ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin resistant Staphylococcus aureus harbouring other virulence factors in food is a serious threat to food safety. Delayed detection of MRSA may lead to fatal consequences due to the challenging treatment methods. To address the issue, early prognosis is prerequisite. We report one step QuEChERS detection method wherein, a media with selective agents such as cefoxitin, cyanidin, lithium chloride and methicillin (CCLM) was formulated for the specific cultivation and detection of MRSA within 5 hours. Direct application of the newly developed method was tested by screening food for MRSA and its comparison with conventional biochemical and molecular characterization. Growth of MRSA in the CCLM media was observed within 4 hours. This highly specific method is sensitive to detect 104 CFU/mL visually and 101 CFU/mL spectrophotometrically. 25% of food samples harboured MRSA which points to an immediate need of consideration by the authorities. The newly designed QuEChERS cyanin based chromogenic detection system is unequivocal with the conventional and molecular method of MRSA detection and can be of great use in diagnostic and in food safety laboratories.
MRSA, QuEChERS, Anthocyanin, CCLM Media, mPCR, Antibiotic Resistant
Staphylococcus aureus is a common opportunistic pathogen found on the skin, nose and respiratory tract of humans and other animals. They are contributory agents of small skin infections to life threatening septic shocks. They are armoured with numerous virulence factors such as staphylococcal enterotoxins (SE), leukocidin, exfoliative toxins, haemolysin, toxic shock syndrome toxins (TSST) and less explored staphylococcal enterotoxin like proteins (SEl) that can trick the human innate and humoral immune responses and cause numerous health hazards in humans.1,2 Highly heat stable and low pH resistant super antigenic enterotoxins are the reason for food poisoning and toxic shocks.3 In 1960’s, the antibiotic resistant strain of S. aureus was first reported, which was nosocomial in origin and termed as HA-MRSA (Hospital associated – MRSA). Later, other lineages were identified which showed higher invasive ability termed as CA-MRSA (Community Associated-MRSA) and LA-MRSA (Livestock associated-MRSA). Presently, MRSA has emerged as a major pathogen that has acquired pan-resistance to all antibiotics used to treat microbial infections.4 Indiscriminate use of antibiotics is identified as the major root cause of antibiotic resistance in various microorganisms leading to increased public health problems, morbidity, mortality and expensive treatment methods.5,6 International travels and the global food trading play a vital role in the transmission of the bacteria across the world.7 Dissemination of USA 300, a CA-MRSA clone to Australia, Europe, South America and Japan is an example.8,9,10 This demands recurrent monitoring and surveillance in every country.
The incidence of LA-MRSA is increasing which can turn into a serious consequence in contaminating the food products.11 The uncontrolled and aggressive use of antibiotics as medicaments, protection and feed additive are the major reason for the achievement of antibiotic resistance in the livestock and colonization with the resistant strains (Joint Inter-agency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report, 2017). Thus, animals reared for food becoming colonized with the MRSA happen to be a favourable ecological niche for the evolution of more invasive strains which can lead to food borne outbreaks.12 The contamination of food with MRSA lineages other than LA-MRSA are reported from various studies across the world.13,7
Based on the observation of Indian Network for Surveillance of Antimicrobial Resistance (INSAR), the prevalence of MRSA throughout the country is calculated to be 42%.14 Incidence of MRSA in India has increased from 29% to 47% in recent years.15 74% of worldwide S. aureus infections are turned out to be MRSA. Till date, for the diagnosis of MRSA; phenotypic methods such as culture and colony counting methods have been in use; the immunological, molecular approaches like polymerase chain reaction (PCR), real time PCR (RT-PCR), flow cytometry, mass spectrometry, microarrays, fluorescence and electrochemical based methods are also currently used. Unfortunately, these methods are laborious and complex, as they require laboratory facilities, sophisticated and expensive instruments as well as specialized personnel to perform the technique. Moreover, these methods are not field deployable. On the other hand, conventional phenotypic methods yield results after 48h with an additional 24h of enrichment. Therefore, there is a high demand to develop new methods for rapid, sensitive and economical detection of MRSA. In the present study, we report QuEChERS (quick, easy, cheap, effective, rugged and safe) method by development of a new chromogenic CCLM (Cyanin, Cefoxitin, Lithium Chloride, Methicillin) media for the rapid detection of Methicillin-Resistant Staphylococcus aureus (MRSA) within 5 hours. The CCLM media contains four essential selective agents; cefoxitin, lithium chloride, methicillin and sodium chloride along with carbon, nitrogen and other nutrient sources for the selective enrichment of MRSA. Moreover, for the chromogenic identification, a purple dye cyanidin-3-O-diglucoside-5-O-glucoside (Anthocyanin) prepared from Brassica oleracea, a red variety of cabbage turns purple to pink on the growth of MRSA.
