ISSN: 0973-7510
E-ISSN: 2581-690X
Oral carcinogenesis is a multi-step process, affecting predominantly elderly males with a long history of smoking and alcohol drinking while the role of infectious agents in its progression has not been fully identified. Human cytomegalovirus (HCMV) has been hypothesized in the oral pathogenesis and carcinogenesis. Usually, p16 protein is absent in oral squamous cell carcinoma (OSCC), either for p16 gene being mutated or deleted or its expression being abrogated while p16 expression is associated with favorable prognosis. Is to analyze the concordant impact of p16 expression and HCMV infection on tissues from a group of patients with Oral squamous cell carcinoma (OSCC) in mid-Euphrates-Iraq. One-hundred formalin-fixed, paraffin- embedded Oral tissues enrolled in this study; 40 oral tissue-biopsies from Oral squamous cell carcinoma (OSCC); 40 tissues obtained from patients with benign oral tumors and 20 apparently healthy oral tissues were used as control group. Detection of HCMV was done by chromogenic in situ hybridization (CISH) technique while the protein expression of p16 gene by using immunohistochemistry (IHC) detection system. In OSCC group, the hCMV-positive CISH and the p16-positive IHC reactions were detected in 42.5% and.52.5% of tissues, respectively. The correlation between HCMV and P16 was highly significant (P= 0.003). The significant co-expression of P16 and HCMV genes in OSCC tissues could point for their possible role in either oral pathogenesis or carcinogenesis.
HCMV, OSCC, P16, CISH, IHC.
Oral cancer is an interchangeably used term with oral squamous cell carcinoma (OSCC), the most common, poor- prognostic, epithelial malignancy, representing up to 90% of all oral neoplasms. This group of neoplasms is affecting any oral and pharyngeal regions as well as salivary glands1.
Complex, reciprocally-affected, environmental and genetic factors, as well as major etiological and predisposing factors for OSCC, have been reported in their multi-step carcinogenesis, include smoking, drinking habits, and ultraviolet radiation, several other factors such as the effects of viral and Candida infections, have been also associated in their development. These risk factors are independent, however, their action seems to be combined2. Human papillomavirus3,4, herpes viruses5, adenoviruses6, and hepatitis C virus7,8, are most the commonly implicated viruses in oral cancer transformation. Human papillomavirus and herpes viruses are the most likely “synergistic viruses” involved in oral carcinogenesis. Epstein-Barr virus (EBV), human herpes virus-8, and cytomegalovirus (CMV) are the most often viruses linked to oral cancers9.
Human cytomegalovirus (HCMV) is a commonly infects the majority of the population and presents in the gingival sulcus fluid (GSF) of many healthy people10. HCMV has characterized tropism for T-lymphocytes with an ability to persist in a latent form in reticuloendothelial cells, salivary cells and kidney cells11. High prevalence of HCMV has been reported in with different tumors, such as brain cancer and salivary gland cancer12. Also, HCMV has been proved to have ability to control host gene expression, oncomodulatory or oncogenic function9,10.
The tumor-suppressor p16INK4a (also called Cdkn2a) is a tumor suppressor protein and an inhibitor of cyclin-dependent kinase (CDK) that blocks CDK4- and CDK6-mediated pRb phosphorylation to inhibit E2F-dependent transcription and cell cycle progression )13. P16 is expressed in a wide variety of SCCs, other than those originating from cervix14. The abrogation / loss of p16 expression occurs frequently in human oral premalignant lesions and oral cancers15. Studies of two studies16,17 have correlated the alteration of p16 levels with improved relapse-free survival and more likely to develop a recurrence, respectively. In addition, the alteration of p16 gene in oral SCC helps to evaluate its early diagnosis, aggression and prognosis.
The aim of this research was to unravel the rate as well as impact of either hCMV or p16 on oral tumors tissues obtained from a group of Iraqi patients.
Tissue samples
This retrospective case- control study included one hundred -twenty formalin-fixed, paraffin embedded, oral squamous cell tissues samples, including 40 oral carcinomas, 40 benign oral tumor tissues and 40 apparently normal healthy oral tissues samples.Beside the medical records for the tissue samples, a consultant pathologist re-examined these tissue samples to confirm their histopathological diagnosis.
The specimens were collected during the period 2010–2018 from major hospitals and private laboratories in the governorates of Iraq including Babylon, Baghdad and AL-Najaf.
The diagnoses were based on the recorded pathological reports of the corresponding patients.
Laboratory methods
Thick-tissue sections (4 mm) were prepared and stuck onto positively charged slides. An Chromogenic In Situ Hybridization (CISH) detection system (Zytovision GmbH) was used to target DNA sequences in tissue specimens using digoxigenin-labelled HCMV- DNA probe .An immunohistochemistry (IHC) detection system technology (Abcam- England) was used to target gene expression of p16 in tissue specimens using monoclonal p16 primary antibody .
