ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.32 | © The Author(s). 2019
One problem faced by chrysant farmer in North Sumatera, Indonesia is to control fungal disease caused by Fusarium oxysproum. The aims of this study were to know antagonistic bacterial isolates to reduce fungal disease caused byF. oxysporum and to know the isolates ability in producing plant growth hormon such as indole acetic acid and in solubilizing phosphate. Eight bacterial strains, collection of Laboratory of Microbiology, Universitas Sumatera Utara were tested against F. oxysporum in dual culture assay. Two bacterial isolates, SP1, closely related to Bacillus amyloliquefaciens strain MPA 1034 and SP7R, closely related to Pseudomonas versuta strain L10.10 showed to reduce fusarium wilt in chrysant along with production of indole acetic acid and phosphate solubilizing properties in vitro. Application of the two antagonistic bacterial isolates resulted in considerably decrease of disease intensity caused by F. oxysporum. The results showed that utilization of bacterial isolate to promote chrysant performance is promising in the next future.
Antagonistic bacteria, biological control, plant growth promoting, Chrysanthemum sp.
Chrysanth (Chrysanthemum sp.) is one of the most popular ornamental plants, with relatively high economic value and future marketing prospect. The domestic and foreign market demand is increasing every year (Arjana et al., 2015). One of the problems in chrysanth cultivation is fungal disease. Disease caused by Fusarium oxysporum or Fusarium wilt may render the cultivation of chrysanths in farms due to the severe disease intensities and attacks. Fusarium oxysporum is a soil-borne fungal pathogen that can survive in the soil for 8-20 years in the form of chlamydospores which are difficult to control. Mechanism of Fusarium wilt is by blocking the significant flow of water in xylem tissue of chrysanths, causing the leaves to turn yellow and wilt, and then the plants die, suffering from inadequate water and nutrients (Singh et al., 2014).
Fusarium oxysporum has been controlled using various chemical and fungicides such ethazol, benomyl, bitertanol, triadimefon, thiabenzadole, which shown effective to decrease Fusarium wilt in plants. However it may has negative effect to the environment. Crop rotation, cultivation, and sterilization are used to control the wilt disease of chrysant. Reduction of F. oxysporum with antifungal and antimicrobial properties of several plants extracts and essential oils were investigated by many workers. Inhibition of Fusarium using extract of Tagetus erecta (Riaz et al., 2008), Menthanasp (Ghorbany et al., 2010), and Lantara camera (Begum et al., 2007) were done. Treatment of plant extracts efficaciously reduced F.oxysporum.
Biological control is considered an environment-friendly approach and it has gradually replaced the application of traditional chemical pesticides to reduce environmental problem (Li et al., 2017). The use of antagonistic microbes generally does not have a negative impact on the environment compared to the use of synthetic fungicides. Trichoderma (Locke et al., 1985), Penicillium sp. (Hussain et al., 2016), Alcaligenes faecalis and Bacillus cereus (Abdallah et al., 2016), and Pseudomonas fluorescens (Mohammed et al., 2011) were antagonistic microorganisms which potential as biocontrol agent for reducing Fusarium wilt in plants.
Local farmers generally perform fertilization and control of pathogenic fungi in chrysanths by using synthetic fertilizers and fungicides. The use of synthetic fertilizers and fungicides may cause soil damage, fungal resistance, resurgence, health and environment problem. An alternative to suppress Fusarium wilt is to utilise a biological control agent. This study examined two bacterial isolates SP1 and SP7R to control Fusarium wilt and to increase chrysant growth performance.
Isolation of fungal pathogen from chrysanth
Fungal pathogens were isolated from diseased plants of relatively cold area of Brastagi, exhibiting Fusarium wilt symptoms. Plant leaves were removed, cleaned with running water, cut into 1 cm2 size, immersed in 70% ethanol for 1 minute, rinsed in sterile distilled water for 1 minute, then drained on filter paper. After drying, the sample pieces were placed on a petridish containing potato dextrose agar(PDA) medium. The culture was incubated for three days at ambient temperature until the fungal mycelium grew. The fungal colony emerging from the plant part is purified to new PDA.
Identification of fungal pathogen
Fungal pathogens were identified by using slide culture method. Isolates were inoculated at four points of block of PDA agar blocks measuring 1 cm2 on the object glass, covered with glass, and then sterile distilled water was dropped on filter paper to keep moisturity. The slide cultures were incubated for 3–4 days, then slide observed under microscope. Fungal identification was performed using standard identification book of Gilman (1975) and Pitt et al., (2009)
In vitro antagonisms of bacterial isolates against F.oxysporum causative agent of fusarium wild in chrysant
Antagonisms of bacterial isolates against F.oxysporum were observed using dual culture assay. Bacterial isolates of rhizospheric bacteria of Brassica rapa pekinensis of relatively cold area of Brastagi (SP7R, SP9R, SP11R), endophytic bacteria of Brassica rapa pekinensis (SP5E, SP4E), and aerobic bacteria from liquid waste of oil palm (SP1, SP14, SP15) of previous study were collections from Laboratory of Microbiology, Universitas Sumatera Utara. Mycelium of F. oxysporum was cut and inoculated in the center of a petri dish containing a medium of PDA + 3% yeast extract. A total of 10mL of bacterial suspension, each with concentration of » 108 CFU.mL-1, was inoculated into blank paper discs (Oxoid), arranged on two side of fungal culture with a distance of 1 cm. The dual culture plates were then incubated for 7 days at amibient temperature. Inhibitory activity is indicated by the formation of inhibitory zones around the bacterial colonies towards fungal culture (Suryanto et al., 2016). Antagonism activity was further characterized microscopically by observing the mycelium at inhibitory zone of dual culture plates, showing some abnormal hyphae characteristics (Lorito et al., 1992).