Another major focus of the study was to evaluate the application of the newly developed CCLM chromogenic system in laboratory settings. Moreover, considering the fact that the transmission of MRSA through food is less studied area, several food items were investigated by conventional biochemical and molecular methods for the incidence of MRSA. The study also focussed on finding potential food poisoning enterotoxin SEB, a CDC categorized bioweapon and pvl gene, a marker of CA-MRSA. The result was then compared with the newly developed chromogenic media. The media reported in the current study was found to have immense importance for the rapid detection of MRSA.
Bacterial strains and growth conditions
Bacterial strains used in the study are Staphylococcus aureus (ATCC 700699) and Methicillin-resistant Staphylococcus aureus (MRSA) from SDM Medical College and Hospital, Sattur, Dharwad, Karnataka (Supplementary data).16 Bacterial cultures were grown in Brain Heart Infusion (BHI) broth (HiMedia, Mumbai, India) at 37°C.
Formulation of new media for one step identification of MRSA
A novel enrichment CCLM media with carbon, nitrogen and other essential nutrients was formulated. Sodium chloride at the concentration of 5% and lithium chloride at 0.7% were used as the selective agents to suppress the growth of other interfering closely related organisms. The reported concentrations of cefoxitin and methicillin (0.6 mg and 0.4 mg in 100 mL) were incorporated to the media as selective agents to exclude MSSA (Methicillin Sensitive Staphylococcus aureus).
Preparation of Anthocyanin dye indicator incorporated CCLM media
Red cabbage (Brassica oleracea) was purchased from BigBasket, Mysore. The red cabbage leaves (100 grams) were taken in 200 mL of acidified methanol (0.1% of HCl) in a 500 mL flask. The mixture was incubated overnight at room temperature. The resulting red cabbage leaves extract was filtered and concentrated to ¼th of volume by rotary evaporation and was stored at -20°C for further use.17
Optimization of the dye concentration and sensitivity of media
To reduce the turnaround time, an experiment was conducted in microtitre plate where 5 ml CCLM media containing 25%, 10% and 5% anthocyanin extract was poured in 8 individual wells. The serially diluted MRSA cultures (10-1-10-8) were then inoculated into the media. The microtitre plate covered and were incubated for 4-18 hours at 37°C for the observation of colour change at every one-hour interval. The absorbance (λ max of 530 nm) was read with a UV-vis spectrometer (TECAN, India) to differentiate the colour change which cannot be visualized by the naked eye.18
The images of dilutions of the CCLM media were digitally analysed. All images were analysed using ImageJ software (National Institute of Health) and the RGB mean was calculated. The values were then used for calculating the ִE values (distance or difference between two colours) using an online platform Colormine.org (http://colormine.org/delta-e-calculator).
Sample collection and Staphylococcus aureus isolation
All food samples used in this study were collected from various parts of Mysore city, Karnataka, India. These samples were aseptically transported to the laboratory in sterile bags or bottles (Table 1). All samples were pre-enriched in BHI broth overnight at 37°C. Serially diluted
(10-1-10-8) overnight cultures in peptone water were then plated onto Baird Parker Agar (BPA) plates supplemented with egg yolk–tellurite emulsion (HiMedia, Mumbai, India) and were incubated at 37°C for 24 – 48 hours.
Biochemical characterization
Further characteristic black centred colonies with opaque zone of S. aureus grown on BPA plates were biochemically characterized. Presumptive colonies were subjected to the Indole, Methyl Red-Voges Proskauer, Mannitol fermentation, Coagulase, Catalase, Phosphatase production and DNase test. The positive colonies were then further screened with MeReSa agar (HiMedia, Mumbai, India), which is a chromogenic agar specific for the MRSA.
Genomic DNA isolation and PCR
Genomic DNA for this study was isolated by DNASure mini kit according to the manufacturer’s instructions (Genetix, India). Extracted DNA was screened for the presence of nuclease gene (nuc), Methicillin resistance determinant gene (mecA), SEB toxin coding gene (seb), and Panton-valentine leukocidin S and F fraction encoding genes (lukF and lukS) (Table 2).19
Multiplex PCR (mPCR) optimization
A total of two multiplex PCR were standardized for this study. First set of mPCR was performed for the determination of MRSA with nuc and mecA primers. It was then followed by the second mPCR which was optimized with lukF, lukS (pvl) and nuc primers for the characterization of CA-MRSA. The reactions were optimized with 1X PCR buffer, 2.5 mmol l-1 MgCl2, 0.2 mmol l-1 dNTP mixture and 1.2 unit of Taq polymerase. The primer concentrations were optimized to 0.5 µ mol l-1 of nuc F and nuc R and 0.6 µ mol l-1of mec A F and mec A R. For the second set of mPCR the primer concentration was optimized to 0.7 µ mol l-1 of both luk F and luk S primers and the concentration of nuc primer was maintained the same as the first set of mPCR.
