ISSN: 0973-7510
E-ISSN: 2581-690X
Hepatitis C virus (HCV) is a blood-borne pathogen that transmits infection via transfusion. Hepatocellular carcinoma is the fifth most common cancer and a major cause of death in patients with chronic HCV infection. Response to treatment is mainly based on the genotypic characterization of HCV. The gold standard for genotyping HCV is by sequencing highly conserved regions such as NS5, core, E1, and 5’UTR. Serum samples of patients who visited the tertiary care hospital with clinical features suggestive of HCV infection formed the study group. HCV genotyping was performed using multiplex Polymerase Chain Reaction in the samples tested positive by Chemiluminescence Immunoassay (CLIA). The viral loads were also performed on selected patient samples. In the present study, Genotype 4 (35.71%), followed by Genotype 3 (17.53%) and 1 & 1b (12.34%) were the common genotypes observed. Genotype 1,1b & 4 mixed type and genotype 4 and 5 mixed type was detected in one sample each (0.65%). The mean measured value of HCV antibody was 11.51 ± 4.57. The viral load was detected in 61 out of 81 samples tested. The mean viral load ranged from 550 to 552769250IU/ml (log 2.74-log 8.74). Genotype 4 was the most common genotype demonstrated in our study as opposed to the other studies were genotype 3 was the dominant one in south India.
Dominant, Genotype, Hepatitis C, Polymerase Chain Reaction, Seropositive
Hepatitis C virus (HCV) infection is a blood-borne viral disease deadlier than HIV.1,2 It belongs to the genus Hepacivirus of the Flaviviridae family and is implicated in the causation of chronic liver disorders.3 HCV is a public health challenge as the infection is often chronic and further disadvantageous due to high mutation rate and non-availability of an effective vaccine.4,5 The HCV RNA (genome) is a single-stranded, positive-sense RNA with a length of around 9,600 nucleotide bases. The HCV genome has a single lengthy open reading frame (3006-3037 codons) flanked by untranslated sections at the 5′ and 3′ ends (UTRs).6
HCV is known to demonstrate high genetic heterogeneity and the International Committee on Taxonomy of Viruses (ICTV) has so far identified eight genotypes, numbered from 1 to 8 with about 90 subtypes and millions of quasispecies. The enzyme RNA-dependent RNA polymerase, responsible for viral replication, does not exhibit ‘proofreading’ activity and thus results in multiple mutations and evolution of numerous quasispecies. The genomic instability that is found by HCV has been termed as quasispecies.7-10
The prevalence of HCV differs between developed and developing countries.11 Globally, genotype – 1 is the most prevalent type with almost 49 % prevalence, followed by genotype-3 (18%), genotype 4 (17%), and around 5 % each of genotypes 5 & 6. Genotypes 4,5,6 and 7 are endemic to certain places while the other types are found all over the world.7 The prominent genotype in India was observed to be Genotype 3 followed by Genotype 1. Region-wise distribution of HCV genotypes suggested that Genotype 3 was noticed in eastern, western, and northern parts of India, whereas Genotype 1 predominated in South India.3 These studies were in concordance with other. Studies by Sadhukhan et al. from West Bengal, eastern India, found Genotype 3 to be the prominent type from this region followed by Genotype 1, 3a and 1b. 12-17
Genotype 3 was associated with increased liver complications when compared to the other genotypes with reduced response to therapy as well.13
Knowledge about the genotypic distribution of HCV in a geographical area may facilitate treatment planning and provide appropriate clues about the consequences of HCV-related liver disease in that geographical area. This study analyzed the various genotypes of HCV using multiplex Real-time PCR in seropositive HCV samples.
This was a retrospective study, conducted in the Department of Microbiology, at a tertiary care hospital. Data from January 2017 to January 2020 were analyzed. The Institutional Human Ethics Committee approval was obtained prior to start of the study (reference ID PSG/IHEC/2021/Appr/ Exp/243). Patients suspected to have Hepatitis C infection based on the clinical presentation, irrespective of age formed the study group. Collection of blood for analysis followed a standard protocol where in 5 ml venous blood was drawn into a plain vial, serum separated and aliquot in different vials and stored at -80°C until tested. During the study period of 3 years around 81163 serum samples were tested for HCV antibodies by Chemiluminescence immunoassay (CLIA) (Abbott, ARCHITECT Analyzer), among which only 154 samples were sent for genotyping. The serum samples, which were icteric, lysed or turbid were excluded.