Screening of plant growth promoting properties
Bacterial isolates were subjected to screen for their plant growth promoting properties i.e. phosphate solubilization and production of exogenous auxinIAA. To determine phosphate solubilizing activity of isolates, Pikovskaya medium was used in this assay (Pikovskaya, 1948). The plates were inoculated with bacterial isolates and incubated for 7 days at ambient temperature. The formation of visible and clear halo zones around bacterial colonies confirmed the positive result of phosphate solubilization. To determine the production of exogenous IAA of isolates, colorimetry method was used in this assay. The assay was done as follows: three millilitres of bacterial suspension (» 108 CFU.mL-1) was inoculated into 27 mL Luria-Bertani broth medium, supplemented with L-tryptophan in 100 ml flask, incubated in ambient temperature with 100 rpm agitation. To observe the trend of IAA production in culture, sufficient volumes of broth were sampled for every 4 hrs, and centrifuged at 5500 rpm for 10 minutes. The resulting supernatants were transferred into sterile reaction tube, filled with Salkowski reagent(4:1, v/v).The mixture was incubated for 20 minutes and measured for its absorbance using a spectrophotometer at a wavelength of 530 nm compared to the standard curve (ppm).
In vivo antagonism of bacterial isolates against F. oxysporum
The assay was done in greenhouse. Ten millilitres of bacterial suspension (» 108 CFU.mL-1) was inoculated into 2 kilograms mixture of soil and compost in pot. A 10 mL of sterile distilled water without bacterial suspension were used as a control. After 7 days chrysanth seeds were planted in the pot. Inoculation of 10 mL per pot of F. oxysporum (H” 106CFU.mL-1) was done after 7 days of seed planting. Observations were conducted until 40 days including disease intensities, number of yellowing and wilting leaves, plant heights, and total number of leaves. Treatments of chrysant in pot were:
Cell number counted
One gram of soil in treatment was taken and diluted in sterile distilled water to make 10-7to 10-8 suspension. One ml of suspension was inoculated into plate count agar and incubated at 31°C for 24-48 hours. Cell number was counted as colony forming unit per gram soil (CFU.g-1).
Molecular identification of potential isolates using 16S rRNA gene
Freeze-thawed method was utilised to break bacterial cell. Bacterial suspension of SP1 and SP7R in microtube was frozen at -10°C and thawed at 90°C for 10 minutes. This was done for five times for cell-breaking efficiency. Cell-breaking solution containing DNA was used for the amplification of the 16S rRNA gene performed using PCR method with a specific primer of 63f (5′-CAG GCC TAA CAC ATG CAC-3) and 1387r (5′-GGG CGG WGT GTA CAA GGC- 3 ‘) (Marchesi et al., 1998). The PCR wascarried out for 40 cycles in a Thermo Cycler (LabCycler gradient) with the initial denaturation at 94°C for 2 min, cyclic denaturation at 92°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final extension of 5 min at 72°C. The PCR products were checked visually by using agarose gel electrophoresis. Purification and commercially sequencing was done to the PCR product. The sequenced data were checked by BLAST analysis.
Isolation and identification of F.oxysporum causative agent of fusarium wild in chrysant
