its similarity with SARS and MERS, origin of the virus, epidemiology, and replication cycle of the virus and comparison of drugs available till date.
Virology and Origin
Betacoronavirus is the genus of the Coronaviridae family in Nidovirales order (“Betacoronavirus ~ ViralZone page,”). Betacoronavirus is single stranded, positive sense (+) RNA viruses of approximately 26-32 kilobases genome length. The word “coronavirus” is derived from Greek word ‘κορwνη’ (korone) which means ‘crown or halo’ referring to the structural appearance of the virus having fringed reminiscent of a solar corona. Coronaviruses have four structural proteins spike (S), membrane (M), envelope (E) and nucleocapsid (N)4,6. Spike proteins are approximately 20 nm in size located on the peripheral region of the virus. Betacoronavirus is the genus which has 2 sub-families(Letovirinae, Orthocoronavirinae), 5 genera(Alphaletovirus, Alphacoronavirus, Betacoronavirus, Deltacoronavirus and Gammacoronavirus), 23 sub-genera and about 40 species8. Coronavirus can cause Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) in mammals and birds. Betacoronavirus is associated with respiratory infections such as common cold and typically mild fever.
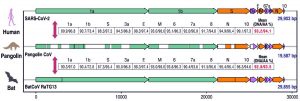
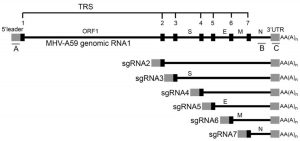
Genome structure of coronaviruse consists 14 Open Reading Frames (ORFs) that encode 27 proteins, ORF1 and ORF2 at the 5’ end encode for proteins important in replication while 3’ region of the genome encodes for structural proteins like S, M, E and N as shown in Fig. (1)9.
Phylogenetic tree analysis showed that the novel coronavirus SARS-CoV-2 is similar to SARS-CoV and Bat SARS-like coronavirus of horseshoe bat from China while MERS-CoV is in different clad of the tree showing lesser similarity which suggests viral evolution in SARS and MERS11. Genomic studies of SARS-CoV-2 and SARS-CoV have shown that there are only 380 amino acids substitutions between them which are concentrated in the nonstructural region that is responsible for the replication of the virus, while 27 mutations are found in the structural region of the genome which encodes for structural proteins essential for the virus entry into the host cell11. As SARS-CoV and MERS-CoV are known to be transmitted from bats to humans via palm civets, SARS-CoV-2 must have an intermediate host between bat and human12. Based on the Simplot analysis, Pangolin genome showed highest similarity to SARS-CoV-2 (91.02%) throughout the genome. There were two preprints supporting this studies saying CoV in pangolins shared 93.2% amino acid and 93.1% DNA identity with the SARS-CoV-2 as shown in Fig. (2).
Fig. 2. Comparison of common genome organization of SARS-CoV-2, Pangolin-CoV and Bat-CoV Reproduced with permission from [13]
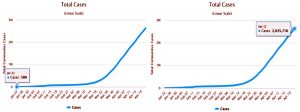
Fig. 3. Number of total cases reported in the starting of year till the date around the globe Source: [16]
Comparison of genome organization of Human SARS-CoV-2, Pangolin CoV and Bat CoV was done. As a result, Bat CoV genome showed 92.8% nucleotide similarity and 93.5% amino acid similarity to human SARS-CoV-2 while pangolin CoV showed 93.2% nucleotide and 94.1% amino acid similarity to SARS-CoV-2. Hence, pangolins were suggested as an intermediate host in the transmission route of SARS-CoV-2 from bats to humans13.
Spike (S) proteins play an important role in entry to the host cell. S-proteins are known to bind with Angiotensin Converting Enzyme 2 (ACE2), the same receptor used by SARS-CoV. The S-protein amino acid sequences of SARS-CoV and SARS-CoC-2 showed 76.47% structural similarity, having same structural confirmation and interaction interface. Binding residue positions are 442, 472, 479, 487, and 491 in spike protein. Binding free energy of SARS-CoV-2 spike protein is -78.6 Kcal mol-1 while binding free energy of SARS-CoVis -50 Kcal mol-1. Relatively high binding affinity of SARS-CoV-2 than SARS-CoV suggests rapid human to human transmission2,14.