Collection of blood for analysis followed a standard protocol where in 5 ml venous blood was drawn into a plain vial, serum separated and aliquot in different vials and stored at -70°C until tested. While biochemical tests are crucial markers for the diagnosis of suspected Hepatitis C cases, as it was out of scope for our study department.
Viral RNA extraction
The extraction of Hepatitis C virus ribonucleic acid (HCV RNA) was performed from plasma using QIA amp R viral RNA mini kit(QIAgen GmbH) RNA kit following the manufacturers’ instructions.
Following extraction, the RNA was eluted in 50µl of elution buffer and stored at -80 °C until further processing. The amplification was done using Sansure Biotech amplification kit. The test is based on real-time one-step polymerase chain reaction (PCR) technology. It includes reverse-transcriptase (RT) reaction to convert RNA into complementary deoxyribonucleic acid (cDNA) followed by multiplex PCR for the amplification of specific genotype sequences i.e., 5’ untranslated region (UTR) region (Genotype2,3,4,5, and 6) and NS5b region (Genotype 1a and 1b) using target specific probes. The assay principle is based on Taqman probes which allow higher specificity and sensitivity. The thermal cycling parameters include reverse transcription at 50°C for 15 minutes followed by RT inactivation at 95°C for 20 seconds and PCR amplification (45cycles) for 30 seconds the fluorescent data is collected after 60°C extension step and analyzed for the genotypes as per manufacturer’s instructions.
The viral load was performed using real-time PCR RGQ (ROTOR GeneQ) analyser. It utilizes the RT-PCR to quantitate HCV RNA. The analytical detection limit of the HCV RG RT-PCR Kit is 0.19 IU/µl. The anti-HCV antibody detection from plasma samples were tested using CLIA Abbott, ARCHITECT Analyzer. It is a two step immunoassay that uses chemiluminescent microparticle immunoassay (CMIA) technology for the qualitative detection of anti HCV antibodies in human serum/plasma. It uses the HCr43 protein composed of two non contiguous coding regions of the HCV genome (33c and core) and c100 3 (putative nonstructural NS3 and NS4). As per the analyzer- a result of greater than 1.0 indicates a reactive sample and is suggestive of the presence of HCV antibody.
For quality purposes, selected samples were subjected to interlaboratory comparison at NABL accredited laboratories whose results were satisfactory.
The data thus obtained were tabulated and statistically analyzed using statistical software R 4.0.3 and Microsoft Excel for Windows 2016. Continuous variables were represented by mean ± SD and categorical variables by a frequency table.
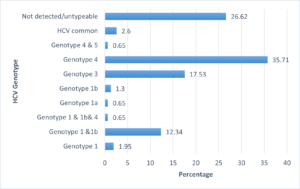
A total of 374 samples were positive for HCV antibodies’ from 81163 samples tested (0.4%) satisfying the inclusion and exclusion criteria. In the present study, 154 out of the 374 samples were sent for genotyping. There were 61 females (39.61%) and 93 males (60.39%) with ages ranging from 24 to 85 years with mean age of 56.47±13.14 years. Out of 154 samples, genotype was detected in 113. The most common HCV genotype identified was that of genotype 4 in 55 samples (35.71%) followed by genotype 3 in 27 samples (17.53%) and only one each of Genotype 1 & 1b & 4 mixed type, Genotype 1a and Genotype 4 & 5 mixed type (0.65%). Table 1 represents the distribution of the various genotypes.
Table (1):
Distribution of HCV genomes in the present study
HCV genotype |
Frequency |
Percentage |
|---|---|---|
Genotype1 |
3 |
1.95 |
Genotype1 &1b |
19 |
12.34 |
Genotype1 & 1b& 4 |
1 |
0.65 |
Genotype1a |
1 |
0.65 |
Genotype1b |
2 |
1.30 |
Genotype3 |
27 |
17.53 |
Genotype4 |
55 |
35.71 |
Genotype4 &5 |
1 |
0.65 |
HCV common |
4 |
2.60 |
Not detected/untypeable |
41 |
26.62 |
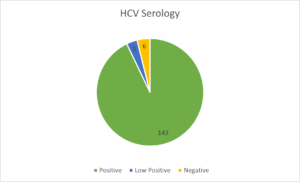
Distribution of genotypes in the samples is represented in Figure 1 and the HCV serology is summarized in Figure 2.