Eight isolates P1, P2, P3, PD1C, PD2, PD3, PD4, and PDX of potential fungal pathogens were isolated from diseased chrysanths were.One strain, P3 was further identified as F.oxysporum based on morphological characters. This isolate d showed white mycelium with pinkish or purplish center of colonies. Microscopy observation confirmed that ‘P3 showed to have microconidia, and an oval-shaped, elongated, and septate macroconidia (Fig. 1). The confirmation of strain P3 as F.oxysporum was also compared to characteristics described by Pitt et al., (2009) and Gilmann (1975).
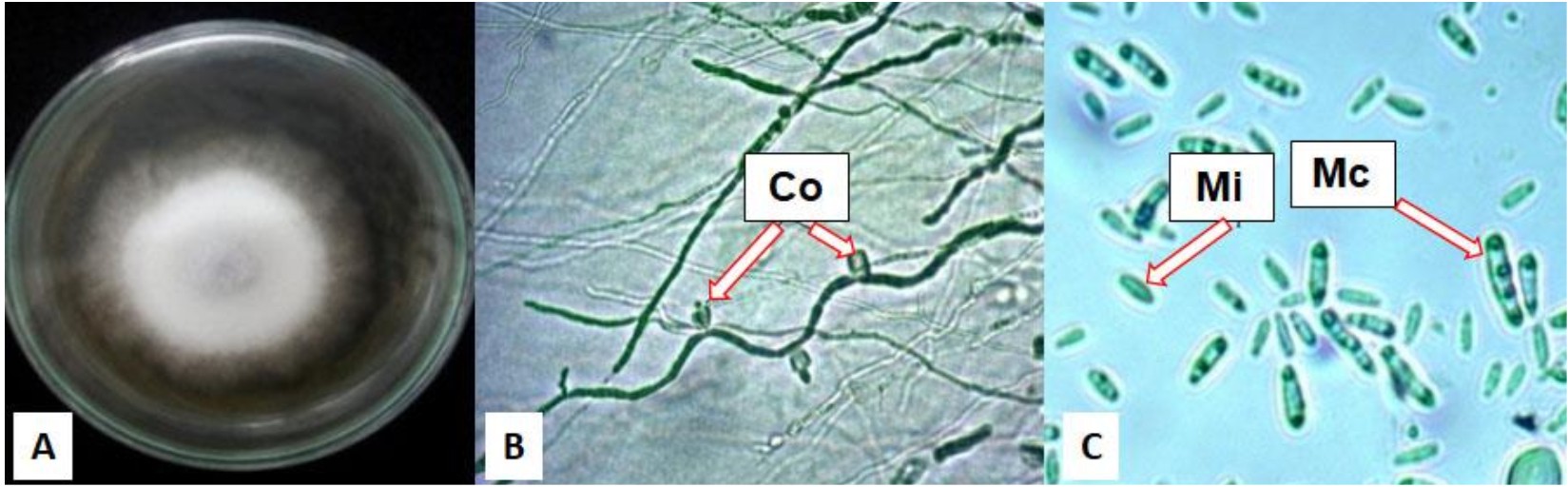
Fig. 1 . F. oxysporum isolated from unhealthy chrysant. (A). Colony on PDA medium after 7 incubation days at 29°C, (B). Hypha with conidiospore (Co), (C). Microconidia (Mi) and Macroconidia (Ma)
Dual culture assay on antagonistic bacterial isolates against F.oxysporum
Four bacterial isolates SP1, SP5E, SP7R, and SP9R showed to inhibit F.oxysporum growth (Fig. 2). It showed that bacterial isolates varied in inhibiting fungal growth (Table 1). The varying degree of antagonism among isolates were assumed due to differences in production of antimicrobe substancies such as extracellular hydrolytic enzymes chitinase and glucanase into the dual culture medium. Fungal cell walls are mainly composed of chitin (4–9% of dry weight), followed by other component of 1,3-glucans. These polymers are the main target of hydrolytic enzymes of antagonistic bacteria (Benhamou et al., 1990; Rajarathnam et al., 1998). Suryanto et al.(2011) also stated that effectiveness of antagonistic bacteria in inhibiting fungal pathogens was affected by composition of fungal cell walls, differences in growth rate between bacteria and fungi, and competing metabolites produced by bacteria to attack fungi.
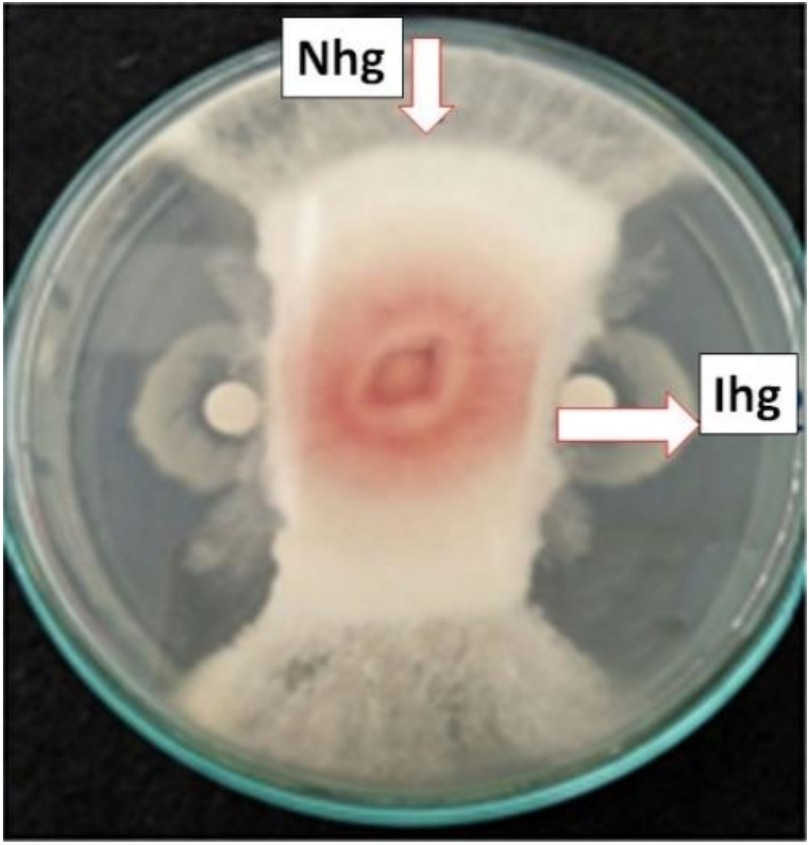
Fig. 2. Antagonistic assay of SP1 to F. oxysporum (Nhg: normal hyphae growth; Ihg: inhibited hyphal growth)
Table (1):
Inhibition zone produced by antagonistic bacteria against F. oxysporum
Isolates |
Inhhibition zone (mm) |
|---|---|
SP7R |
7.0 |
SP9R |
5.5 |
SP11R |
0 |
SP5E |
3.30 |
SP4E |
0.00 |
SP1 |
15.5 |
SP14 |
0 |
SP15 |
0 |
Antagonistic activity of antagonistic bacteria against F. oxysporum was observed through microscopical examination of abnormal hyphae. The antagonistic activity of four bacterial isolates caused F.oxysporum hyphae to develop abnormal growth such as stunted, curved, distorted, lysed, and dwarf hyphae when compared to normal hyphae (Fig. 3). Inhibition of fungal growth by antagonistic bacteria was shown by the inhibition zone around fungal radial growth. The fungal mycelium did not increase in length hence the tip of the mycelium looks short and flat. The abnormalities of the hyphae were assumed to be caused by hydrolytic enzymes produced by antagonistic bacteria within inhibition zones. Meanwhile the results of the research Asnita et al., (2012) stated that the antagonistic activity of chitinolytic bacteria causes F.oxysporum hyphae to be damaged, lysed, distorted and curve. According to Gohel et al., (2005), the amount of bacterial chitinase and glucanase will increase when pathogenic fungi attack. Both enzymes attack the cell wall causing hypha lysis, fragmentation and fungal hypha growth is inhibited. Chitinase enzyme is a hydrolytic enzyme that can hydrolyze b-1,4 bonds between N-acetylglucosamine subunits in chitin polymer. Damage to chitin polymer which is an important component of the cell wall of fungal hyphae can inhibit hyphae growth.
