ISSN: 0973-7510
E-ISSN: 2581-690X
Rhizobacteria and endophytic bacteria are popular for its abilities in influencing plant growth and development. The strategy employed these bacteria as biofertilizer for planting is believed to bring several benefits such as low cost, eco-friendly, and feasible. One of the remarkable products for plant growth promoting provided by rhizobacteria and endophytic bacteria were the advantageous enzymes such as 1-aminocyclopropane-1-carboxylate deaminase, phosphatase, and cellulase. These biocatalysts then involve in several direct or indirect pathways of nutrient, growth factor, and/or defense factor synthesizes. From five different essential leafy vegetables in Thailand, this study aimed to investigate the plant growth promoting potentials of endophytic bacteria and rhizobacteria isolated from root tissue and rhizosphere, respectively, via IAA quantitative and enzyme activity assays. The selected bacterial strains were further identified using 16S rRNA gene sequencing and observed their interaction with plant root using scanning electron microscope method. Our study, thus far, has isolated two bacterial strains of Bacillus subtilis MSE5 and Bacillus cereus AVR1, respectively, with multifunctional traits of potential on the plant growth. Importantly, these two strains of MSE5 and AVR1 had shown the capacity to advance root colonization. Therefore, MSE5 and AVR1 are recommended for further studies in developing eco-friendly biofertilizer. In addition, some novel cellulose-degrading bacterial strains with significant potential on hydrolysis capacity were also isolated that might be valuable for industrial applications.
Rhizobacteria, Endophytic Bacteria, Plant Growth Promotion, IAA Quantitative, Biofertilizer, Vegetable, Thailand
Root colonization is defined as the capability of bacteria to proliferate and multiply in the rising root under the soil.1 Rhizobacteria are members of rhizosphere bacteria group that have the ability of root colonization as well as other impacts on plant growth and health through various mechanisms.2,3 In 1986, beneficial rhizobacteria has been mentioned as plant growth promoting rhizobacteria (PGPR).3 The PGPR plays an essential role in crop production as bio fertilizer with its advantages including low cost, environmentally friendly and easy practical application,4 whereas endophytic bacteria can infiltrate the plant’s interior tissue without causing harm to the host.5,6 Both endophytes and rhizobacteria have an great influence on plant growth and development through different benefits including biological nitrogen fixation, growth hormone secretion (cytokinin and gibberellins), nutrient uptake and assimilation (phosphate solubilization, ammonia and siderophores production). Moreover, they also play key role in regulating plant stress responses in order to cope with biotic and abiotic stresses.7–10 Given that the potentiality of endophytes as agrochemicals is steadily increased, diverse microbial endophytes associated with medicinal plants have been studied and found to exhibit antimicrobial activity against phytopathogens and human pathogenic microbes, produce several extracellular enzymes, effect on plant growth parameters including root elongation, length and weights.11-16 Bacterial endophytes are considered to have a closer relationship with the host and live in a more sustainable environment than rhizobacteria.17,18 Moreover, endophytes habitually initiate from the soil which may related to rhizosphere or phyllosphere and infect the plant through root cracks as capable root colonizers.6,19 Thus, research on the relationship of rhizobacteria and endophytic bacteria with the host plant and the study area can implement aspects of microbial population diversity. Since then, the effect of those bacteria on crops and soil has been further clarified to contribute to the sustainable production of vegetable crops.
In Thailand, leafy vegetables have been chosen as one of the main ingredients of the meal due to its high nutritional value, low cost of planting and easy growth. Some reports have shown the ability of leafy vegetables in reducing the risk of some diseases relating to diet.20 In addition, they contain large amounts of antioxidants, beta-carotene, retinol, and other nutrients that lead to the application of them as elements for medicines.21-23
Endophytic bacteria have important roles in a variety of plant growth processes, promoting both direct and indirect methods. The concept is similar to how rhizobacteria involve in the microbial synthesis of phytohormones, furthermore is the potential to produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase, phosphatase, and resistant environmental stress via endospores. In addition, several growth-related processes in plant are manipulated based on the presence and concentration of ethylene. Belong to ethylene, IAA and other enzymes may have agonistic or antagonistic effects on metabolism pathways that promote plant growth, highlighting the necessity of extremely precise hormone localisation and homeostasis in various cells and plant tissues.24 For instance, they are involved in stimulating root hair production while suppressing main root elongation, or lateral root creation and hypocotyl elongation.25,26
Both endophytic bacteria and rhizobacteria have the ability for producing ACC deaminase, which converts ammonia and a-ketobutyrate into nitrogen sources for beneficial bacteria. Moreover, ACC works as a precursor of ethylene production, resulting in root and shoots elongation as well as protection from the inhibitory effects of ethylene on the plant. Therefore, plants with endophytic ACC deaminase-producing bacteria become more stress resistant.27 Hence, the use of low doses of ACC may enhance ethylene biosynthesis, increased the initiation of lateral root primordial, the reverse showed that initiate lateral root primordial were inhibited and the growth of already existing lateral roots was favored. Especially, under constant flooded conditions, the synthesis of ACC deaminase was discovered to be a helpful characteristic for elongation and endophytic colonization of rice roots.28
Enzymatic production is one of the plant growth promotion activities that produce from plant growth promoting microorganism. Cellulase is an enzyme that can degrade cellulose that contains in the cell wall of plant.29 Cellulase is produced by fungi or bacteria, but bacteria have significant potential in cellulase production. Although the synthesis of cellulase by bacteria is not frequently used, Cellulomonas, Cellvibrio, Pseudomonas, Bacillus, and Micrococcus have been shown to have cellulolytic properties.30 Cellulolytic microbial inoculant is demanded for composting process. Also, they can convert agricultural wastes to become organic fertilizer.