The mean measured value of the HCV antibody was 11.51 ± 4.57. The viral load was done in 81 samples for which there was a requisition for the same. The viral load was detected in 60 samples and in the remaining 21 samples, no viral load was detected. The mean viral load was 21439255 ± 80468277IU/ml (log7.33-log7.9), ranging from 550 to 552769250 IU/ml. When the antibody levels were compared with sex of the participants, there was no statistical difference (p=0.089). The standard treatment guidelines for each genotype has been summarized in Table 2.18
Table (2):
Recommendations for treatment of HCV genotypes(18).
| Genotype 1Ledipasvir-sofosbuvir |
|
|---|---|
| Ombitasvir-paritaprevir-ritonavir with or without ribavirin | |
| Sofosbuvir plus simeprevir with or without ribavirin | |
| Daclatasvir plus sofosbuvir with or without ribavirin. | |
|
For retreatment of patients with genotype 1 who had no improvement with peg-interferon and ribavirin |
Ledipasvir-sofosbuvir with or without ribavirin |
| Ombitasvir-paritaprevir-ritonavir and dasabuvir with or without ribavirin | |
| Sofosbuvir plus simeprevir with or without ribavirin | |
| Daclatasvir plus sofosbuvir with or without ribavirin | |
| For patients who had no improvement with sofosbuvir plus ribavirin, with or without peg-interferon | Ledipasvir-sofosbuivr plus ribavirin |
| Genotype 2 | Sofosbuvir with ribavirin |
| Daclatasvir plus sofosbuvir | |
| For retreatment of patients with genotype 2 who had no improvement with peg-interferon and ribavirin | Sofosbuvir plus ribavirin |
| For retreatment of patients with genotype 2 who had no improvement with sofosbuvir plus ribavirin | Daclatasvir plus sofosbuvir (with or without ribavirin) |
| Sofosbuvir plus ribavirin plus peg-interferon | |
| Genotype 3 | Daclatasvir plus sofosbuvir |
| Daclatasvir plus sofosbuvir, with or without ribavirin | |
| Sofosbuvir plus ribavirin plus peg-interferon | |
| For retreatment of patients with genotype 3 who had no improvement with peg-interferon and ribavirin | Daclatasvir plus sofosbuvir for along with Ribavirin in those with cirrhosis |
| Sofosbuvir plus ribavirin plus peginterferon (with or without cirrhosis) | |
| For retreatment of patients with genotype 3 who had no improvement with sofosbuvir | Daclatasvir plus sofosbuvir plus ribavirin |
| Sofosbuvir plus ribavirin plus peg-interferon | |
| Genotype 4 | Ledipasvir-sofosbuvir |
| Ombitasvir-paritaprevir-ritonavir plus ribavirin | |
| Sofosbuvir plus ribavirin | |
| Sofosbuvir plus ribavirin plus peg-interferon | |
| Grazoprevir | |
| Elbasvir | |
|
Genotype 5 and 6 |
Ledipasvirsofosbuvir |
| Peg-interferon plus ribavirin | |
| Grazoprevir-elbasvir | |
| For retreatment of patients with genotype 5 and 6 who had no improvement with peg-interferon plus ribavirin | Ledipasvir-sofosbuvir |
| Sofosbuvir plus ribavirin plus peg-inteferon |
HCV infection can be acute or chronic. Acute HCV infections are frequently asymptomatic, and the majority of them do not progress to a life-threatening condition. It is observed that 60-80% of patients with acute infection progress to chronic HCV infection in their lifetime. HCV is the major cause of liver cirrhosis, hepatic decompensation, and/ or hepatocellular carcinoma and is also a common cause of liver transplantation.