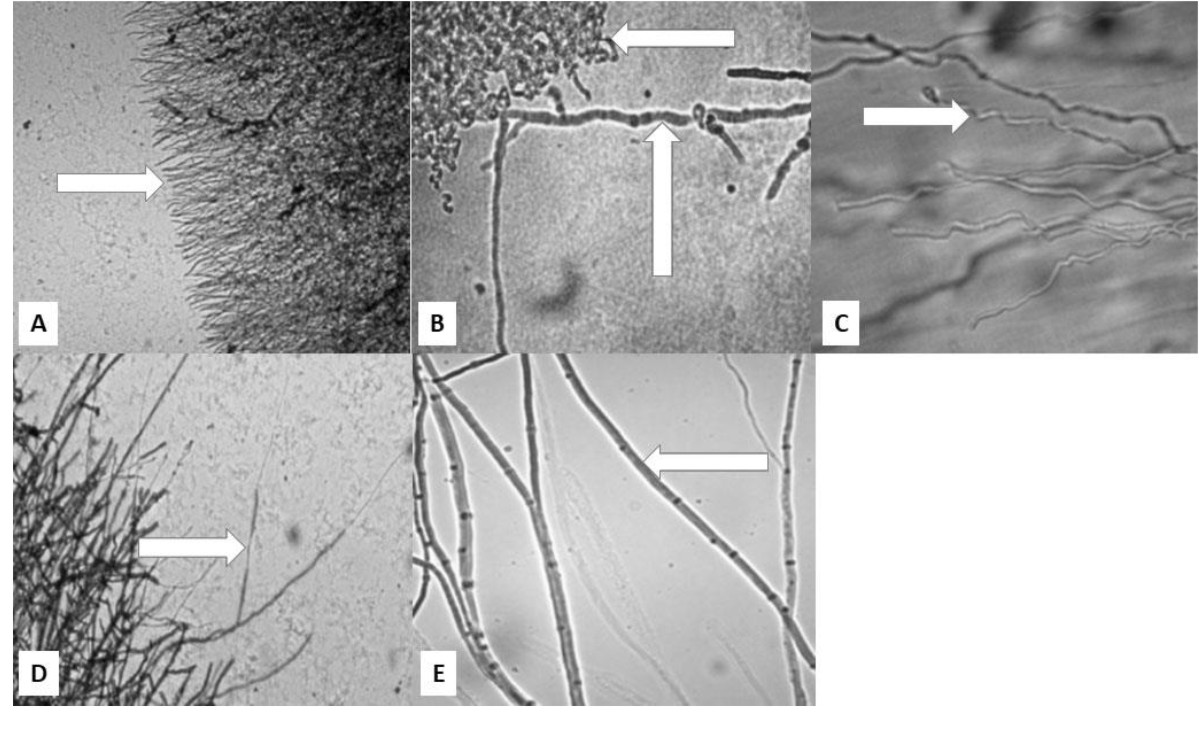 Fig. 3. Microscopical image of hyphal abnormalitiesshowed by arrow: A. stunted, B. curved, C. distorted, D. lysed and dwarfed, and E. normal hyphae.
Fig. 3. Microscopical image of hyphal abnormalitiesshowed by arrow: A. stunted, B. curved, C. distorted, D. lysed and dwarfed, and E. normal hyphae.Plant growth promoting properties of antagonistic bacterial isolates
Four antagonistic bacteria were subjected to phosphate solubilization assay in agar medium supplemented with insoluble tricalcium phosphate on Pikovskaya medium. The four isolates showed different ability in solubilizing phosphate as shown in Table 2. The formation of halo zones around colonies indicated the synthesis of extracellular organic acids which are able to bind ion Ca3+ bound into insoluble form of Ca3(PO4)2 and free the ion H2PO4, forming a more clear zone than surrounding zone with bound P element in agar medium (Awais et al., 2017). Another mechanism to solubilize bound P element is by producing extracellular phosphatase that enzymatically hydrolyze P from its bound compounds (Parhamfar et al., 2016). Both mechanism is triggered genetically in which may explain the different ability among bacteria in effectively utilizing phosphate in environment (Khan et al., 2009).
Table (2):
Plant growth promoting ability produced by antagonistic bacteria against F. oxysporum
| Isolates | Plant Growth Promoting Properties | |
|---|---|---|
| Phosphate Solubilization Index | IAA Production (ppm) | |
| SP7R | 0.96 | 83.50 |
| SP9R | 0.75 | – |
| SP11R | 0 | – |
| SP5E | 0.62 | – |
| SP4E | 0 | – |
| SP1 | 0.49 | 23.33 |
| SP14 | 0 | – |
| SP15 | 0 | – |
Two bacterial isolates SP1 and SP7R capable to solubilise phosphate were chosen and subjected to IAA production assay. It was shown that SP7R produced more IAA copared to that of SP1 in 24 hrs of incubation. Higher production of IAA demonstrated during late logarithmic and early stationary phase of bacterial growth. Ji et al. (2014) reported the yield of IAA reaching 24.6 ppm during stationary phase of ten bacterial strains used in their study. Limited nutrients and low pH contributed to transcription of certain genes involved in catalysis of potential intermediet compounds into IAA. Addition of L-tryptophan into medium may also enhance synthesis of IAA (Spaepen et al., 2007). Our result might be considered higher due to the nature of bacterial isolate used in this assay. SP7R sampled from rhizosphere of B.rapa pekinensis showed to have high IAA production in this study. Rhizospheric bacteria are known as potential IAA producers because of their interaction with plant roots. Plant roots secrete exudates such as L-tryptophan to bacteria, in order to enhance the root growth by incorporating exogenous IAA synthesized by rhizospheric bacteria into plant (Khalid et al., 2004; Rosenblueth et al., 2008).