Epidemiology
Species of Betacorona virus are known for public health menaces after they are discovered in Republic of China, Canada, Singapore and the Middle East. Among them, three of the species outbreaks caused severe respiratory syndromes in humans in the very first two decades of the 21st century: (i) Severe Acute Respiratory Syndrome (SARS-CoV) in the year 2002-2003 (ii) Middle East Respiratory Syndrome (MERS-CoV) in the year 2012 and (iii) Severe Acute Respiratory Syndrome related coronavirus (SARS-CoV-2) which has caused COVID-19 pandemic since December 2019 and escalated in the beginning of the year 20202,15. Initially there were 580 confirmed cases and 1781 daily reported cases of SARS-CoV-2 around the globe which raised public health concerns and declared as pandemic16.
From the day of first reported case to April 22, 2020 there were approximately 2,635,716 reported cases of COVID-19 including symptomatic and asymptomatic patients and 79,956 daily reported cases for the same16.
R0 indicates the reproductive number of the infectious virus that represents contagiousness of the infected disease. If R0 is >1 then infection is likely to be increased and if R0 is <1 then the infection is likely to decline within a short period. Estimated R0 of SARS-CoV-2 is ranged from 1.4 to 6.49 with a mean of 3.28 which is higher than both SARS-CoV(3) and MERS-CoV (<1). Since reproductive number of SARS-CoV-2 is higher than both viruses, it is more likely to infect people17.
Incubation period of the virus also plays an important role in transmission. Incubation period counts from the day of infection to the day it starts developing symptoms of the disease. Initial 425 cases of Wuhan, China showed mean incubation period of 5.2 days18. Case studies of 4021 patients showed 4.75 days of incubation period while 88 people with the travel history who got infected had shown 6.4 days of incubation period19,20. From the latest data of 1099 patients from 552 hospitals in China showed median incubation period of 3.0 days ranging from 0 to 24 days. Hence, 14 days of observation period is decided based on data analysis after the infection. SARS-CoV-2 has a longer incubation period than SARS-CoV (4.0 days) and MERS-CoV (4.5 to 5.2 days)20,21.
Difference in intrinsic virulence properties can explain the capacity of the transmission of the viruses22. SARS-CoV-2, SARS-CoV and MERS-CoV use receptor in both upper and lower respiratory tract. Where SARS-CoV-2 and SARS-CoV binds to the ACE 2 receptors and MERS-CoV binds to the dipeptidyl peptidase 4 (DPP4) receptors to enter into the host cell. According to the Chinese Center of Disease Control and Prevention (CCDC), fatality rate of SARS-CoV-2 is 2.3, which is lower than SARS-CoV (9.5%) and MERS-CoV (34.4%). MERS-CoV has higher mortality rate than SARS-CoV-2 but lower transmissibility2.
Replication of SARS-CoV-2
Coronavirus replicates into the host cells having fairly unique mechanisms like ribosome frame-shifting in genome translation for initial production of polyproteins to generate replisomes or transcriptosomes and other is synthesis of genomic and subgenomic RNAs23. Genomic RNA of coronavirus contains ~30000 nucleotides that encode for both nonstructural and structural proteins. 5’ end of the genomic RNA region is responsible for encoding nonstructural proteins that are essential in replication process of RNA while 3’ end encodes for the structural proteins like S, M, E and N9.
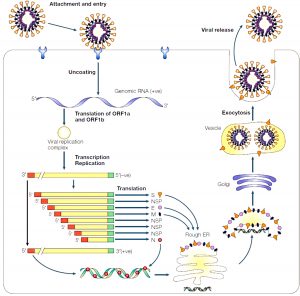
Fig. 5. Schematic diagram of coronavirus replication inside the cell Reproduced with permission from [10]
Fig. 5 illustrates how genomic RNA of the virus gets attached to the plasma membrane of the host cell and chemical reaction occurring into the cell once it entered.
Virus gets attached to the receptor of the plasma membrane that influences the genomic RNA to enter into the host cell. As spike proteins on the peripheral region of the virus binds the receptor protein, conformational changes facilitate receptor mediated endocytosis that pulls the RNA inside the cytoplasm of the host cell24. Detachment of virus and (+) sense RNA translation takes place as shown in the Fig. 6.