Genotyping of HCV infections helps predict the severity of the infection and also determines the response to therapy initiated accordingly.19 The major goal of treatment is to reduce the side effects by attaining total viral eradication, which is defined as undetectable HCV RNA 3 months after completion of antiviral therapy.20 Among 154 samples analyzed only 113 (73%) samples were typeable. In our study, Genotype 4 was found to be the most common variant (35.71%) followed by genotype 3(17.53%). A study conducted in South India by Rooby et al. showed Genotype 3 as the dominant type followed by type 1 and type 4. Genotype 1 showed subtype 1b as the dominant type (75%) followed by 1a and 1c. They found a higher percentage of genotype 4 which was previously dominant in the Middle East and Africa. The result of the present study is in concordance with study by Rooby et al, which is also from South India. Higher percentage of individuals taking up travel for various reasons and migration of population from different parts of the world or India might have caused this kind of a variation in the HCV genotypes, as has been suggested in literature.21
The studies by Christdas et al. from Tamil Nadu, South India, revealed the most common HCV genotype was identified to be type 3 accounting for 63.85%. Genotype 1 (25.72%), genotype 2 (0.002%), genotype 4 (7.5%), and genotype 6 (7.5%) were among the other genotypes discovered. The studies also confirmed that genotype 3 was predominant, whereas patients from the South were more likely to have genotype 1 or 4. Patients from North-Eastern India were the only ones with genotype 6. Recombinant variants of genotypes 1 and 2 were observed in two patients in their study.9
The geographical isolation of HCV genotypes has led to the traditional belief that only the Middle East and Africa are home to genotype 4, which is believed to be resistant to treatment. It is noteworthy that there has been an incidence of increasing Genotype 4 mainly in South India.22 Also, studies done by Christdas et al. also highlighted the increasing prevalence of Genotype 4 in Andhra Pradesh, South India.9 The present result from South India also is in concordance with the above observation where an increased percentage of type 1 and type 4 were detected, along with prominent genotype 3.
A new Genotype 7 was isolated in patients originating from the Democratic of Congo, Central Africa.23
Khan et al. found genotype 3a to be the predominant type followed by mixed types as Genotype 1 and 3 (0.4%), Genotype 2 and 3 (1.4%) and Genotype 3 and 4 (0.2%) along with a significant being ‘un-typeable’ genotypes.24 The present study also found a mixed 4 and 5 genotypes, and a mixed type – 1, 1b, and 4, in one sample each. In addition, about 26% of the samples were not genotyped and about 2 % of them were common HCV types, like the previous study. The likely possibilities for untypeable genotypes could be due to low HCV RNA level (<500 IU/mL), probe reactivity with multiple HCV genotype, and variation in patient’s HCV target sequences with mismatches to PCR primers and/or probes.25 For those samples which could not be genotyped, it can be subjected to sequencing which is the gold standard method for further analysis.
In the study by Sadhukhan et al, the mean viral load of HCV samples was 6.28±0.89 log10IU/ml and a statistically highly significant association of viral load was found in Genotype 1.17 Similar association between genotype 1 and high viral load was found in the study by Riaz et al.26 In the observations of the present study, the mean viral load was much higher by 21439255 ± 80468277 with a range 550 to 552769250IU/ml. (log 2.74-log 8.74). The high viral load of log 5.3 (>200000IU/ml) was also seen in Genotype 1 and in Genotype 4.
Patients having Genotype 2-a show a better response to Interferon-a compared to type 1b. Genotype 4 is a difficult strain to treat even with a combination of interferon and ribavirin. Genotype 2 and 3 are relatively easier to treat whereas genotypes 1 and 4 are difficult to treat using interferons alone or in combination with ribavirin.9,27
Introduction of direct-acting antivirals (DAA) has rendered treatment of HCV relatively easier when compared to interferons as they can cause significant complications. DAA provides better pharmacological safety and now are considered standard care for the management of HCV but on the other hand, is very expensive.17,28 Interestingly, Genotype 3 has been found to show reduced response to even DAA.17,29 Sofosbuvir has been noted to inhibit all genotypes of HCV and high efficacy is noted with genotypes 1 to 6 and to a limited extent in genotypes 7 and 8. A combination of sofosbuvir, velpatasvir, and voxilaprevir has provided a better response across all genotypes, even in those who had failed treatment with other DAA.30 Treatment for each genotype has been summarized in table 2.31
Early diagnosis of HCV infection and initiation of treatment is of utmost importance as it delays and precludes complications. Further, it adds to the prevention of spread of infection in the community.32
Limitation
The present study did not include the patient’s clinical details as history of blood transfusion, intravenous drug abuse, sexual history, and geographic origin or serum markers. Also, the treatment undertaken by the patients was not recorded in this study. Future studies should include all these parameters to obtain a robust information system that can provide a thorough insight into HCV in South Indians.