In recent years, scientists became aware of the frightening impact of agricultural waste on the environment. The previous studies have reported that a wide number of fertilizers and pesticides were not utilized by plant and lost to the environment such as lake, river, and marine systems. The use of chemical fertilizers causes air and groundwater pollution as a result of eutrophication of water bodies.31 Research on microorganisms in agriculture, namely that endophytic bacteria and rhizobacteria plays an essential role in the biocontrol of plant pathogens, as well as decreasing the environmental repercussions of chemical fertilizers and pesticides. This study aimed to isolate, identify bacterial species, and demonstrate the potential abilities of endophytic bacteria and rhizobacteria screened from root and rhizosphere of five different types of vegetables in stimulating plant growth. In this research, five kinds of vegetable have been selected to study including Ipomoea aquatica (Morning Glory), Basella alba (Ceylon Spinach), Cymbopogon citratus (Lemon Grass), Amaranthus viridis (Amaranth) and Piper sarmentosum Roxb. (Wild Betel Leaf Bush). Moreover, the interaction between root and rhizosphere and inner root and endophytic bacteria were investigated by scanning electron microscope method. The results were recommended optimal bacterial strains to be used as commercial biofertilizer strains to improve the quality of soil and environment to further studies.
Sampling
Rhizosphere soil and roots of five healthy vegetables were randomly collected at two various field plots in Khon Kaen, Thailand, namely, Ipomoea aquatica (Morning Glory), Basella alba (Ceylon Spinach), Cymbopogon citratus (Lemon Grass), Amaranthus viridis (Amaranth) and Piper sarmentosum Roxb (Wild Betel Leaf bush).
Isolation of Bacteria
Rhizobacteria were isolated from soil adhered to roots, 10 g soil sample and 90 ml sterile distilled water were mixed, then incubated at 30 ± 2°C for 2 days under shaking condition. Serial dilution was made; 0.1 ml of soil suspensions of 10-3 to 10-5 were spread on plate count agar (PCA). Endophytes were isolated from the root portions of the plant samples by using the modify method of Dobereiner et al.32 The samples were surface sterilized with running tap water to remove the soil, immersed in commercial detergent for 5 min, dipped in 70% ethanol for 30 sec, submersed in 5% NaOCl for 15 min, and washed again with sterile distilled water. The final wash solution was spread on PCA plate, incubated at 30 ± 2°C for 1-2 days to verify the surface sterilization was uncontaminated. The sterile sample was cut into small pieces and homogenized in sterile distilled water. Dilution series from sample were prepared, 0.1 ml of each dilution of 10-2 were spread on tryptic soy agar (TSA), subcultured and incubated at 30 ± 2°C to collect the pure isolates. Isolated colonies of rhizobacteria and endophytic bacteria were counted; endophytes and rhizosphere populations were performed as CFU/g of sample, observed, and grouped on the basis of morphology characteristics (shape, motility, color) based on naked eyes and under light microscope. The Gram stain reaction was performed as followed by Vincent et al.33 The purified isolates were preserved in 20% glycerol solution at 4°C for further study.
Indole-3-Acetic Acid (IAA) Assay
All isolated pure cultures were tested for the ability of IAA production. Firstly, 0.5 mL of 24 hours bacterial suspension was inoculated in 10 ml of nutrient broth (NB) containing 1g/L of L-tryptophan that acts as a physiological precursor for the production of auxins in microorganisms and plants, then incubated at 30 ± 2°C under shaking condition for 48 hours.34 After centrifuged at 8000 rpm for 20 min, 1 ml of supernatant was mixed with 2 ml of Salkowski’s reagent and kept stable for 25 min in the dark room. The blank sample consisted of 1 ml of NB, tryptophan, and 2 ml of Salkowski’s reagent. The IAA production was estimated from the absorbance at 530 nm by spectrophotometer and evaluated data by using IAA standard (10-100 µg/ml). The high IAA production isolates were selected to be used in further investigations.
Enzyme Activity Assays
Cellulase Enzyme Assay
The cellulolytic activity of selected bacterial from IAA assay was estimated using method described by Ariffin et al.35 The bacteria were cultured on carboxymethyl cellulose (CMC) agar and incubated at 30°C for 5 days. Congo red dye (1% w/v) was added until flooded to the plate, drained, and rinsed with 1M NaCl for 15 min each part. The clear zones formed by the isolates that showed cellulose degradation were recorded to calculate hydrolysis capacity. Further, to select the high potential on cellulase activity producer, the hydrolysis capacity (HC) of bacteria was calculated from the ratio between the diameter of the clear zone and bacterial colony.36
ACC Deaminase Enzyme Assay
The ACC deaminase activity was determined followed by Kumar et al. with some changes. First, bacterial suspension was centrifuged at 8000 rpm, cell pellet was washed and re-suspended in Dworkin and Foster (DF) mineral medium (7.5 mL) containing 3 mM ACC.37 The samples were incubated at 30 ± 2°C for 24 hours, re-suspended in 1 ml of 0.1 M Tris-HCl buffer (pH 7.6), and centrifuged it again. The pellet was added 0.6 mL of 0.1 M Tris–HCl buffer, vortex and added 30 μl of toluene. Samples were measured by spectrophotometer at wavelength 540 nm, using a blank as non-inoculated and ACC solution.