In vivo antagonism of two selected antagonistic bacterial isolates against F.oxysporum
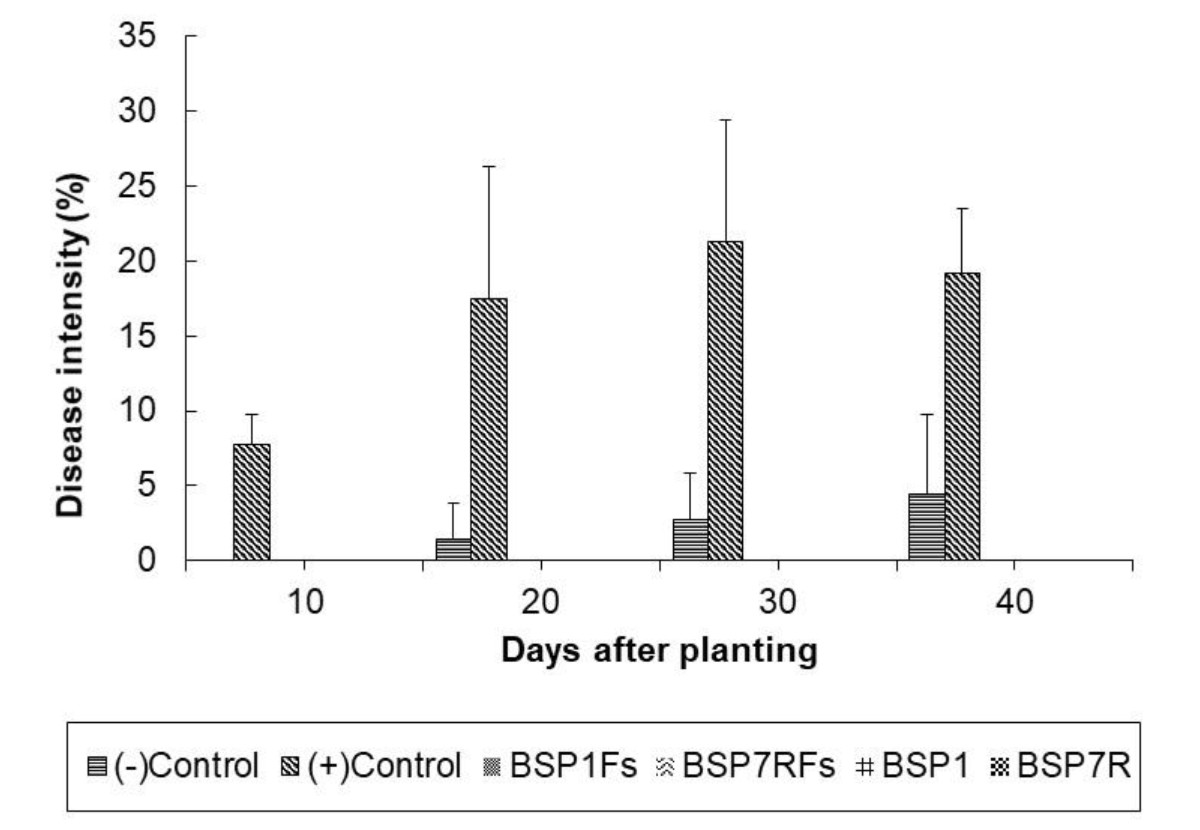
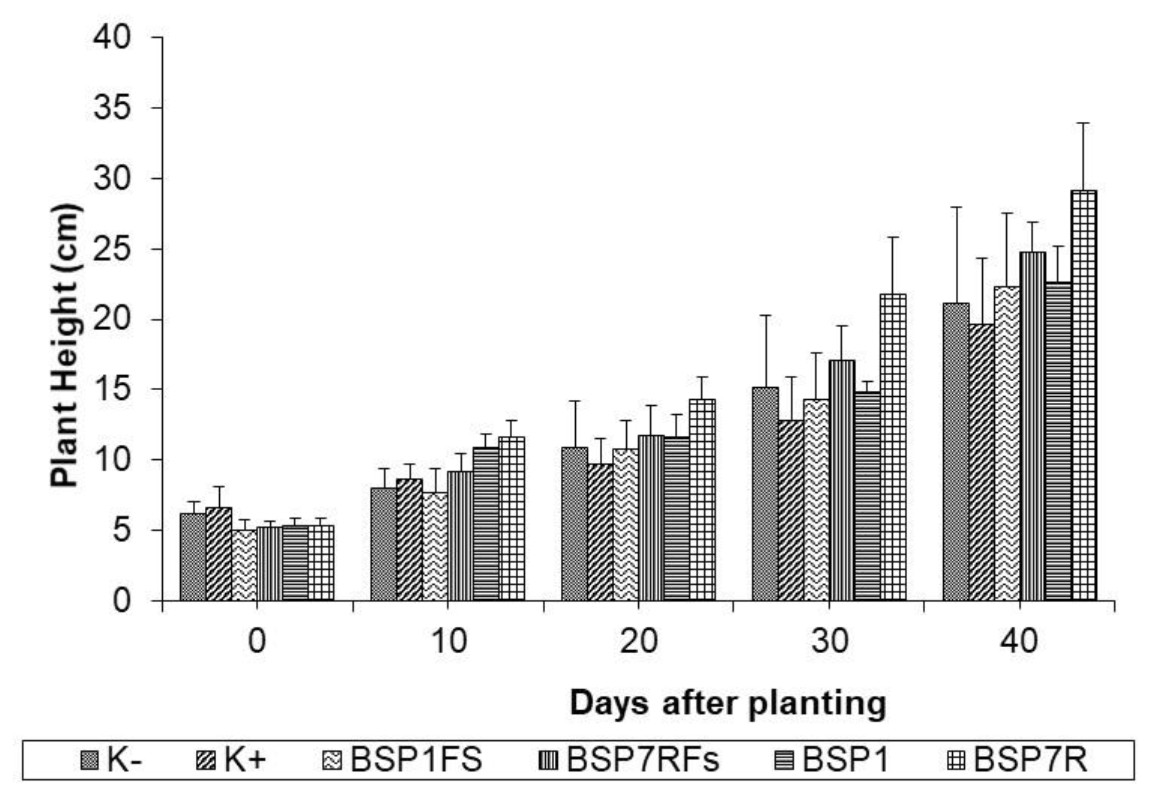
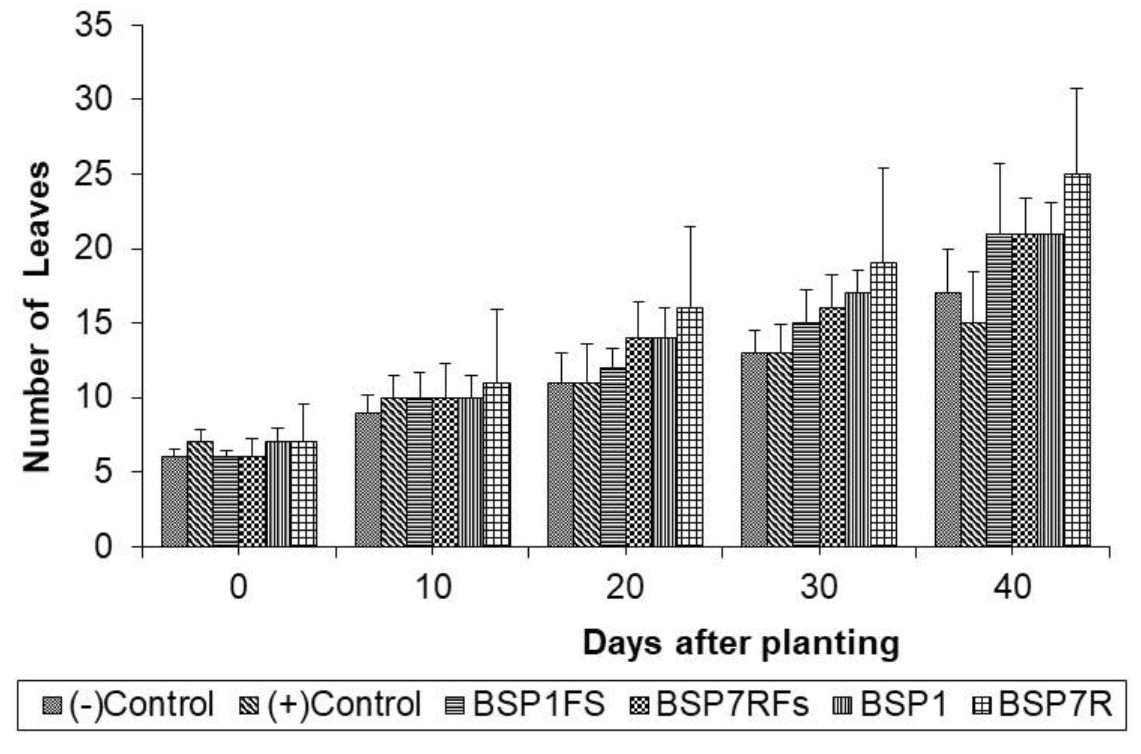
It was observed that treated chrysanth with antagonistic bacteria did not show any Fusarium wilt symptoms during 40 days of planting as compared to that of (+) control, chrysant infested with fungal conidia (Fig. 4). Overall profile of plant parameters in terms of plant height and number of leaves were also improved in general (Fig. 5 and 6). Other studies have reported the potential of antagonistic bacteria in treating F.oxysporum infection in several plants. Paenibacillus sp. 300 and Streptomyces sp. 358 were reported to reduce Fusarium wilt on cucumber plants (Singh et al., 1999). Other study reported the potential of three antagonists, Streptomyces setonii, Bacillus cereus, and Serratia marcescens to reduce Fusarium wilt symptoms in the stem of tomato plants by F.oxysporum (Ferraz et al., 2014). Other factor besides hydrolytic enzymes produced by antagonistic bacteria in reducing Fusarium wilt were siderophores, cyanide, and antibiotics (Cantelan et al., 1999). Those products may be evaluated in future study to gain deeper understanding of antagonistic mechanisms from our isolates.
It showed that SP7R increased more plant performance rather than that of SP1. The ability of SP7R to improve plant growth seemed correlating with high production of IAA. Bacterial strains capable of producing IAA might accelerate the growth of plants (Bolero et al., 2007). Several IAA-producing bacteria such as Acinetobacter, Alcaligenes, Arthrobacter, Azospirillium, Azotobacter, Bacillus, Beijerinckia, Burkholderia, Enterobacter, Erwinia, Flavobacterium, and Rhizobium were reported (Esitken et al., 2010). IAA functions as essential signal molecule to regulate plant growth, such as accelerating root growth, increasing resistance against pathogens, with final outcome on overall plant growth promotion (Shaharoona et al., 2006). Chrysant growth might also be supported by phosphate solubilization ability from isolates to provide sufficient plant needs of nutrients (Thakuria et al., 2004).