This study looked at the genotypic variations of HCV in the South Indian population visiting a tertiary care hospital. Genotype 4 was found to be the most common type, as opposed to genotype 3 which was previously reported as common. This shows that there could be changing patterns of genotypic distribution probably because of migration or travel. A seropositivity of 0.4% was detected in the present study setting. It can be reiterated that genotyping and viral load estimation is of utmost importance for diagnosis, treatment planning along with follow up and the current study has been effective in achieving the objectives.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee,PSG Institute of Medical Sciences & Research with reference number PSG/IHEC/2021/Appr/ Exp/243.
- Audu RA, Okwuraiwe AP, Ige FA, Adeleye OO, Onyekwere CA, Lesi OA. Hepatitis C viral load and genotypes among Nigerian subjects with chronic infection and implication for patient management: a retrospective review of data. Pan Afr Med J. 2020;37:335.
Crossref - Cacoub P, Comarmond C. Considering hepatitis C virus infection as a systemic disease. Semin Dial. 2019;32(2):99-107.
Crossref - Borgia SM, Hedskog C, Parhy B, et al. Identification of a Novel Hepatitis C Virus Genotype From Punjab, India: Expanding Classification of Hepatitis C Virus Into 8 Genotypes. J Infect Dis. 2018;218(11):1722-1729.
Crossref - Bhumbla U, Shekhawat L, Kothari A, Rao J. Detection and distribution of genotypes of Hepatitis C in a tertiary care hospital. J Family Med Prim Care. 2020;9(10):5249-5251.
Crossref - Arashkia A, Rouhvand F, Memarnejadian A, Alizadeh S, Motevalli F, Ebrahimi M. Immunoinformatics modeling, construction of DNA plasmids Carrying CTL epitopes of hepatitis C virus and their preliminary immunological analysis. Iran J Med Microbiol. 2011;4(4):30-40.
- Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13(2):223-235.
Crossref - Bhattacharjee C, Singh M, Das D, Chaudhuri S, Mukhopadhyay A. Current therapeutics against HCV. Virusdisease. 2021;32(2):228-243.
Crossref - Colomba GME, Urone N, Di Marco V, Ferraro D. Phylodynamic Analysis and Implication of HCV Genotype 4 Variability on Antiviral Drug Response and T-Cell Recognition. Viruses. 2020;12(12):1363.
Crossref - Christdas J, Sivakumar J, David J, Daniel HDJ, Raghuraman S, Abraham P. Genotypes of hepatitis C virus in the Indian sub-continent: a decade-long experience from a tertiary care hospital in South India. Indian J Med Microbiol. 2013;31(4):349-353.
Crossref - Zeisel MB, Crouchet E, Baumert TF, Schuster C. Host-Targeting Agents to Prevent and Cure Hepatitis C Virus Infection. Viruses. 2015;7(11):5659.
Crossref - Hussein N, Al-Obeidy E S, Naqid I, Abd K H AS. The Distributions of HCV Genotypes in Hemodialysis Patients in Iraq. Iran J Med Microbiol. 2019;13(4):232-236.
Crossref - John M, Oommen S, Jagan O, George S, Pillai S. A study on the circulating genotypes of hepatitis C virus in a tertiary care hospital in Central Kerala. Indian J Med Microbiol. 2018;36(4):532-536.
Crossref - Tsukiyama-Kohara K, Kohara M. Hepatitis C Virus: Viral Quasispecies and Genotypes. Int J Mol Sci. 2017;19(1).