Phosphatase Enzyme Assay
First, 38 isolates were point inoculated on TSA medium and incubation for 24-48h at 30°C. Then, 0.5% phenolphthalein diphosphate solution was prepared by using filtration (0.22 m) and was added until flooded to the plate. The disclosure was performed by dropping NH4OH (8.4%) on the Petri dish and reading after 15 min. The positive result was indicated by the formation of a pink zone which was scored at 1+ to 4+ from a pale pink to a deep red.38
Identification Selected Isolates
The bacterial strains exhibiting the highest multifunctional traits were identified on the basic of 16S rRNA gene sequence. Bacterial genomic DNA was extracted from the precipitate using the TIANAMP Bacterial DNA kit of Tiangen biotech (Beijing) Co., Ltd., China. The 16S rRNA gene was then amplified with universal primer pair 8F (5’-AGA GTT TGA TCM TGG CTC AG-3’) and 1512R (5’-ACG GYT ACC TTG TTA CGA CTT-3’), which were carried out in a FlexCycler2 PCR thermal cycler (Analytik Jena, Germany).39 The PCR products were purified using gel extraction kit, then sequenced using the 3500 genetic analyzer. After that, Basic Local Alignment Search for Nucleotide software to match the gene sequences of various bacterial strains to bacteria sequences was conducted using National Center for Biotechnology Information data banks.40
Root Colonization Assay
Scanning Electron Microscopy
The Morning Glory seed was prepared by surface sterilized with 5% NaOCl and 70% alcohol, the final wash solution was dropped on NA medium to verify the surface sterilization was uncontaminated. The sterilized seed was soaked in bacteria suspension for 2 hours, and aseptically transferred to sterile bottles of Hoagland’s nutrient agar, then kept in the dark room for 7 days. Four kinds of treatment, MSE5 bacteria suspension, AVR1 bacteria suspension, the mixture of both bacterial suspension and distilled water were prepared. The rooting of 7 days old of each treatment were randomly selected from plant growth bottles and were separately cut the root and shoot. After washed two times by phosphate saline buffer (PSB), tissue samples were fixed in 2.5% glutaraldehyde in the refrigerator (4°C) for 2 hours. The fixed roots were washed three times in PSB for 10 min and dehydrated in a serial alcohol from 50 – 100% ethanol for 15 min each. For endophytic bacteria and rhizobacteria observation, a protocol of Altschul et al. was followed with some changes.40 All samples were fixed critical point drying by CO2 dryer and metalized with 10 mÅ of gold-palladium (Sputter Coater Cressington 108auto). The samples were assessed under a field emission scanning electron microscope (1450VP SEM, Zeiss, England) operated at 13kV.
Isolation of Rhizobacteria (RB) and Endophytic Bacteria (EB) from Rhizosphere and Root of Vegetable Samples
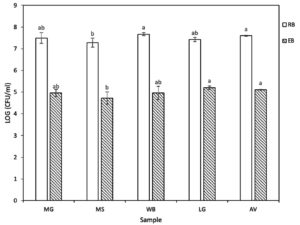
From soils and interior root of five different vegetable samples within the areas of Khon Kaen province, a total of 38 isolates with different character of colony morphologies in each treatment were obtained. Twenty-six of rhizobacteria isolates were found whereas 12 isolates of endophytic bacteria were obtained from root samples. The number of viable rhizobacteria in each sample ranged from 3.53 x 107 to 9.43 x 107 CFU ml-1 while the number of viable endophytic bacteria was considerably lower which ranged from 1.23 x 105 to 2.14 x 105 CFU ml-1. As shown in Figure 1, the data showed that there were not significant differences in both rhizobacteria and endophytic bacteria in all kinds of vegetable. However, the amount of both kinds of bacteria in MS sample was showed significantly different from AV sample. The CFU of WB and AV were showed highest results groups of rhizobacteria while LG and AV were recorded with quite high results of endophytic bacteria. Among 38 isolates, 29 isolates were gram positive and bacterial endospores; 9 isolates were gram negative.
Figure 1. The colony-forming unit (CFU) of rhizobacteria (RB) and endophytic (EB) bacteria at 24 hours of incubation. The values were log-transformed from CFU ml-1 to stabilize the variance. At each sample, bars with the same letter do not differ significantly by Tukey’s multiple range test at P ≤ 0.05. Ipomoea aquatica (Morning Glory – MG), Basella alba (Ceylon Spinach – MS), Cymbopogon citratus (Lemon Grass – LG), Amaranthus viridis (Amaranth – AV) and Piper sarmentosum Roxb (Wild Betel Leafbush – WB).
The plant-microbe interaction through the root system was studied to advance good aspects of this relationship such as enhancing plant growth and development, stimulating disease resistance.1 The impact of these microbes on the ecology when applying to the natural environment, particularly, on different soil types is challenged that need to be considered. Applied microbes can modify soil ecosystem utility, hence influencing on soil microflora and other plant microbial community.41 The CFU results of both rhizobacteria and endophytic bacteria did not represent significant characteristics or differences between five kinds of vegetables. In addition, these 38 strains showed similarity between them when more than three quarters of Gram and endospore staining results were positive. Thus, those bacteria are predicted to be native strains at the local province. Notably, inoculation using native microbes, which are referred to as plant growth promoters, have shown the effect in different plant species without disordering to the rhizosphere microbial diversity until one year after inoculation under drought stress in greenhouse conditions.41 Locally isolated strains may be a critical source for plant growth promoting as it limits unwanted environment impacts in practical usage on multiple crops.