Multiplex PCR conditions were optimized as an initial denaturation for 4 minutes at 94°C followed by 30 cycles of denaturation for 1 minute at 94°C, annealing for 1 minute at 56°C, extension for 1 minute at 72°C and a final extension for 8 minutes at 72°C.
Another PCR reaction was optimized for the detection of seb gene. The amplification of gene was found good at the primer concentration of 0.6 µ mol l-1 of seb F and R. The reaction condition was maintained as mentioned above.
Automated Identification and antibiotic susceptibility (AST) studies
All the PCR positive isolates were then loaded onto the BD PHOENIX M50 system installed in Defence Food Research Laboratory, Mysore, Karnataka. Positive isolates were identified and studied for their antibiotic resistance pattern using this system. Various biochemical tests were performed in the ID panel of appropriate Gram stain. ID panels are embedded with several chromogenic and fluorogenic substrates which help in the identification of the cultures used. This system determines the antibiotic susceptibility by minimal inhibitory concentration (MIC). It uses two-fold dilutions of various antibiotics embedded in the Gram Positive/Negative panels to determine the MIC.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism (GraphPad Software, Version 8.0.2). A two-way ANNOVA test was preformed and a p-value < 0.05 was considered statistically significant.
In this study, we reported a novel natural dye incorporated media; CCLM media, which can reduce the turnover time of the MRSA detection. Anthocyanin is a Food and Drug Administration approved pigment, which is used as pH indicator due to its ionic nature that changes colour based on the pH.20,21 They exist as red in acidic, blue/ purple in basic and green/yellow at higher pH conditions.22 The advantages of using anthocyanin are its low cost of production, easy preparation, bio-compatibility, production of wide-ranging colour spectrum and high stability at harsh experimental conditions.23
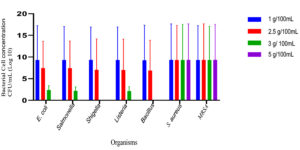
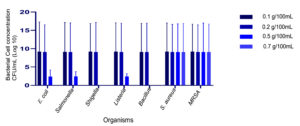
In the current study, a new media was formulated with the optimized concentrations of NaCl and LiCl. Lower concentrations of NaCl in the range of 1.0 – 2.5% could not inhibit most of the non-specific test organisms. Increasing the concentration of NaCl to 3.0% and 0.5% of LiCl also could not suppress the growth of Listeria monocytogens, Salmonella typhi and Escherichia coli. Faint growth was observed in all these. Complete suppression of other organism and selective growth of S. aureus (both MSSA and MRSA) was observed at the concentration of 5% NaCl and 0.7% LiCl (Figure 1). Further, cefoxitin and methicillin were used to suppress the growth of MSSA.
Figure 1. A: Optimization of NaCl concentrations. Growth of different microbial isolates in varying concentration of NaCl. The cultures used are Escherichia coli, Salmonella typhimurium, Shigella sonii, Listeria monocytogens, Bacillus cereus, Staphylococcus aureus and MRSA. B: Optimization of LiCl concentrations: Growth of different microbial isolates in varying concentration of LiCl. The cultures used are Escherichia coli, Salmonella typhimurium, Shigella sonii, Listeria monocytogens, Bacillus cereus, Staphylococcus aureus and MRSA
Optimization of Anthocyanin concentration
To reduce the detection time of MRSA, anthocyanin was incorporated in the CCLM media. In order to obtain an optimal colorimetric signal, the optimal concentration of anthocyanin was investigated. A series of concentrations ranging from 5% to 25% were tested with known cell concentration of 105 CFU/ml of MRSA in the CCLM media.
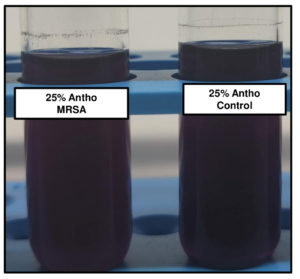
Anthocyanin gets protonated as the organism grows and utilizes mannitol, producing acid in the medium. At acidic pH, purple anthocyanin turns to pink. As the growth increased with time, the protonation of anthocyanin will give the colour change. At 25% anthocyanin concentration the colour couldn’t be differentiated (Figure 2A). The colour differentiation was prominent in 10% anthocyanin tubes. In 5% of the tubes, the colour was very faint to differentiate between the positive and negative control (Figure 2B). Hence 10% of anthocyanin concentration was employed for further studies.