Chromogenic In-Situ Hybridization (CISH)
The CISH signals were detected as BLUE discolouration at the site of sequence complementarity as nuclear signals by proper use of the Zytovision system.
In order to detect Chromogenic In-Situ Hybridization (CISH) for HCMV, 4µm paraffin embedded tissue sections were tested by CISH kit (according the instructions of the manufacturing companyZyto Vision GmbH. Fischkai, Bremerhaven. Germany), where a digoxigenin-labeled, oligonucleotide probe was used to target the HCMV-DNA.
The main steps for CISH procedure were: incubation of slides for 18 hr at 60°C in oven, then rehydration process was done at room temperature which include: slides immersion in two changes of absolute xylene for 15 min then absolute ethanol for 5 minute each, then immersion in ethanol (95%) for 5 minute each, after that immersed in ethanol (70%) for 5 minute each, finally immersion in a distilled water for 5 minutes to remove residual alcohol. After that, slides were allowed to dry completely by incubating them at 37°C for 5 minutes. Then application (drop-wise) pepsin solution to the tissue/cell section and incubate for 45 min at 37°C in a humidity chamber. After that we immersed slides in distilled water and drain off the water, air dried sections. Then addition of the probe to the center of a cover slip and placing cover slip upside- down on target area. Denaturation of the slides at 75°C for 5 min on hot plate, then transferred the slides to a humidity chamber and hybridize for 18 hrs at 37°C for DNA-targeting probes and the post-hybridization and detection process that included removing the cover slip by submerging in 1x wash buffer TBS, then washed for 5 min in 1x wash buffer TBS at 55°C. Then the slides were allowed not dry out at any time during the hybridization and staining. All reagents used during hybridization and detection were warmed to room temperature. Then application of AP-Streptavidin drop- wisely to the slides and incubate for 30 min at 37°C in a humidity chamber. Then washed in wash buffer TBS and then twice times for 5 min in distilled water and application. Then one to two drops of Slides were rinsed in detergent wash buffer for 5 minutes and then drained. After that one to two drops of 5-bromo3-chloro3-indoly/phosphate/nitro blue tertrazolium substrate chromogen solution (BCIP/NBT) were placed on tissue sections. Slides were incubated at 37°C for 30 minutes or until color development was developed completed. Color development was monitored by viewing the slides under the microscope. A dark blue colored precipitate forms at the complementary site of the probe in positive cells. Then the slides were rinsed in distilled water for 5 minutes, then counter staining process by immersion of the slides in Nuclear Fast Red stain for 120 seconds, then washing process was followed by immersion the slides for 2 minute in tap water. After that Sections were dehydrated by ethyl alcohol, (95%, once for one minute then, 100% twice times for 2 minutes each); cleared by Xylene, then mounted with permanent mounting medium (DPX).
Immunohistochemistry Technique
The P16 protein was detected using immunohistochemistry detection system technology (Abcam- England), the latter utilizing specific monoclonal antibodies to detect the production of gene expression (protein) in the normal and infected tissues. The dewaxing protocols routinely used in immunohisto-chemistry procedures, e.g. 5 min xylene, 5 min 100% ethanol, 5 min 96% ethanol, 5 min 70% ethanol, can be used. Air drying of sections. Then application (drop-wise) pepsin solution to the tissue/cell section and incubate for 45 min at 25°C in a humidity chamber. After that we immersed slides in phosphate buffer solution (PBS) and drain off the buffer, air dried sections. Primary antibody for specific epitope (usually mouse anti-human monoclonal antibody), which binds to nuclear targeted protein 25°C for over night. The bound primary antibody is then detected by secondary antibody (usually rabbit or goat anti mouse), which contains specific label (in this context we used peroxidase labeled polymer conjugated to goat anti-mouse immunoglobulin). After that streptavidin-alkaline phosphatase conjugate reagent was added to tissue sections. Then slides were kept in a humid chamber at 25°C for 60 minutes.
The substrate is DAB in chromogen solution, positive reaction will result in a browning color precipitate at the antigen site in tested tissues. Then the slides were dehydrated by immersing them sequentially in the following solution at room temperature for the indicated times, distilled water for 1 minute, 70% ethanol for 1 minute, 95% ethanol for 1 minute and 100% by incubating them at 37% for 5 minutes. Then mounted with permanent mounting medium (DPX).
Statistical analysis have used Chi–square tests to calculate the statistical significance (p value) for the studied parameters, where SPSS-21 package has been performed to analyze complex relationships between a large numbers of variable. A p value less than 0.05 indicates a significant relationship between the studied parameters.