Crossref - Mehta V, Mahajan R, Midha V, et al. Impact of Direct Acting Antiviral Therapy for Treatment of Hepatitis C Genotypes 1, 3 and 4: A Real Life Experience from India. J Clin Exp Hepatol. 2018;8(1):7-14.
Crossref - Chakravarti A, Ashraf A, Malik S. A study of changing trends of prevalence and genotypic distribution of hepatitis C virus among high risk groups in North India. Indian J Med Microbiol. 2013;31(4):354-359.
Crossref - Jindal N, Bansal R, Grover P, Malhotra R. Risk factors and genotypes of HCV infected patients attending tertiary care hospital in North India. Indian J Med Microbiol. 2015;33(1):189-190.
Crossref - Sadhukhan P, Saha K, Firdaus R, Biswas A, Mukherjee A. An Increasing Trend of Hepatitis C Virus Genotype 1 among High Risk Group Populations in Eastern India. J Clin Exp Hepatol. 2014;4(2):S8.
Crossref - Keikha M, Eslami M, Yousefi B, et al. HCV genotypes and their determinative role in hepatitis C treatment. Virusdisease. 2020;31(3):235-240.
Crossref - Goolsby Hunter A, Rosenblatt L, Patel C, Blauer-Peterson C, Anduze-Faris B. Clinical characteristics, healthcare costs, and resource utilization in hepatitis C vary by genotype. Curr Med Res Opin. 2017;33(5):829-836.
Crossref - Irekeola AA, Malek NA, Wada Y, Mustaffa N, Muhamad NI, Shueb RH. Prevalence of HCV genotypes and subtypes in Southeast Asia: A systematic review and meta-analysis. PLoS One. 2021;16(5):0251673.
Crossref - Rooby EH, Peter G, Mashhood PV, et al. Genotype Distribution and Time Trend of Hepatitis C In South India. J Clin Exp Hepatol. 2014;4(Suppl 2):S7-S8.
Crossref - Puri P, Anand AC, Saraswat VA, et al. Consensus Statement of HCV Task Force of the Indian National Association for Study of the Liver (INASL). Part I: Status Report of HCV Infection in India. J Clin Exp Hepatol. 2014;4(2):106-116.
Crossref - Guntipalli P, Pakala R, Gara SK, et al. Worldwide prevalence, genotype distribution and management of hepatitis C. Acta Gastroenterol Belg. 2021;84(4):637-656.
Crossref - Khan MU, Sadia H, Irshad A, et al. Detection, quantification and genotype distribution of HCV patients in Lahore, Pakistan by real-time PCR. Afr Health Sci. 2020;20(3):1143-1152.
Crossref - Islam N, Krajden M, Shoveller J, et al. Hepatitis C cross-genotype immunity and implications for vaccine development. Sci Rep. 2017;7(1):12326.
Crossref - Riaz S, Bashir MF, Haider S, Rahid N. Association of genotypes with viral load and biochemical markers in HCV-infected Sindhi patients. Braz J Microbiol. 2016;47(4):980-986.
Crossref - Hedskog C, Parhy B, Chang S, et al. Identification of 19 Novel Hepatitis C Virus Subtypes-Further Expanding HCV Classification. Open Forum Infect Dis. 2019;6(3):ofz076.
Crossref - Wada N, Ikeda F, Mori C, et al. Mixed HCV Infection of Genotype 1B and Other Genotypes Influences Non-response during Daclatasvir + Asunaprevir Combination Therapy. Acta Med Okayama. 2018;72(4):401-406.
- McPhee F. Developments in the treatment of HCV genotype 3 infection. Expert Rev Anti Infect Ther. 2019;17(10):775-85.
Crossref - Kumar A, Rajput M, Paliwal D, Yadav A, Chhabra R, Singh S. Genotyping & diagnostic methods for hepatitis C virus: A need of low-resource countries. Indian J Med Res. 2018;147(5):445-455.
Crossref - Bhattacharya PK, Roy A. Management of Hepatitis C in the Indian Context: An Update. J Liver. 2015;4.
Crossref - Gupta V, Kumar A, Sharma P, Arora A. Newer direct-acting antivirals for hepatitis C virus infection: Perspectives for India. Indian J Med Res. 2017;146(1):23-33.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.