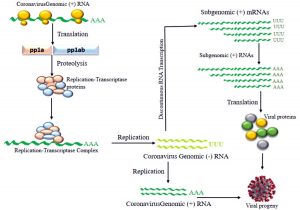
Fig. 6. Schematic diagram of RNA translation Proteolysis of pp1a and pp1ab forms replication-transcriptase complex (RTC) which is involved in replication of genomic RNA and viral proteins (S, M, E and N) encoded by discontinuous transcription forms a viral progeny
Transcriptase proteins encoded in ORF1a (Open Reading Frame 1a) and ORF1b are initially synthesized as large polyproteins pp1a and pp1ab respectively. These polyproteins are synthesized by programmed frame-shifting mechanism in this RNA(+) translation. As proteins come from these polyproteins are involved in replication, they are also referred as replicase polyproteins. As translation progresses, pp1a is generated from ORF1a. When translation reaches to the point called ribosomal shifting site it shifts to ORF1b in the middle of the translation which eventually encodes for the hybrid protein called pp1ab24. After synthesis of these polyproteins, they are cleaved by virus-encoded proteinases with papaine like (PLpro) and chymotrypsin like (3Lpro) into 16 smaller proteins which is called RTC (Replication Transcription Complex)25.
These RTC then bind with the genomic RNA(+) as shown in Fig. (6). When genomic RNA(+) replicate, it will be resulting in RNA(-) antisense strand. Now this RNA(-) can again be replicated by the RTC and get back into RNA(+) sense or this RNA(-) can be transcribed in discontinuous manner and can transcribe bunch of different RNA that code for different proteins25.
If RNA(-) gets replicated into (+) sense then it gets packaged into their viral offsprings and then released to infect other cells. If it undergoes for discontinuous transcription then RNA Dependent RNA Polymerase that will bind to the RNA and initiates transcription from different sites which will eventually result in producing different lengths of genomic RNAs. These RNAs are known as subgenomic RNAs as they are transcribed from the same RNA(-). These sub-genomic RNA of different length will encode for different proteins which eventually packaged into viral offsprings and released to infect other cells. TRS is the Transcription Regulatory Sequences on m-RNA having different body TRS that codes for different structural proteins. As soon as transcription reaches to the point where body TRS corresponds, it jumps to the site where leader TRS is present right after ORF sites as shown in Fig. (7) and finishes transcription. Everything between body TRS and leader TRS is emitted and generates different length of subgenomic RNAs25. All of these subgenomic RNA codes for structural proteins. These translated viral proteins then combine with RNA(+) strand to make a progeny. This translation process occurs in rough Endoplasmic Reticulum (ER) as this involves a secretory pathway which includes rough ER, Golgi apparatus and exocytosis. Proteins made in rough ER are sent to Golgi apparatus with RNA(+) where they put into the Golgi vesicles and packaged as viral progeny. These vesicles will exocytose that release viral progeny as fully matured coronavirus to infect other cells as shown in Fig. (5)24.
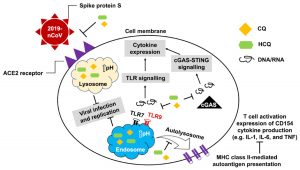
Fig. 8. Graphical illustration of mechanism of action of CQ and HCQ that not only reduce the binding efficiency of ACE2 on host cell and spike protein of the coronavirus and prevents replication but CQ and HCQ also attenuates the possibility of cytokine storm. Reproduced with permission from [33]
In-silico analysis of COVID-19 drugs
Looking at the current scenario of coronavirus, a rapid drug application strategy is necessary. For current situation the only way is to repurpose the drugs which are available commercially. Some drugs like remdesivir, lopinavir/ritonavir have been reported to reduce pneumonia like symptoms in COVID-19 patients. Certain group of scientists performed docking studies and found out the common potent site of interaction for COVID-19 that could be used as drug targets. Autodock vina gave 3 drugs which showed best results namely saquinavir, nelfinavir and grazoprevir. Autodock vina also predicted drugs which could bind to 3C like proteinase receptor of SARS-CoV-2 like purmorphamine, lumacaftor and verrucarin A27.