IAA Content by Bacteria Strains
The result revealed that all isolates had ability to produce the IAA and this result was consistent with the study of Zheng et al. that more than 60% of endogenous bacteria had ability to synthesizing IAA.42 IAA production level by rhizospheric bacteria, it ranged from 2.175±0.25 µg/mL to 8.397±0.46 µg/mL, which was significantly higher than that from endophytic bacteria, varied from 3.043±0.15 µg/mL to 4.242±0.35 µg/mL. The group of rhizospheric bacteria with highest IAA level was observed in the results of isolate MSR7 (8.397±0.46 µg/mL), isolate LGR5 (7.8914±1.62 µg/mL) and isolate AVR1 (6.8687±0.49 µg/mL). Whereas isolate MGE4 (4.242±0.35 µg/mL) and isolate MSE5 (4.0909±0.59 µg/mL) were the highest IAA-producing endophytic bacteria.
Although all evaluated isolates have ability to produce plant hormone IAA in this study, the amounts of IAA produced in vitro similar greatly due to insignificant differences at P < 0.05 by Tukey’s test. The previous study of Datta et al. revealed that IAA production almost by Gram-negative bacteria while the present study showed that 29/38 Gram positive strains and 9/38 Gram negative strains were positive on IAA biosynthesis.43 IAA concentrations of Gram-positive bacteria ranged from 2.715±0.25 µg/mL to 6.869±0.49 µg/mL, while the general IAA concentration of all isolates ranged from 2.175±0.25 µg/mL to 8.397±0.46 µg/mL. Therefore, it can be concluded that Gram-positive bacteria produce a significant amount of IAA in this investigation. Tryptophan is known as the primary precursor of IAA in plants, thus leading the availability of tryptophan to influence the generation of IAA by numerous bacteria in culture.44 In this case, the result of IAA production was true for 1 g/L of tryptophan. According to the research of Pant et al., in the presence of 1 g/L L-tryptophan the concentration of IAA produced by the rhizobacteria ranged from 6 to 8 µg/mL, this is consistent with the data of this study (2.175±0.25 µg/mL to 8.397±0.46 µg/mL.45 In the study of Swain et al., B. subtilis was expanded in IAA production was watched up to concentration of tryptophan 1 g/L and there was a slight diminish at higher concentrations, and its reach maximum value on the 10th day.46 This opens the prospect that if the bacterial strains are culture in more than 2 days, the amount of IAA obtained will be higher or achieve maximum value.
Plant Growth Promoting Related Enzymes Production by Rhizobacteria and Endophytic Bacteria
After inoculation, the bacterial strains were biochemically experiments for three plant growth promotion properties including ACC deaminase, cellulase and phosphatase. Among them, evidence of cellulase decomposition activity was observed in 27 strains while there were 14 and 24 isolates that revealed to produce phosphatase and ACC deaminase, respectively. According to Florencio et al., the enzymatic index (HC value) by strains that are higher than 1.50 indicated the potential of cellulase producing.47 The HC value results were recorded of 24 cases with values greater than 1.50 which varied between 1.83±0.19 to 20.33±0.58. In general, 7 of these 38 isolates were exhibited results at all three enzyme assays (Table 1). Combine with the result of IAA, two growth promoting strains, namely MSE5 and AVR1 showed stable and potential results in most investigation. Consequently, they were selected for further examination of seed germination and root colonization, which are important in assessing the practical applicability of them (Table 2).
Table (1):
The detection of 3 enzymes activities involved in plant growth promoting of seven multifunctional isolates.
Isolate |
Cellulase activity (HC value) |
Phosphatase activity |
ACC deaminase activity |
|---|---|---|---|
MSE5 |
1.83±0.19cd |
++ |
✓ |
LGR3 |
2.26±0.31bc |
+ |
✓ |
LGR7 |
2.04±0.34cd |
+ |
✓ |
LGE1 |
1.38±0.32d |
+ |
✓ |
AVR1 |
2.80±0.08b |
++ |
✓ |
AVR2 |
5.17±0.29a |
+ |
✓ |
AVE4 |
2.17±0.27bc |
+ |
✓ |
‘HC’ refer to hydrolysis capacity which calculated from the results of cellulase enzyme assay, ‘✓’ refer to the positive result of ACC deaminase producing activity, ‘+’ correspond to positive responses of phosphatase producing activity. Values are displayed as means ± standard deviations from three replications followed by the same letter do not differ significantly by Tukey’s multiple range test at P ≤ 0.05.
In agricultural industries, cellulases have a major impact on plants for supporting plant disease control, improving soil quality and being as a microbial inoculum for composting process, however, there are some studies on cellulolytic fungi such as Trichoderma sp., Gelidium sp. and Penicillium sp. showing the ability of these species to improve plant growth as well as root system, seed germination and flowering. The study of Sharma et al. suggested that phosphatase enzyme could be a key factor in supporting germinating seeds adapt to abiotic stresses.48 Thus, enzymatic assays provide a straightforward yet effective method to evaluate the plant growth promoting traits of microorganisms in plant.