Figure 2. A: Tubes with 25% of Anthocyanin + MRSA and Anthocyanin alone respectively. B: 1) 5% Anthocyanin Control, 2) 5% Anthocyanin + 5gm Nacl, 3) 5% Anthocyanin + 7.5gm NaCl, 4)10% Anthocyanin Control, 5)10% Anthocyanin + 5gm NaCl, 6)10% Anthocyanin + 7.5gm NaCl; tubes 2, 3, 5 & 6 have MRSA. All the tubes were incubated at 37⁰C overnight for the observation of color change
Cell concentration versus time of protonation of anthocyanin
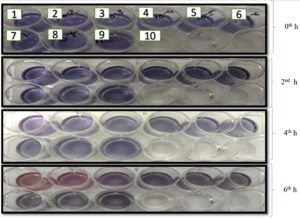
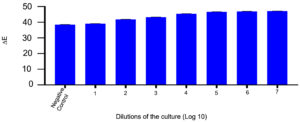
101 CFU/mL to 108 CFU/mL of MRSA cells were inoculated in CCLM media with 10% of anthocyanin. At the time of inoculation, the colour of the media was purple. As the organism reaches to its logarithmic stage of growth phase, the change in the colour could be visualized which took around 4 hours with cell concentration of >106 CFU/mL and 6 to 7 hours with lower cell concentration of 102 CFU/mL (Figure 3). The colour change is due to production of volatile compounds by MRSA that reduces pH of the media. The colour changes are due to the protonation of anionic quinonoidal base to quinonoidal base.24,25 This system is quick as changes can be visualized with the naked eye within 4 hours of incubation.
Figure. 3. A: Microtitre plate with 5 ml CCLM media in each well inoculated with 10 µl MRSA culture. The plate was incubated at 37⁰C and was observed at 2nd, 4th and 6th hour. The overnight grown MRSA culture used was serially diluted from 10-1 to 10-8 (L-R) plus 2 control (CCLM without culture & media without anthocyanin). A well with CCLM control was also used to compare the color change observed. B: ∆E value is plotted in x-axis against the dilution of culture used in Log 10 to evaluate the color change.
Specificity of the CCLM media
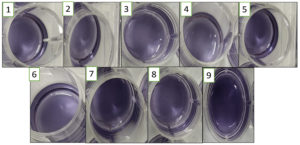
Different closely related organisms and common food spoilage and food borne disease causing agents were inoculated in the CCLM media incorporated with the natural dye (Figure 4, Supplementary data). These non-specific test organisms did not grow in these tubes and no change in the colour of the broth was observed in these tubes. The media remained purple even after 48 hours of incubation indicating that, the CCLM incorporated with anthocyanin will not support the growth of other organisms except S. aureus and the pigment is stable.
Figure 4. The overnight incubated cultures of 1. Bacillus cereus, 2. Escherichia coli, 3. Enterococci, 4. Klebsiella pneumonia, 5. Listeria monocytogens, 6. Staphylococcus aureus, 7. Shigella sonii, 8. Salmonella typhimurium, 9. Control (No culture added) in CCLM
Comparison of CCLM media with the traditional and molecular methods of MRSA cultivation and confirmation
A total of 70 food samples were screened for the presence of MRSA. Presumptive colonies from BPA plates were subjected to different biochemical tests. Of 70 samples 35 samples were found to be harbouring S. aureus and the results are depicted in the Table 1. The confirmed S. aureus colonies were then streaked onto MeReSa agar plates supplemented with MeReSa supplement and cefoxitin. The light pink coloured colonies were selected as it is ascribed to be distinctive morphology of MRSA on this medium.
Table (1):
Samples used and the characterization with traditional and CCLM media
| Sample | No.of sample | Biochemical positive (S. aureus) | Culture positive ( S. aureus) | Molecular positive | CCLM positive (MRSA) | |
|---|---|---|---|---|---|---|
| S. aureus | MRSA | |||||
| Pastries | 10 | 7 | 7 | 7 | 5 | 5 |
| Chicken | 10 | 6 | 6 | 6 | 5 | 5 |
| Mutton | 8 | 5 | 5 | 5 | 1 | 1 |
| Fish | 6 | 4 | 4 | 4 | 1 | 1 |
| Milk | 6 | 4 | 4 | 4 | 1 | 1 |
| Curd | 2 | 0 | 0 | 0 | 0 | 0 |
| Cheese | 2 | 1 | 1 | 1 | 0 | 0 |
| Prawns | 1 | 1 | 1 | 1 | 0 | 0 |
| Egg | 3 | 1 | 1 | 1 | 0 | 0 |
| vegetables | 12 | 1 | 1 | 1 | 0 | 0 |
| Ready to eat foods | 10 | 5 | 5 | 5 | 5 | 5 |
| Total | 70 | 35 | 35 | 35 | 18 | 18 |
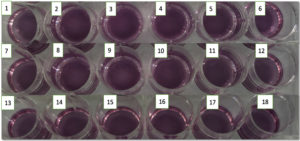
All these isolates were inoculated into CCLM anthocyanin media and observed for the growth and colour change (Figure 5). The detection of MRSA by traditional culturing method takes 2-3 days but the CCLM media is quick as it can detect MRSA in 6-7 hours and thus the delay in prognosis can be avoided. Thus, the patients can get immediate attention and further infections can be prevented.