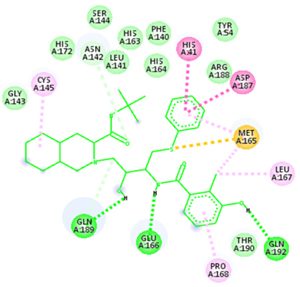
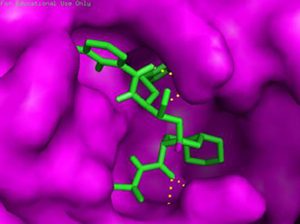
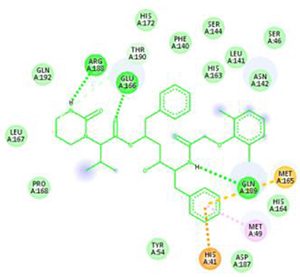
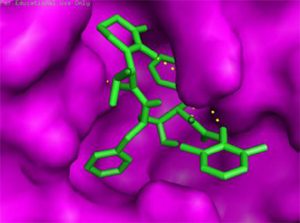
The main protease (Mpro)/chymotrypsin-like protease (3CLpro) from COVID-19, act as potential target for the inhibition of CoV replication. There are certain receptors in the respiratory tract which play a crucial role in attachment of virus which can also be targeted like Angiotensin Converting Enzyme 2 (ACE 2). The structure of the protease reveals that there is 96.1% sequence similarity between SARS-CoV and SARS-CoV 2(Bolcato, Bissaro & Pavan et al.). Structure Based Drug Discovery (SBDD) can be applied to identify the MPro inhibitors and could help in the repurposing process to find a potential drug. Many of the studies have found out the inhibitors of Human Immunodeficiency Virus (HIV) as possible drugs for anti-COVID candidates. Supervised Molecular Dynamics (SuMD) is an emerging technique to study detailed recognition process and the binding confirmation which could supplement the drug discovery process (Bolcato, Bissaro & Pavan et al.).It is found that the urea moiety governs the interaction of hydrogen bond with the side chain of Gln189. Residues like His164, Glu166, Gln189, Thr190 and Gln196 mediated the interaction of water bridged hydrogen ions (Bolcato, Bissaro & Pavan et al.). The drug like properties of any probable compound can be studied by Lipinski rules and the determination of the active sites can be done by Computed Atlas for Surface Topography of Proteins (CASTp) and Biovia Discovery Studio 4.529. Some people used idock docking software and got lower score in the catalytic pocket of SARS-CoV-2(-9.11 kcal/mol), SARS-CoV(-8.03 kcal/mol) and MERS-CoV(-8.26 kcal/mol). Regarding the contact modes by idock it was concluded that there is a significant contribution of hydrogen bonds in binding and there were some additional hydrogen bonds found between theaflavin and Asp452, Arg553 and Arg624 of SARS-CoV-2 RdRp, and between theaflavin with Thr440, Ser566, Ala569, and Asp644 of SARS-CoV RdRp, and between theaflavin with Arg294, Thr292, Gln291, Leu427, Asn 390, Leu427, and Asp728 of MERS-CoV RdRp. As theaflavin has the lowest idock score with SARS-CoV-2(-9.11 kcal/mol) blind docking server was used to confirm the result and found that theaflavin has a lower docking score of -8.8 kcal/mol against the catalytic pocket of SARS-CoV-2. Studies have also shown that extracts from Pu’er (pu-erh) tea and black tea, and theaflavin-3,3′-digallate and 3-isotheaflavin-3-gallate, in the theaflavins family, have strong activity to inhibit SARS against SARS-CoV 3CLpro activity30. Pentoxyfylline (PTX) is another compound which can be used as a drug against SARS-CoV-2. PTX has some characteristic features to reduce the blood thickness, increase the Red blood cell flexibility so that RBC could migrate through capillaries more rapidly and as a result blood circulation becomes easier. Due to the anti-inflammatory and anti-viral activity of PTX, it can be used as drug to cure SARS-CoV-2. PTX also down regulates the proinflammatory cytokines and proliferating cells in lungs. So PTX can be used as a preventive drug against the COVID-1931. Some of very commonly used drugs for COVID-19 treatment are Chloroquine and Hydroxychloroquine (HCQ). Chloroquine is a potent anti-malarial drug but it shows the results of inhibiting many other micro-organisms including coronaviruses also. The anti-viral and anti-inflammatory activity of Chloroquine helped people to recover from COVID-19. Interestingly Chloroquine inhibits the MAP-Kinase pathway that interferes the molecular crosstalk with Mprotein32. SARS-CoV-2 utilizes the surface receptor ACE2 and is believed that Chloroquine obstructs ACE2 receptor glycosylation as a result it prevents SARS-CoV-2 attachment with target cells.