Table (2):
Multifunctional traits displayed by selected isolates MSE5 and AVR1 for in vivo assays.
Isolate |
Gram |
Endospore |
IAA Content (µg/mL) |
Cellulase activity (HC value) |
Phosphatase activity |
ACC deaminase activity |
|---|---|---|---|---|---|---|
MSE5 |
+ |
+ |
4.0909 ±0.592 |
1.83±0.19 |
++ |
✓ |
AVR1 |
+ |
+ |
6.8687 ±0.493 |
2.80±0.08 |
++ |
✓ |
‘HC’ refer to hydrolysis capacity which calculated from the results of cellulase enzyme assay, ‘✓’ refer to the positive result of ACC deaminase producing activity, ‘+’ correspond to positive responses of phosphatase producing activity. Values are displayed as means ± standard deviations from three replications.
In cellulase assay, the results of WBR5, AVR2 and AVE1 make them a promising strain to be developed as cellulolytic microbial inoculants for biofertilizer production. The two isolates WBR5 and AVR2 had the HC value over 5.0, AVE1’s HC value was over 6.0 in particular (Figure 2). The clearing zone and HC value are in the range between 11.0 to 21.0 mm and 5.08 to 6.56 respectively. This outcome is quite different when compared to previous studies shown in Table 3. Cellulases has found applications in various industries other than agriculture such as bioconversion, detergents, fermentation, composting, food, pulp and paper, textile, and others.49 Therefore, these three strains were able to continue further research on microbial inoculants of cellulase activity.
Table (3):
The comparison of cellulase enzyme activity results through clearing zone and enzymatic index with other studies.
Isolates/References |
Clear zone (mm) |
Enzymatic index (HC value) |
|---|---|---|
WBR5 |
21a |
5.08±0.14b |
AVR2 |
11c |
5.17±0.29b |
AVE1 |
20a |
6.56±0.19a |
Hatami et al.51 |
– |
2.8c |
Behera et al.52 |
– |
2.5d |
Shailendra et al.53 |
– |
2.7c |
Rasul et al.54 |
9.5d |
– |
Chaiaharn et al.55 |
12.164b |
– |
‘HC’ refer to hydrolysis capacity which calculated from the results of cellulase enzyme assay, ‘-’ indicate the information was not mentioned in corresponding references. In the same column, values followed by the same letter do not differ significantly by Tukey’s multiple range tests at P ≤ 0.05.
Figure 2. Three isolates (WBR5, AVR2, and AVE1) showed outstanding cellulase production results after staining with Congo red dye, as indicated by the clear white zones formed on the CMC agar plate.
Identification of Selected Bacteria Strains
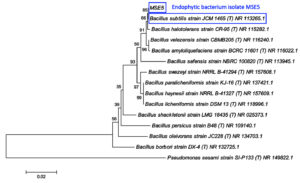
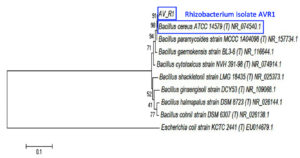
Endophytic bacterium isolate MSE5 and rhizobacterium isolate AVR1 had been characterized by 16S rRNA gene sequences. After the sequencing of the 16S rRNA gene segment, the attained sequences were aligned and analyzed with other bacterial sequencing information in the GenBank database (NCBI) using BLAST. The results showed the 100% similarity of MSE5 with Bacillus subtilis (accession number: NR113265.1) whereas AVR1 highly similar to Bacillus cereus (accession number: NR074540.1) with 100% similarity. The morphology features of the two bacterial strains also corresponded to the characteristics of the respective groups. Phylogenetic analysis was established to assess the evolutionary relationship of MSE5 and AVR1 with other species (Figure 3 & 4).
Figure 3. The phylogenetic tree of MSE5 was inferred using the Neighbor-Joining method of MEGA6. The number beside the branches shows bootstrap values created after doing 1000 replicates. The analysis involved 15 nucleotide sequences with a total of 1211 positions in the final dataset. Accession number of each sequence is written next to taxon names. Pseudomonas sesami SI-P133 was used as the outgroup.
Figure 4. The phylogenetic tree of AVR1 was inferred using the Neighbor-Joining method of MEGA6. The number beside the branches shows bootstrap values created after doing 1000 replicates. The analysis involved 9 nucleotide sequences with a total of 1372 positions in the final dataset. Accession number of each sequence is written next to taxon names. Escherichia coli KCTC was used as the outgroup.