Figure 5. M1, B. M2, C. M3, D. M4, E. M5, F. M6, G. M7, H. M8, I. M9, J. M10, K. M11, L. M12, M. M13, N. M14, O. M15, P. M16, Q. M17 and R. M18 are the 18 isolates cultured in 5 ml CLMN media (after 8-12 hours of incubation)
The traditional method of identification of the MRSA includes overnight pre-enrichment of the samples with the brain heart infusion broth and then streaking on to Baird parker agar supplemented with egg yolk emulsion. Then followed by the biochemical characterization like DNase test, mannitol fermentation etc. Then Latex agglutination is done for the characterization as coagulase positive or negative S. aureus. Then finally going for confirmation by PCR amplification. MIC tests of different antibiotics are done in case of MRSA. It takes almost 3-4 days of tedious work.
Table (2):
List of primers
Genes Targeted |
Primer Sequence 5′-3′ |
Amplicon size (bps) |
Accession No. |
|---|---|---|---|
mec A |
For-GGATTGCTTCACTGTTTTG |
800bps |
JF778650.1 |
Rev-GAGTAGCACTCGAATTAGGC |
|||
Nuc |
For-GCTGGCATATGTATGGCAATT |
385 bps |
DQ50738.1 |
Rev-GCTTCAGGACCATATTTCTCTAC |
|||
Luk-S |
For-GCAGCTCAACATATCACACC |
191 bps |
AB532027.1 |
Rev-TGTTTCCAGCAGCTTTGAG |
|||
Luk-F |
For-CAGCTCAACATATCACACCT |
623 bps |
AB532027.1 |
Rev-GAAGTTTTGTCCAGCATTTAAG |
|||
Seb |
For-CCAGATCCTAAACCAGATGAGTT |
325 bps |
AB479119.1 |
Rev-GTTTTTCGTTTGTCAGTTTGATG |
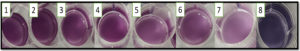
Whilst in a short span of 6-8 hours the change in colour was observed indicating the growth of MRSA. This newly formulated broth is of immense help for the easy and early detection of MRSA. This can even be used by a lay man under field condition. Similar results were observed when the spiked food samples after homogenization and filtration was inoculated into CCLM anthocyanin broth (Figure 6).
Figure 6. Spiked food samples after 8-12 hours of incubation. Wells 1-3:-Cake, 4:-Mysore Pak, 5- Chicken, 6:- Peda, 7-Milk, 8:- Negative Control ( Without any sample)
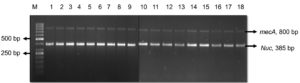
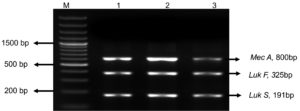
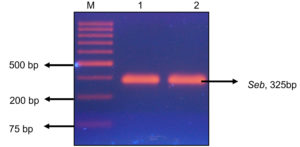
Further the genomic DNA was isolated from the positive colonies and was amplified using nuc, mecA, seb and pvl genes. Out of 18 MRSA isolates, all were found to be nuc and mecA positive (Figure 7A). Five isolates were found to be harbouring pvl gene indicating they were CA-MRSA as pvl genes are markers of the CA-MRSA.26 (Figure 7B). Two isolates possessed seb gene which specified they were potent food poisoning agents (Figure 7C). This is highly alarming with the existing COVID-19 situation as the immuno-compromised/geriatric patients can get the infection through food and the treatment if delayed can turn fatal.27 The pvl positive S. aureus were isolated from chicken, ready to eat foods and confectioneries and seb was detected in ready to eat food and pastry. Thus, the study gave us a warning that both HA and CA MRSA are wide spread in the community and the colonization is getting transmitted to different host species.
Figure 7. A: Agarose gel electrophoresis image of nuc and mec A gene duplex PCR: M-50bp DNA ladder (GeneRuler, Thermo Scientific, India) 1-18:- M1-M18 isolates. B: Agarose gel electrophoresis image of mPCR product of pvl and nuc gene in M3, M12 and M16:- M- 1 Kb molecular marker (GeneDirex, India), 1-M3,2- M12, 3- M16. C: Agarose gel electrophoresis image of seb gene (325bp) in isolate M5 and M13:- M- 1kb molecular marker, Gene (Ruler, Thermo Scientific, India) 1- M5, 2- M13.