Chinese researchers also stated that Chloroquine helps in reducing viral replication that can be easily achieved with standard dosing due to its favourable penetration in tissues including the lungs32. Chloroquine is also an antimalarial drug and an immune modifier that can be distributed throughout the body including lungs. HCQ has shown in-vitro activity against COVID-19 and may possess immunomodulatory properties. Both chloroquine and HCQ may have a common mode of inhibition like viral protein glycosylation, virus assembly, virus transport etc. Preclinical trials suggest HCQ to be more potent as compared to Chloroquine34. Certain plants having anti-viral properties have also been used. Eucalyptol (1,8 cineole) is the principal component found in eucalyptus oil from all eucalyptus plants. Eucalyptol can be used as a drug to inhibit the activity of MPro in COVID-19 patients. Mpro/eucalyptol complex forms hydrophobic interactions with ALA7, PRO52, TRP207, LEU29, TRY126, PRO184; hydrogen bond interactions with M4, V18, L30, D10, T16; and ionic interactions with LYS3, ASP34, ARG38, HIS163 hence these sites can act as target to play a major role in protein-protein interaction and help to inhibit the function of MPro. Flavonoids are important plant components having a phenolic group can also be used to inhibit the COVID-19 pathway. Particularly apigenin, luteolin, quercetin, amentoflavone28, quercetin, daidzein, puerarin, epigallocatechin, epigallocatechin gallate, gallocatechin gallate29 and kaempferol30 were suggested to inhibit the proteolytic activity of SARS-CoV 3CLpro35. The seeds of Annona muricara can also be used to inhibit the COVID-19 pathway. Anti SARS-2019 nCoV nsp12 activity was studied using docking softwares like Discovery studio 2017, Autodock tool 1.5.6, Autodock vina 1.1.2 and Edupymol version 1.7.4.4., and obtained the affinity scores from -5.6 to -4.4 kcal/mol and showed a very promising inhibitory behaviour36. Nelfinavir and Lopinavir are the inhibitors of protease which can be used as drugs, who are affected with HIV, but they can also be used to treat SARS and MERS as all the 3 have similar mechanisms. Nelfinavir showed the best result of docking in Autodock version 4.2 with the lowest docking score and is now used as a drug as well as it is used as drug standards for comparison29. Studies have also shown that kaempferol, quercetin, luteolin-7-glucoside, demethoxycurcumin, naringenin, apigenin-7-glucoside, oleuropein, curcumin, catechin, epicatechingallate, zingerol, gingerol, and allicin act as potential inhibitors of the COVID-19 Mpro as shown in table below. An in-silico analysis revealed that the compounds share a similar pharmacophore as nelfinavir. Studies also say that these phenolic compounds are present in high numbers in the medicinal plants found worldwide.
Table (1):
Comparative analysis of potent drug compounds and their binding free energy (ΔG) with targeted protein.
Drug and its Binding Affinity (Free Energy ΔG) |
Chemical Structure of the compound |
Binding of drug molecule with the targeted protein |
|---|---|---|
Nelfinavir C32H45N3O4S (ΔG = -10.72) |
||
Lopinavir C37H48N4O5 (ΔG = -9.41) |
||
Kaempferol C15H10O6 (ΔG = -8.58) |
||
Quercetin C15H10O7 (ΔG = -8.47) |
||
Luteolin-7-glucoside C21H20O11 (ΔG = -8.17) |
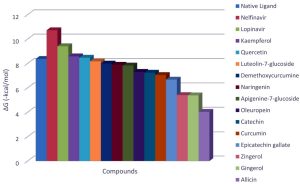
Docking results of some potent drugs with the best score and higher binding energy where binding affinity of Nelfinavir is highest. Figures in the table are reproduced with permission from[29]
The docking analysis showed that inhibition affinity of several active compounds present in medicinal plants, ranked by affinity (ΔG); Nelfinavir(-10.72)> lopinavir(-9.41)> kaempferol(-8.58)> quercetin(-8.47)> luteolin-7-glucoside(-8.17)> demethoxycurcumin(-7.99)> naringenin(-7.89)> apigenine-7-glucoside((-7.83)> oleuropein(-7.31)> catechin(-7.24)> curcumin(-7.05)> epigallocatechin(-6.67)> zingerol(-5.40)> gingerol(-5.38)> allicin(-4.03). Kaempferol, quercetin, luteolin-7-glucoside, apigenin-7-glucoside, naringenin, oleuropein, demethoxycurcumin, curcumin, catechin, and epigallocatechin were the most potent inhibitors of COVID-19 Mpro29. High binding affinity of the drug compound depends on the amount of bonding that occurred in the active site of protein.