Several researches have mentioned various aspects of the wide application of Bacillus spp. as elicitor of PGP. Bacillus subtilis, Bacillus cereus and many members of the genus were characterized as an effective PGPR in plant by both direct and indirect pathways. Mechanisms of direct plant growth promotion of Bacillus spp. include production of phytohormones, degradation of cellulose, nitrogen fixation and others.55 In addition, the biological control mechanisms by which Bacillus spp. can promote plant growth indirectly have been also remarked in many studies such as antibiosis, induced systemic resistance in host plant, and inhibited pathogens by competing for nutrients.56
Scanning Electron Microscopy (SEM)
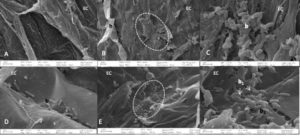
Interaction between Morning glory roots and shoots from endophytic bacteria and rhizobacteria, namely AVR1 and MSE5, respectively, was observed by SEM. Four treatments were used: (1) inoculated seed plants with distilled water. (2) inoculated seed plants with MSE5 bacteria suspension. (3) inoculated seed plants with AVR1 bacteria suspension. (4) inoculated seed plants with mixture suspension of both bacteria. The reason for the treatment (4) was to check the antagonism between two different bacterial strains on the same plant. No bacteria were observed on plants of treatment (1) (Figure 5A). The major MSE5 bacteria were observed to be concentrated in large areas on the epidermal surface (Figure 5B). The ovoid shape was recorded (Figure 5C) and corresponded to the morphology features of MSE5. Bacteria mostly in pairs covering the root and shoot surface. The fact that both root and shoot epidermal surfaces were damaged in strongly infested places shows that bacteria are using an active invasion strategy. A large presence of bacteria was identified in root samples infected by the AVR1 bacteria extraction, primarily in the root hair zone, where bacteria appeared to cling to the root epidermal cell, and rod-shaped bacteria were observed (Figure 5D). This is true when compared with the AVR1’s morphology observed under the microscope. No rhizosphere bacteria were detected in the tissues of the shoot plants. The plants seed inoculated with treatment (4) showed clusters of these bacteria in the zones of the epidermal cells, which entered the host tissue through injuries and mostly in pair covering (Figure 5E). At higher magnification, two different strains could be seen clearly, it is true when compared with morphology of MSE5 and AVR1 (arrowhead, Figure 5F). For the interaction between endophytic bacteria and rhizobacteria, this result presents clear support in this respect. The amount of AVR1 bacteria in treatment (1) is greater than the treatment (2) but only appears in the root zone. On the other hand, MSE5 bacteria have appeared in the roots and can move up the shoot; the number of these bacteria was relatively high and homogeneous that proved its ability to infect the root and shoot plant. This is consistent with the isolation site of AVR1 bacteria and MSE5 bacteria. Altogether, this evidence indicated that AVR1 belonged to rhizosphere and MSE5 bacteria were suitable for the characteristic of endophytic bacteria.
Figure 5. SEM of Morning Glory by 4 treatments. Note that no bacteria are present on the distilled water treatment plants (A). The pointed ellipse shows the area invaded by MSE5 extraction, and mixture of MSE5 and AVR1, respectively (B, E). Detailed view of zone, cell of the MSE5 bacteria was detected in the root and shoot while cell of AVR1 bacterium was detected only in the root (C, D). The arrow indicates the location of the 2 kinds of bacteria can be observed in the root and shoot of the co-inoculated plants. EC: epidermal cells. h: root hair. b: bacteria.
Examination of SEM revealed that MSE5, AVR1 or mixture of them were able to colonize in great extent the morning glory root surface. The images of cross sections from roots after inoculation with mixture of MSE5 and AVR1 confirmed the presence of both strains; however, AVR1 showed more extensively and abundantly than MSE5. The observations revealed that all the three treatments formed micro-colonies on the root surface by way of clusters of cells linked to fibrillar material. The spatial distribution of individual cells as well as those microcolonies of MSE5 and AVR1 was concentrated in the area of the epidermal cells at the root. These marked the effect of bacterial colonization at the time of the survey and subsequent higher bacterial population development. In addition, the co-inoculation might provide nutrient competition condition that leads to enhanced interactions between microorganisms and plants, thereby optimizing the promotion of plant growth. As proposed by Shaharoona et al., some PGPR were more effective when nutrients became limited and thus could save fertilizers.57 Results from several previous studies have reported the effectiveness of PGPR using mixture of strains on many different plants.55,58
This study has given an account of initial results on development potential as multifunctional bio-fertilizer of two strains MSE5 and AVR1. The strains have the characteristics to create root colonization through SEM results and were identified as Bacillus subtilis and Bacillus cereus. The results of co-inoculation of two strains, along with predictions of “native strains”, could possibly support to practical applicability. Moreover, we have found some novel cellulose-degrading bacterial strains with the high potential on hydrolysis capacity and that might be valuable in further microbial cellulase studies.
ACKNOWLEDGMENTS
The authors would like to thank Ms. Ngo My Ngan and Mr. Natthawat Sritongon (Microbiology laboratory, Faculty of Science, Khon Kaen University) for their assistance in the Laboratory work. The authors also thank Prof. Dr. Piyada Theerakulpisut, Faculty of Science, Khon Kaen University for her support and encouragement.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NR Conceptualized the study. NYM, PNH and PS performed methodology. NYM and PNH performed formal analysis. NYM, PNH wrote the original draft. NR reviewed the manuscript. NYM, PNH and NR edited the final manuscript.
FUNDING
This study was funded by Research and Graduate Affairs, performed by Salt-Tolerant Rice Research Group, Department of Biology, Faculty of Science, Khon Kaen University, Thailand with research program number Rp64-11-001.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Kloepper JW, Lifshitz R, Zablotowicz RM. Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 1989;7(2):39-44.
Crossref - Kloepper JW. Effects of rhizosphere colonization by plant growth-promoting rhizobacteria on potato plant development and yield. Phytopathology. 1980;70(11).
Crossref - Kloepper JW, Scher FM, Laliberte M, Tipping B. Emergence-Promoting rhizobacteria: description and implications for agriculture. Iron, Siderophores, Plant Dis. 1986;155-164.