Bacterial cell number in pot soil
Bacterial cell number after 40 days of planting reached » 1013 CFU.g-1 (Table 3). This could be assumed that bacterial cell of SP1 and SP7R survived in all treatment pots and tend to thrive in the soils. Hence, cell densities between BSP1-BSP7R (promoting groups) and BSP1FR-BSP7RFS (antagonistic groups) did not differ. The abundance of soil organic matters in soil might support growth of most soil bacteria until reaching adequate cell number to execute their phenotypic roles in nutrient cycling and niches.
Table (3):
Bacterial cell number among treatments before and after application
| Treatments | Number of cell densities (CFU.g-1) | |
|---|---|---|
| Before application | After application | |
| B0 | 7.0 × 107 | 1.2 × 108 |
| B0FS | 8.0 × 107 | 9.0 × 108 |
| BSP1 | 1.7 × 108 | 5.2 × 1013 |
| BSP7R | 1.1 × 108 | 3.6 × 1013 |
| BSP1FS | 9.0 × 107 | 2.9 × 1013 |
| BSP7RFS | 1.4 × 108 | 3.7 × 1013 |
Molecular identification of antagonistic bacterial isolates based on their 16S rRNA gene
Two bacterial isolates, SP1 and SP7R were choosen to be identified. Isolation of gene fragment, 16S rRNA, followed by amplification in PCR using primers 63f and 1387r for 40 cycles, was successfully done. Sequence analysis using Basic Local Alignment Search Tool (BLAST) using database of 16s rRNA genes, showed considerable similarity of SP1 and SP7R toBacillus amyloliquefaciens MPA 1034 by 90% and to Pseudomonas versuta L 10.10 by 99%, respectively.
Arias et al. (2009) reported that B. amyloliquefaciens was a promising biocontrol agent and plant growth promoting bacteria based on genetic and biochemical characteristics. Application of B. amyloliquefaciens in reducing severity of plant diseases and controlling microbial pathogens was reported in several plant disease in several plant species such as bacterial wilt caused by Ralstonia solanacearum in tobacco plants, fruit rot caused by Botryosphaeria berengeriana in pear, and stem rot caused by Sclerotinia sclerotiorum in canola plants (Pingping et al., 2007; Wu et al., 2014; Wu et al., 2016). The bacteria had ability to synthesize antifungal lipopeptides, volatile organic compounds, and cell wall degrading enzymes (Wu et al., 2014). In this study, newly isolated P.versuta of relatively cold area of Brastagi for the first time was reported as biocontrol agent or plant growth promoting bacteria. The species was described as novel species by See-too et al., (2017), isolated from Antarctic soil. The prospect on evaluating further characteristics of both isolates as biocontrol and plant growth promoting agents is still needed for field application and deeper understanding of their natures.
Antagonistic bacteria can be used as an alternative to control plant diseases and to increase plant growth. The ability of antagonistic bacteria to inhibit the growth of plant pathogenic fungi is thought to be due to their ability to produce chitinase and glucanase. According to Adams (2004), Chitinase and glucanase are hydrolase enzymes that can hydrolyze chitin and b-glucan which are the main components of fungi cell walls. Hydrolysis of chitin and b-glucan in the walls of fungi cells can reduce the integrity of the walls of fungal cells so that they cannot infect plants. Chitinase and 1.3 glucanase are enzymes that are key to inhibiting the growth of fungi.
Bacteria can increase plant growth by dissolving phosphate and producing IAA. Phosphorus (P) is one of the nutrients that are important for plant growth, but its availability is very low in the soil because most phosphorus is bound by calcium (Ca) and magnesium (Mg) in calcareous soils and by iron (Fe) and aluminum (Al) in acidic soils, so there is very little available in plants (Liu et al. 2015), therefore phosphate solubilizing bacteria are needed in increasing the availability of phosphorus (P) in plants.
The ability of phosphate dissolving bacteria is related to the enzyme phosphatase and organic acids synthesized by these microorganisms. The phosphatase enzyme can decide which phosphate is bound by organic compounds to be the form available to plants. The presence of organic acids can directly dissolve phosphate which is the result of changes in PO43- anions by acid anions or the binding of Fe and Al ions which previously bind P elements so that they can release phosphates which are bound and can be absorbed by plants (Khan et al. 2009). Meanwhile, the ability of bacteria to produce auxin depends on the phase of bacterial growth and the production of auxin is usually maximal in the stationary phase (Ji et al. 2014)
The direct mechanism of bacteria in increasing plant growth includes producing growth hormones, auxin, cytokinin, gibberellins, inhibiting the production of ethylene hormones, solubilizing phosphate , and nitrogen fixation. Plant growth is increased indirectly by bacteria through its ability to produce antimicrobial pathogens, siderofors and enzymes that can suppress the growth of fungi that cause plant diseases (Esitken et al. 2010).
Eight bacterial strains, collection of Laboratory of Microbiology, Universitas Sumatera Utara were tested against F.oxysporum in dual culture assay. Two bacterial isolates SP1 and SP7R closely related to Bacillus amylolique-faciens strain MPA 1034 and to Pseudomonas versuta strain L10.10 reduced fusarium wilt in chrysant. They produced indole acetic acid and capable to solubilise phosphate in vitro. Application of the two antagonistic bacterial isolates resulted in considerably decreased of disease intensity caused by F.oxysporum and increased chrysant performance.