From binding affinity of the compounds to the targeted molecule we can clearly see that ΔG of the nelfinavir is the highest. Hence, it can be used as potent drug to treat the disease.
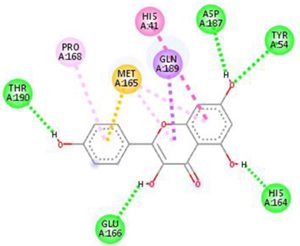
The novel COVID-19 pandemic is rapidly spreading throughout the world without limiting boundaries of weather, temperature and other climate conditions. Till the date, this disease has infected millions of people and taken thousands of lives in almost all the countries and continents of the world. Rapid development of drugs and vaccines are initiated in all the research oriented institutes. With the availability of the data regarding phylogenetic relation, genomic constitution, transmission, replication researchers could find few potent drugs by in-silico analysis. Considering the limits of in-vitro and in-vivo testing of drugs, in-silico study is better for finding potent drugs that can be used for the treatment of COVID-19. The main protease (Mpro)/chymotrypsin-like protease (3CLpro) from COVID-19, act as potential target for the inhibition of CoV replication. There are certain receptors in the respiratory tract which play a crucial role in attachment of virus which can also be targeted like Angiotensin Converting Enzyme 2 (ACE 2). The structure of the protease revealed that there is 96.1% sequence similarity between SARS-CoV and SARS-CoV 2. Docking results based on their binding affinity showed best scores for nelfinavir, lopinavir, kaempferol, quercetin and luteolin-7-glucoside, in which nelfinavir has highest binding ability according to pharmacophore studies. Amino acids like Tyr54, Phe140, Cys145, His163, His164, Glu166, Asp187, Arg188, Gln189, Thr190, and Gln192 of 6LU7 (The crystal structure of COVID-19 main protease in complex with an inhibitor N3) are involved in the interaction with potent drugs that are being used to treat SARS-CoV-2. Since nelfinavir is being considered as drug standard, these amino acids can be considered as target sites for newly developed drugs.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declares that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All the listed author(s) have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
Not applicable.
- AE Gorbalenya et al. “The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2,” Nat Microbiol. 2020;5(4):536–544,
Crossref - M Xie and Q Chen. “Insight into 2019 novel coronavirus — an updated intrim review and lessons from SARS-CoV and MERS-CoV,” Int J Infect Dis. 2020,
Crossref - N Petrosillo, G Viceconte, O Ergonul, G Ippolito and E Petersen. “COVID-19, SARS and MERS: are they closely related?,” Clin Microbiol Infect. 2020,
Crossref - YS Malik et al. “Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments,” Vet. Q. 2020;40(1):68–76.
Crossref - K Ramanathan et al. “Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study,” no. January, 2020;19–21,
Crossref - BW Neuman et al. “A structural analysis of M protein in coronavirus assembly and morphology,” J Struct Biol. 2011;174(1):11–22.
Crossref - “Betacoronavirus ~ ViralZone page.” [Online]. Available: https://viralzone.expasy.org/764?outline=all_by_protein. [Accessed: 25-Apr-2020].
- “International Committee on Taxonomy of Viruses (ICTV).” [Online]. Available: https://talk.ictvonline.org//taxonomy/p/taxonomy-history?taxnode_id=201851847. [Accessed: 25-Apr-2020].
- A Wu et al. “Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China,” Cell Host Microbe. 2020;27(3):325–328.
Crossref - K Stadler, V Masignani, M Eickmann and S Becker et al. “SARS — BEGINNING TO UNDERSTAND A NEW VIRUS,” 2003,
Crossref - D Benvenuto, M Giovanetti, A Ciccozzi, S Spoto, S Angeletti and M Ciccozzi. “The 2019-new coronavirus epidemic: Evidence for virus evolution,” J. Med. Virol. 2020;92(4):455–459, 2020,
Crossref - Tommy Tsan-Yuk Lam et al. “Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins,” Nature, 2020,
Crossref - T Zhang, Q Wu and Z Zhang et al. “Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak,” Curr Biol. no. January, 2020,
Crossref - X Xu et al. “Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission,” Sci China Life Sci. 2020;63(3):457–460, 2020,
Crossref - The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team China CDC, “The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronaviurs Diseases,” Vital Surveillances. 2020; 2(8):113–122,2020, doi: 10.3760/cma.j.issn.0254-6450.