Crossref - Zaidi A, Ahmad E, Khan MS, Saif S, Rizvi A. Role of plant growth promoting rhizobacteria in sustainable production of vegetables: current perspective. Sci Hortic. 2015;193:231-239.
Crossref - Schulz B, Boyle C. What are Endophytes? Microb Root Endophytes. 2006;1-13.
Crossref - Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278(1):1-9.
Crossref - Backer R, Rokem JS, Ilangumaran G, et al. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 2018;871.
Crossref - Fouda A, Hassan SED, Eid AM, El-Din Ewais E. The interaction between plants and bacterial endophytes under salinity stress. Ref Ser Phytochem. 2019;591-607.
Crossref - Eid AM, Fouda A, Abdel-rahman MA, et al. Harnessing bacterial endophytes for promotion of plant growth and biotechnological applications: An overview. Plants. 2021;10(5).
Crossref - Eid AM, Salim SS, Hassan SE-D, Ismail MA, Fouda A. Role of endophytes in plant health and abiotic stress management. Microbiome Plant Heal Dis. 2019;119-144.
Crossref - Fouda AH, Hassan SED, Eid AM, Ewais EED. Biotechnological applications of fungal endophytes associated with medicinal plant Asclepias sinaica (Bioss.). Ann Agric Sci. 2015;60(1):95-104.
Crossref - Alkahtani MDF, Fouda A, Attia KA, et al. Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy. 2020;10(9):1325.
Crossref - Khalil AMA, Hassan SED, Alsharif SM, et al. Isolation and characterization of fungal endophytes isolated from medicinal plant Ephedra pachyclada as plant growth-promoting. Biomolecules. 2021;11(2):140.
Crossref - Fouda A, Eid AM, Elsaied A, et al. Plant Growth-Promoting Endophytic Bacterial Community Inhabiting the Leaves of Pulicaria incisa (Lam.) DC Inherent to Arid Regions. Plants (Basel). 2021;10(1):76.
Crossref - Ismail MA, Amin MA, Eid AM, et al. Comparative study between exogenously applied plant growth hormones versus metabolites of microbial endophytes as plant growth-promoting for Phaseolus vulgaris L. Cells. 2021;10(5).
Crossref - Mahgoub HAM, Fouda A, Eid AM, Ewais EED, Hassan SED. Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol Rep. 2021;15(6):819-843.
Crossref - Abedinzadeh M, Etesami H, Alikhani HA. Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater with biotechnological potential in agriculture. Biotechnol Rep. 2019;21:e00305.
Crossref - Moronta-Barrios F, Gionechetti F, Pallavicini A, Marys E, Venturi V. Bacterial microbiota of rice roots: 16s-based taxonomic profiling of endophytic and rhizospheric diversity, endophytes isolation and simplified endophytic community. Microorganisms. 2018;6(1).
Crossref - Hardoim PR, Overbeek LS, Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16(10):463-471.
Crossref - Johnson GI, Weinberger K, Wu MH. The vegetable sector in tropical Asia: Importance, issues and a way ahead. Acta Hortic. 2009;809:15-34.
Crossref - Sungpuag P, Tangchitpianvit S, Chittchang U, Wasantwisut E. Retinol and beta carotene content of indigenous raw and home-prepared foods in Northeast Thailand. Food Chem. 1999;2(64):163-167.
Crossref - Thalang VN, Trakoontivakorn G, Nakaharab K. Determination of antioxidant activity of some commonly consumed leafy vegetables in Thailand. JIRCAS J. 2001;9:39-46.
- Turreira-Garcia N, Vilkamaa AM, Byg A, Theilade I. Diversity, knowledge, and use of leafy vegetables in Northern Thailand-Maintenance and transmission of ethnobotanical knowledge during urbanisation. Nat Hist Bull Siam Soc. 2017;62(1). https://so04.tci-thaijo.org/index.php/nhbss/article/view/170101
- Muday GK, Rahman A, Binder BM. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 2012;17(4):181-195.
Crossref - Ruzcka K, Ljung K, Vanneste S, et al. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19(7):2197-2212.
Crossref - Lewis DR, Negi S, Sukumar P, Muday GK. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development. 2011;138(16):3485-3495.
Crossref - Jaemsaeng R, Jantasuriyarat C, Thamchaipenet A. Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci Rep. 2018;8(1):1950.
Crossref - Shen FT, Yen JH, Liao CS, Chen WC, Chao YT. Screening of rice endophytic biofertilizers with fungicide tolerance and plant growth-promoting characteristics. Sustain. 2019;11(4):1133.
Crossref - Sethi S, Datta A, Gupta BL, Gupta S. Optimization of cellulase production from bacteria isolated from soil. ISRN Biotechnol. 2013;2013:1-7.
Crossref - Immanuel G, Dhanusha R, Prema P, Palavesam A. Effect of different growth parameters on endoglucanase enzyme activity by bacteria isolated from coir retting effluents of estuarine environment. Int J Environ Sci Technol. 2006;3(1):25-34.
Crossref - Mma Y, Mfm E. Biofertilizers and their role in management of plant parasitic nematodes. A review. E3 J Biotechnol Pharm Res. 2014;5(1):1-006.