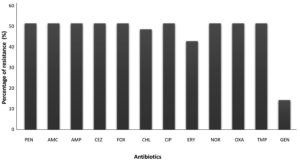
The cultures from CCLM Anthocyanin media were identified with BD PHOENIX M50. The ID results showed all the 18 samples were positive for Staphylococcus aureus. The AST report showed that all were MRSA and most of them showed resistance to Amoxicillin, Ampicillin, Cefazolin, Cefoxitin, Chloramphenicol, Ciprofloxacin, Norfloxacin, Oxacillin, Penicillin G, Erytromycin and Trimethoprin (Figure 8).
Figure 8. Resistance pattern in the MRSA isolates: Percentage is calculated by multiplying the fraction of N/T to 100, where N is number of MRSA isolates resistant to particular antibiotic and T is the total number of MRSA isolates. PEN- Penicillin, AMC- Amoxicillin-clavulanic acid, AMP-Ampicillin, CEZ-Cefazolin, FOX- Cefoxitin, CHL-Chloramphenicol, CIP- Ciprofloxacin, ERY- Erythromycin, NOR- Norfloxacin, OXA- Oxacillin, TMP- Trimethoprim, GEN- Gentamicin
Anthocyanin is now widely investigated as an indicator for spoilage in various food sources.28 The growth of many organisms releases acidic compounds leading to the protonation of the hydroxyl groups associated with the phenolic rings of anthocyanin leading to the change in conjugation pattern all the way through the molecules resulting in the colour change.29,25 The pigment is safe, i.e. does not cause any harm to the food products and to the environment, as they are not toxic or are not involved in production of any harmful by-products. The growth of non-specific organisms is meagre in this medium. As only with one medium we can confirm the presence of MRSA, the method is Cheap. Another advantage of the media is the ease of preparation; the components are environment friendly, involvement of less machinery and sophisticated techniques, i.e. it is quick, easy, cheap, effective, rugged and safe (QuEChERS).
In our study, almost 25% of the total samples harboured MRSA although the sample size was small. This is quite a frightening outcome as superbug load was significantly higher to the size of area chosen for the study. Approximately 28% (5/18) of the total MRSA positive food samples harboured pvl gene and 11% (2/18) carried seb gene. These were all protein rich foods like chicken, sea food, mutton and dairy products consumed daily. A recent study reviewed the reported MRSA isolation from different pig, milk, beef and poultry products both raw and processed.30 The findings agree with the report of EFSA, 2009 that the protein rich foods are reservoirs of MRSA. The domesticated animals and other animals used for food or even handling of these animals harbouring the antibiotic strains can get transferred or colonized in humans. Horizontal gene transfer can happen from the resistant strains of LA-MRSA genes to methicillin sensitive Staphylococcus aureus (MSSA). Several studies have even reported about Linezolid-resistant S. aureus (LRSA). Though its prevalence is low compared to other strains its risk of transmission to humans through the animal handling and animal derived foods cannot be neglected. This also points to the significance of intensive monitoring of the livestock and food.31,32,33
When discussing about the presence of MRSA with pvl, mecA and seb genes in the ready to eat foods and processed food are mainly due to the unhygienic handling post process practices during packing, storage till it reaches the consumer and is a major public health concern.34 The prevalence of S. aureus strains capable of producing enterotoxins and multidrug resistance together could pose a major threat to human kind in near future and food has become a major and easy vehicle of antimicrobial resistance transmission which is difficult to control and treat as well.35-38
In India, from past two decades transmission of MRSA of various lineages (HA-MRSA, CA-MRSA and LA-MRSA) through food has been noticed.39-41 Although in India, the screening of food for the presence of MRSA is not mandatory, the potential role of food in diffusing MRSA lineages cannot be overlooked. Whilst in European countries, EFSA (European Food Safety Authority) recommends continuous monitoring of animals reared for food especially intensively reared ones accompanied with surveillance in humans to study the inclinations of transmission and evolution of zoonotically acquired MRSA in humans.42-45
The outcome of our study was startling and rings alarm for scrutiny and proper characterization of the infections. The unceasing screening of food and food producing animals also is obliged to be made mandatory to comprehend and study the expansion of resistance in MRSA. The lack of awareness of the MRSA infection and transmission may help these superbugs in mounting new virulence characteristics and transmissibility and the currently available treatments may not work as these superbugs have gained more armours.
In conclusion, the study is an effort to screen the presence of MRSA and MSSA and their virulence potential in food. The study has identified 18 samples as MRSA isolates, all were found to be nuc and mecA positive. Five isolates were found to be CA-MRSA and two isolates carried seb gene. This study also focused on formulation of a natural dye cyanidin-3-O-diglucoside-5-O-glucoside incorporated CCLM single step media to cultivate and confirm the presence of MRSA within 6 to 8 hours. The volatile acidic compounds released by the MRSA during growth will change the purple colour of the media to pink. The method is rapid and also simple as even a layman can detect the presence of MRSA by observing the colour change.