- Worldometer, “Coronavirus Cases,” Worldometer, 2020. [Online]. Available: https://www.worldometers.info/coronavirus/coronavirus-cases/#daily-cases. [Accessed: 25-Apr-2020].
- Y Liu, AA Gayle, A Wilder-Smith and J Rocklov, “The reproductive number of COVID-19 is higher compared to SARS coronavirus,” J Travel Med. 2020;27(2):2020,
Crossref - Q Li et al. “Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia,” N Engl J Med. 2020;382(13):1199–1207, 2020,
Crossref - Yang Yang et al. “Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China,” 2020;100071: 2019–2020,
Crossref - JA Backer, D Klinkenberg and J Wallinga. “Incubation period of 2019 novel coronavirus (2019- nCoV) infections among travellers from Wuhan, China, 20 28 January 2020,” Eurosurveillance. 2020;25(5):1–6.
Crossref - Justin Lessler, Nicholas G Reich, Ron Brookmeyer, et al. “Incubation periods of acute respiratory viral infections: a systematic review,” no. January 2020;19–21.
Crossref - VJ Munster, M Koopmans, N van Doremalen, D van Riel and E de Wit. “A novel coronavirus emerging in China – Key questions for impact assessment,” N Engl J Med. 2020;382(8):692–694.
Crossref - SG Sawicki, DL Sawicki and SG Siddell. “A Contemporary View of Coronavirus Transcription,” J Virol. 2007;81(1):20–29.
Crossref - PS Masters. “The Molecular Biology of Coronaviruses,” Adv. Virus Res. 2006;65(no. January):193–292.
Crossref - EJ Snijder et al. “Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex,” J. Virol. 2006;80(12):5927–5940.
Crossref - M Raaben, P Whitley, D Bouwmeester, RA Setterquist, PJM Rottier and CAM de Haan, “Improved microarray gene expression profiling of virus-infected cells after removal of viral RNA,” BMC Genomics. 2008;9:1–12.
Crossref - BR Beck, B Shin, Y Choi, S Park and K Kang. “Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model,” Comput. Struct. Biotechnol. J. 2020;18(no. April):784–790.
Crossref - G Bolcato, M Bissaro, M Pavan, M Sturlese and S Moro. “Targeting the Coronavirus SARS-CoV-2: computational insights into the mechanism of action of the protease inhibitors Lopinavir, Ritonavir, and Nelfinavir,” Res. Sq. 1–19.
Crossref - S Khaerunnisa, H Kurniawan, R Awaluddin and S Suhartati.“Potential Inhibitor of COVID-19 Main Protease ( M pro ) from Several Medicinal Plant Compounds by Molecular Docking Study,” Preprints. 2020;2020030226(March):1–14, 2020,
Crossref - J Lung et al. “The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase,” J. Med. Virol. 2020;92.
Crossref - JF Bermejo Martin, JL Jimenez and MA Munoz-Fernandez. “Pentoxifylline and severe acute respiratory syndrome (SARS): A drug to be considered,” Med. Sci. Monit. 2003;9(6).
- K Ramanathan et al. “Chloroquine and Hydroxychloroquine in the treatment of COVID-19 with or without Diabetes: A systematic search and a narrative review with a special reference to India and other developing countries,” 2020; (no. January): 19–21.
Crossref - D Zhou et al. “COVID-19 : a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression,” 2020; (no. February): 4–7.
Crossref - C-Y Cheng et al. “Lopinavir/ritonavir did not shorten the duration of SARS CoV-2 shedding in patients with mild pneumonia in Taiwan,” J Microbiol Immunol Infect. 2020.
Crossref - S Jo, S Kim, DH Shin and MS Kim. “Inhibition of SARS-CoV 3CL protease by flavonoids,” J Enzyme Inhib Med Chem. 2020;35(1):145–151, 2020,
Crossref - AK Oyebamiji, EH Awoyelu and B Semire. “In-vitro Investigation on Selected compounds in Annona Muricata Seed : A Potential SARS-CoV nsp12 Polymerase Inhibitors down Regulating 2019-nCoV,” no. January, 2020.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.