- Döbereiner J, Reis VM and Lazarini AC. New N2-fixing bacteria in association with cereals and sugarcane. In: Bothe H, de Bruijn FJ and Newton WE (Eds), Nitrogen fixation: hundred years after. Stuttgart: Gustav Fischer. 1988;717–722. https://cir.nii.ac.jp/crid/1573668924117220480
- Vincent JM, Humphrey B. Taxonomically significant group antigens in Rhizobium. J Gen Microbiol. 1970;63(3):379-382.
Crossref - Kumari S, Prabha C, Singh A, Kumari S, Kiran S. Optimization of Indole-3-Acetic Acid Production by Diazotrophic B. subtilis DR2 (KP455653), Isolated from Rhizosphere of Eragrostis cynosuroides. Int J Pharma Med Biol Sci. 2018;7(2):20-27.
Crossref - Ariffin H, Abdullah N, Kalsom MSU, Shirai Y, Hassan M. Production and characterization of cellulase by Bacillus pumilus EB3. Int J Eng Technol. 2006;3(1):47-53. http://www.ijet.feiic.org/journals/J-2006-V1005.pdf
- Poznanski, S. The analysis of mixtures of ethyl alcohol, ethyl acetate, acetic acid and water. J Am Chem Soc. 1928;50:981-988.
Crossref - Kumar A, Kumar A, Pratush A. Molecular diversity and functional variability of environmental isolates of Bacillus species. Springerplus. 2014;3(1):1-11.
Crossref - Smith RF, Blasi D, Dayton SL. Phosphatase activity among candida species and other yeasts isolated from clinical material. Appl Microbiol. 1973;26(3):364-367.
Crossref - Kane MD, Poulsen LK, Stahl DA. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59(3):682-686.
Crossref - Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-3402.
Crossref - Armada E, Leite MFA, Medina A, Azcon R, Kuramae EE. Native bacteria promote plant growth under drought stress condition without impacting the rhizomicrobiome. FEMS Microbiol Ecol. 2018;94(7):fiy092.
Crossref - Zheng M, Tao Y, Hussain S, et al. Seed priming in dry direct-seeded rice: consequences for emergence, seedling growth and associated metabolic events under drought stress. Plant Growth Regul. 2016;78(2):167-178.
Crossref - Datta C, Basu PS. Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanus cajan. Microbiol Res. 2000;155(2):123-127.
Crossref - Brandl M, Clark EM, Lindow SE. Characterization of the indole-3-acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can J Microbiol. 2011;42(6):586-592.
Crossref - Pant G, Agrawal PK. Isolation and characterization of indole acetic acid producing plant growth promoting rhizobacteria from rhizospheric soil of Withania Somnifera. J Biol Sci Opin. 2014;2(6):377-383.
Crossref - Swain MR, Naskar SK, Ray RC. Indole-3-acetic acid production and effect on sprouting of yam [Dioscorea rotundata L.] minisetts by Bacillus subtilis isolated from culturable cowdung microflora. Polish J Microbiol. 2007;56(2):103-110. http://www.pjmonline.org/wp-content/uploads/2015/12/vol5622007103.pdf
- Florencio C, Couri S, Farinas CS. Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by Trichoderma strains. Enzyme Res. 2012;2012.
Crossref - Sharma AD, Thakur M, Rana M, Singh K. Effect of plant growth hormones and abiotic stresses on germination, growth and phosphatase activities in Sorghum bicolor (L.) Moench seeds. African J Biotechnol. 2004;3(6):308-312.
Crossref - Kuhad RC, Gupta R, Singh A. Microbial cellulases and their industrial applications. Enzyme Res. 2011;2011(1).
Crossref - Hatami S, Alikhani HA, Besharati HN, et al. Investigation on aerobic cellulolytic bacteria in some of north forest and farming soils. Biotechnol Bioeng Symp. 2008;3(5):713-716. https://www.idosi.org/aejaes/jaes3(5)/8.pdf
- Behera BC, Parida S, Dutta SK, Thatoi HN. Isolation and identification of cellulose degrading bacteria from mangrove soil of mahanadi river delta and their cellulase production ability. Am J Microbiol Res. 2014;2(1):41-46.
Crossref - Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh V. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J Microb Biochem Technol. 2015;07(02).
Crossref - Rasul A, Balzter H, Smith C. Spatial variation of the daytime Surface Urban Cool Island during the dry season in Erbil, Iraqi Kurdistan, from Landsat 8. Urban Clim. 2015;14(2):176-186.
Crossref - Chaiharn M, Chunhaleuchanon S, Kozo A, Lumyong S. Screening of rhizobacteria for their plant growth promoting activities. Kmitl Sci Tech J. 2008;8(1):18-23. https://www.thaiscience.info/journals/Article/KLST/10424521.pdf
- Upadhyay SK, Singh JS, Saxena AK, Singh DP. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol (Stuttg). 2012;14(4):605-611.
Crossref - Kumar A, Prakash A, Johri BN. Bacillus as PGPR in crop ecosystem. Bact Agrobiol Crop Ecosyst. 2011;37-59.
Crossref - Shaharoona B, Naveed M, Arshad M, Zahir ZA. Fertilizer-dependent efficiency of Pseudomonads for improving growth, yield, and nutrient use efficiency of wheat (Triticum aestivum L.). Appl Microbiol Biotechnol. 2008;79(1):147-155.
Crossref - Han HS, Supanjani, Lee KD. Effect of co-inoculation with phosphate and potassium solubilizing bacteria on mineral uptake and growth of pepper and cucumber. Plant Soil Environ. 2006;52(3):130-136.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.