Additional file: Additional supplementary data.
ACKNOWLEDGMENTS
The authors are grateful to the Director of Defence Food Research Laboratory, Mysore, Karnataka, for supporting this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clinical microbiology reviews. 2000;13(1):16-34.
Crossref - Grumann D, Nübel U, Bröker BM. Staphylococcus aureus toxins–their functions and genetics. Infection, Genetics and Evolution. 2014;21:583-92.
Crossref - Al-Tarazi YH, Albetar MA, Alaboudi AR. Biotyping and enterotoxigenicity of Staphylococci isolated from fresh and frozen meat marketed in Jordan. Food research international. 2009;42(3):374-9.
Crossref - Mehndiratta PL, Bhalla P. Use of antibiotics in animal agriculture & emergence of methicillin-resistant Staphylococcus aureus (MRSA) clones: need to assess the impact on public health. The Indian Journal of Medical Research. 2014;140(3):339-3448.
- Geneva: 2003. FAO/OIE/WHO. Non-human antimicrobial usage and antimicrobial resistance. Available from http://www.who.int/foodsafety/publications/micro/en/amr.pdf [Accessed on June 20, 2011]
- Kluytmans JA. Methicillin-resistant Staphylococcus aureus in food products: cause for concern or case for complacency?. Clinical microbiology and infection. 2010;16(1):11-5.
Crossref - Rodríguez-Lázaro D, Ariza-Miguel J, Diez-Valcarce M, Fernández-Natal I, Hernández M, Rovira J. Foods confiscated from non-EU flights as a neglected route of potential methicillin-resistant Staphylococcus aureus transmission. International Journal of Food Microbiology. 2015;209:29-33.
- Nadig S, Raju SR, Arakere G. Epidemic meticillin-resistant Staphylococcus aureus (EMRSA-15) variants detected in healthy and diseased individuals in India. Journal of medical microbiology. 2010;59(7):815-21.
Crossref - Shibuya Y, Hara M, Higuchi W, Takano T, Iwao Y, Yamamoto T. Emergence of the community-acquired methicillin-resistant Staphylococcus aureus USA300 clone in Japan. Journal of infection and chemotherapy. 2008;14(6):439-41.
Crossref - Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. Journal of Antimicrobial Chemotherapy. 2009;64(3):441-6.
Crossref - D’Souza N, Rodrigues C, Mehta A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. Journal of clinical microbiology. 2010;48(5):1806-11.
Crossref - Verhegghe M, Crombé F, Luyckx K, et al. Prevalence and genetic diversity of livestock-associated methicillin-resistant Staphylococcus aureus on Belgian pork. Journal of food protection. 2016;79(1):82-9.
Crossref - Ogata K, Narimatsu H, Suzuki M, Higuchi W, Yamamoto T, Taniguchi H. Commercially distributed meat as a potential vehicle for community-acquired methicillin-resistant Staphylococcus aureus. Applied and environmental microbiology. 2012;78(8):2797-802.
Crossref - India SJ, Ray P, Manchanda V, et al. Methicillin resistant Staphylococcus aureus (MRSA) in India: prevalence & susceptibility pattern. The Indian journal of medical research. 2013;137(2):363.
- Center For Disease Dynamics, Economics & Policy, https://cddep.org/research-area/mrsa/
- Nagaraj S, Ramlal S, Sripathy MH, Batra HV. Development and evaluation of a novel combinatorial selective enrichment and multiplex PCR technique for molecular detection of major virulence-associated genes of enterotoxigenic Staphylococcus aureus in food samples. Journal of applied microbiology. 2014;116(2):435-46.
Crossref - Chandrasekhar J, Madhusudhan MC, Raghavarao KS. Extraction of anthocyanins from red cabbage and purification using adsorption. Food and bioproducts processing. 2012;90(4):615-23.
Crossref - Lee J, Durst RW, Wrolstad RE. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: collaborative study. Journal of AOAC international. 2005;88(5):1269-78.
Crossref - Poojary NS, Ramlal S, Sripathy MH, Batra HV. A novel mPCR for the detection of prominent toxins in MRSA strains of S. aureus recovered from diverse sources International Journal of Research in Biological Sciences. 2013;1(10):129-38.
- Chigurupati N, Saiki L, Gayser Jr C, Dash AK. Evaluation of red cabbage dye as a potential natural color for pharmaceutical use. International journal of pharmaceutics. 2002;241(2):293-9.
Crossref - Pourjavaher S, Almasi H, Meshkini S, Pirsa S, Parandi E. Development of a colorimetric pH indicator based on bacterial cellulose nanofibers and red cabbage (Brassica oleraceae) extract. Carbohydrate Polymers. 2017;156:193-201.