We would like to express our deep appreciation to Universitas Sumatera Utara for partly supporting this study.
The author declares that there are no conflict of interest.
- Shakibaie, M., Forootanfar, H., Golkari, Y., Mohammadi-Khorsand, T., and Shakibaie, M. R. Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. J Trace Elem Med Biol. 2015; 29:235-241.
- Mohammed, G. J., Kadhim, M. J., and Hameed, I. H. Proteus species: Characterization and herbal antibacterial: A review. International J Pharmacog and Phytochem Res, 2016; 8(11):1844-1854.
- Stankowska, D., Kwinkowski, M., and Kaca, W. Quantification of Proteus mirabilis virulence factors and modulation by acylated homoserine lactones. J Microbiol Immunol Infect, 2008; 41(3):243-253.
- Howery, K. E., Clemmer, K. M., and Rather, P. N. The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr Genetics, 2016; 62(4):775-789.
- Ouda, S. M. Some nanoparticles effects on Proteus sp. and Klebsiella sp. isolated from water. Am J Infect Dis Microbiol, 2014; 2(1):4-10.
- Cestari ,S.E., Ludovico,M.S., Martins,F.H., Rocha,S.P.D., Elias,W.P. and Pelayo. J.S. Molecular Detection of HpmA and HlyA Hemolysin of Uropathogenic Proteus mirabilis. Curr Microbiol., 2013; 67:703–707.
- Rozalski, A., Torzewska, A., Moryl, M., Kwil, I., Maszewska, A., Ostrowska, K., .and Staחzek, P. Proteus sp.–an opportunistic bacterial pathogen–classification, swarming growth, clinical significance and virulence factors. Folia Biologica et Oecologica, 2012; 8(1):1-17.
- Uphoff, T. and Welch, R. Nucleotide sequencing of the Proteus mirabilis calcium independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with Serratia marcescens hemolysin genes (shlA and shlB). J Bacteriol., 1990; 172:1206–1216.
- Jumaily, E. F., and Zgaer, S. H. Characterization and molecular detection of purified Proteus mirabilis pmbs 41 alpha-hemolysin. Basrah J Veter Res, 2016; 15(3).
- Liaw, S. J., Lai, H. C., Ho, S. W., Luh, K. T., and Wang, W. B. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J Med Microbiol, 2003; 52(1):19-28
- Swihart, K. G., and Welch, R. A. The HpmA hemolysin is more common than HlyA among Proteus isolates. Infect and Immun, 1990; 58(6):1853-1860.
- Sosa, V.; Schlapp, G.and Zunino, P. Proteus mirabilis isolates of differentorigins do not show correlation with virulence attributes and can colonize the urinary tract of mice. Microbiol, 2006; 152(7):2149-2157.
- AL-Jumaa, M. H.; Bnyan, I. A. and Al-Khafaji, J. K. Bacteriological and Molecular Study of Some Isolates of Proteus mirabilis and Proteus vulgaris in Hilla Province. A thesis for the Degree of Master of Science in Microbiology. College of Medicine, University of Babylon, 2011.
- Mishara, M.; Thakar, Y.S. and Pathak, A.A. Haemagglutination, haemolysin production and serum resistance of Proteus and related species isolated from clinical sources, Indian J Med Microbiol. 2001; 19(2):5-11
- Abdul-Lateef, L. A. Sequencing of Proteus Toxic Agglutinin (Pta) Gene in Proteus Mirablis and Cytotoxic Effect of Pta on Human Colon Cancer Cell and Human Kidney Cell. J Global Pharma Technol, 2017; 9(9).
- Mordi, R. M., and Momoh, M. I. Incidence of Proteus species in wound infections and their sensitivity pattern in the University of Benin Teaching Hospital. African J Biotechnol, 2009; 8(5).
- Fraser GM, Claret L, Furness R, Gupta S, Hughes C. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiol, 2002; 148:2191–2201.
- Strauss, E. J., Ghori, N., and Falkow, S. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect and Immun, 1997; 65(9):3924-3932.
- Plomin, R., DeFries, J. C., Knopik, V. S., and Neiderheiser, J. Behavioral genetics. Palgrave Macmillan, 2013.
- Li, W., Raoult, D., and Fournier, P. E. Bacterial strain typing in the genomic era. FEMS Microbiol Revs, 2009; 33(5):892-916.
- Hadfield, J. D., and Nakagawa, S. General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi trait models for continuous and categorical characters. J Evolution Biol, 2010; 23(3), 494-508.
- Case, R. J., Boucher, Y., Dahllof, I., Holmstrצm, C., Doolittle, W. F., and Kjelleberg, S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environmen Microbiol, 2007; 73(1):278-288.
- Ghebremedhin, B., Layer, F., Konig, W., and Konig, B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J Clinic Microbiol, 2008; 46(3):1019-1025.
- Michelim, L., Muller, G., Zacaria, J., Delamare, A. P. L., Costa, S. O. P. D., and Echeverrigaray, S. Comparison of PCR-based molecular markers for the characterization of Proteus mirabilis clinical isolates. Brazilian J Infect Dis, 2008; 12(5), 423-429.
- Ma, R., Wu, X., Wang, R., Wang, C., and Zhu, J. Identification and phylogenetic analysis of a bacterium isolated from the cloaca of Chinese alligator. African J Biotechnol, 2008; 7(13).
- Brenner, D. J., Staley, J. T., and Krieg, N. R. Classification of Procaryotic Organisms and the Concept of Bacterial Speciation. Bergey’s Manual of Systematics of Archaea and Bacteria, 2015; 1-9.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.