Crossref - Turturica M, Oancea A, Rapeanu G, Bahrim G. Anthocyanins: naturally occuring fruit pigments with functional properties. Annals of the University Dunarea de Jos of Galati Fascicle VI–Food Technology. 2015;1;39(1):9-24.
- Celik C, Can Sezgin G, Kocabas UG, et al. Novel anthocyanin-based colorimetric assay for the rapid, sensitive, and quantitative detection of helicobacter pylori. Analytical chemistry. 2021;93(15):6246-53.
Crossref - Celik C, Ildiz N, Kaya MZ, Kilic AB, Ocsoy I. Preparation of natural indicator incorporated media and its logical use as a colorimetric biosensor for rapid and sensitive detection of Methicillin-resistant Staphylococcus aureus. Analytica Chimica Acta. 2020;1128:80-9.
Crossref - Roy S, Rhim JW. Anthocyanin food colorant and its application in pH-responsive color change indicator films. Critical Reviews in Food Science and Nutrition. 2021;61(14):2297-325.
Crossref - Darboe S, Dobreniecki S, Jarju S, et al. Prevalence of Panton-Valentine leukocidin (PVL) and antimicrobial resistance in community-acquired clinical Staphylococcus aureus in an urban Gambian hospital: a 11-year period retrospective pilot study. Frontiers in cellular and infection microbiology. 2019:170.
Crossref - Kevin K. Taking Aim at Antibiotic-Resistant Bacteria During COVID. Infection Control Today, 2022;24(9).
- Choi I, Lee JY, Lacroix M, Han J. Intelligent pH indicator film composed of agar/potato starch and anthocyanin extracts from purple sweet potato. Food chemistry. 2017;218:122-8.
Crossref - Khoo HE, Azlan A, Tang ST, Lim SM. Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food & nutrition research. 2017;61(1):1361779.
Crossref - Krumova-Valcheva GL. The role of food in the spread of the Methicillin – resistant Staphylococcus aureus (MRSA). Food Science and Applied Biotechnology, 2023, 6(1), 87-94.
Crossref - Leao C, Clemente L, Cara d’Anjo M, Albuquerque T, Amaro A. Emergence of cfr-mediated linezolid resistance among livestock-associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) from Healthy Pigs in Portugal. Antibiotics, 2022;11(10):1439.
Crossref - Lienen T, Grobbel M, Tenhagen BA, Maurischat S. Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany. Antibiotics, 2022; 11(12): 1802.
Crossref - Timmermans M, Bogaerts B, Vanneste K, et al. Large diversity of linezolid-resistant isolates discovered in food-producing animals through linezolid selective monitoring in Belgium in 2019. Journal of Antimicrobial Chemotherapy, 2022;77(1): 49-57.
Crossref - Mahros M, Mohamed S, Khalid AE, Sallam I. Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health,concern. International Journal of Food Microbiology, 2021, 346(5):109165.
- Argudín MA, Mendoza MC, González-Hevia MA, Bances M, Guerra B, Rodicio MR. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Applied and environmental microbiology. 2012;78(8):2930-5.
Crossref - Castro A, Santos C, Meireles H, Silva J, Teixeira P. Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. Journal of Infection and Public Health. 2016;9(2):153-60.
Crossref - Wang W, Baloch Z, Jiang T, et al. Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Frontiers in Microbiology. 2017; 8:2256.
Crossref - Yang X, Zhang J, Yu S, Wu Q, Guo W, Huang J, Cai S. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Frontiers in microbiology. 2016;7:816.
Crossref - Sivakumar M, Dubal ZB, Kumar A, et al. Virulent methicillin resistant Staphylococcus aureus (MRSA) in street vended foods. Journal of food science and technology. 2019;56(3):1116-26.
Crossref - Murugadas V, Joseph TC, Lalitha KV. Tracing contamination of Methicillin-resistant Staphylococcus aureus (MRSA) into seafood marketing chain by staphylococcal protein A typing. Food Control. 2017;78:43-7.
Crossref - Zehra A, Gulzar M, Singh R, Kaur S, Gill JP. Prevalence, multidrug resistance and molecular typing of methicillin-resistant Staphylococcus aureus (MRSA) in retail meat from Punjab, India. Journal of global antimicrobial resistance. 2019;16:152-8.
Crossref - European Food Safety Authority. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008-Part A: MRSA prevalence estimates. EFSA Journal. 2009;7(11):1376.
Crossref - European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA Journal. 2015;13(1):3991.
Crossref - European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA Journal. 2017;15(2):e04694.
Crossref - European Food Safety Authority (EFSA). The community summary report on food-borne outbreaks in the European Union in 2007. EFSA Journal. 2009;7(5):271r.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.