Analysis
Statistical analysis have used Chi–square tests to calculate the statistical significance (p value) for the studied parameters, where SPSS-21 package has been performed to analyze complex relationships between a large numbers of variable. A p value less than 0.05 indicates a significant relationship between the studied parameters.
Archival specimens enrolled in this study were related to patients with oral cancer whom mean age was (57.28 years) while the mean age of those who have benign oral tumor & apparently healthy tissues were 45.0 40.0 and 50.8 years, respectively (Table 1). Also, Table (1) shows the distribution of gender frequency in this study where 50% (60 out of 120) were males and 50% (60 out of 120) were females. In malignant oral tumors group, 40% (24 out of 60) were males and 26.6% (16 out of 60) were females. In benign oral tumors group, 30% (18 out of 60) were males and 36.7% (22 out of 60) were females. Lastly, 30% (18 out of 60) were males and 36.7% (22 out of 60) were females in those with oral healthy tissues group.
Table (1):
The Study Group Characteristics.
| Malignant Oral tissues, N= 40(%) | Benign oral tissues, N = 40 (%) | Healthy tissues N=40(%) | ||
|---|---|---|---|---|
| Age | Mean of Age (%) | 57.28 | 45.40 | 50.80 |
| Range of Age | 32 – 88 | 7 -75 | 14 – 91 | |
| ≤ 50 | 14 (35) | 26 (65) | 22 (55) | |
| 51 – 91 | 26 (65) | 14 (35) | 18 (45) | |
| Gender | Male | 24 (40) | 18 (30) | 18 (30) |
| Female | 16 (26.6) | 22 (36.7) | 22(36.7) | |
| Grade | Poorly | 4 (10 ) | ||
| Moderately | 7 (17.5 ) | |||
| Well | 29 (72.5 ) | |||
| Site of tumor | buccal mucosa | 2 (5) | 2 (5) | 2 (5) |
| Cheek | 3 (7.5) | 0 (0) | 0 (0) | |
| Floor | 2 (5) | 2 (5) | 2 (5) | |
| Gingiva | 1 (2.5) | 0 (0) | 0 (0) | |
| Hard palate | 2 (5) | 0 (0) | 0 (0) | |
| Upper lip | 1 (2.5) | 4 (10) | 4 (10) | |
| Lower lip | 2 (5) | 0 (0) | 0 (0) | |
| Maxilla | 3 (7.5) | 0 (0) | 0 (0) | |
| Oral cavity | 10 (25) | 24 (60) | 24 (60) | |
| Tongue | 14 (35) | 8 (20) | 8 (20) |
The most affected site in the group of malignant oral tumors was tongue (35%; 14 out of 40); followed by oral cavity (25%; 10 out of 40), 7.5% ( 3 out of 40) in both maxilla and cheek, tumors in buccal mucosa, lower lip of mouth, hard palate and floor of mouth each constituted (5% ; 2 out of 40) and lastly, tumors in both lips & gingiva (2.5%; 1 out of 40). The most affected site of benign oral tumors was oral cavity (60%; 24 out of 40); followed by tongue (20%; 8 out of 40), lips (10%; 4 out of 40) and lastly, 5% (2 out of 40) in both buccal and floor of mouth. In the oral healthy tissues group, oral cavity tissues constituted 60% (24 out of 40),followed by 20% (8 out of 40) tongue tissues, 10% (4 out of 40) in lips, and lastly, 5% (2 out of 40) in each buccal and floor of mouth. On distributing oral squamous cell carcinoma (OSCC) group according to their grading,the results of well differentiated (grade I) OSCC constituted 72.5% (29 out of 40), whereas moderately OSCC (II) grade and poorly differentiated OSCC grade (III) constituted 17.5% (7 out of 40) and 10% (4 out of 40), respectively.
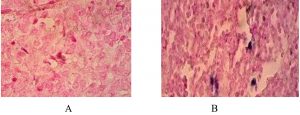
Fig. 1. Oral sections in OSCC: CISH reaction for hCMV detection ׳40. (A): Negative reaction; (B) positive reaction with low score and strong signal intensity.
Fig. 1 shows the HCMV-DNA which was detected in tissue blocks obtained from patients with oral squamous cell tumors using the Chromogenic In Situ Hybridization technique.
HCMV – CISH signals were seen as blue signals / discoloration at the sites of complementary sequences, where 42.5% (17 out of 40 tissues) of malignant oral squamous cell tumors gave positive signals. The benign oral squamous cell group had 25% positive signals which represented 10 out of 40 tissues in this group. The healthy tissues control group had 7.5% positive signals which represented 3 out of 40 tissues. Statistically, highly significant differences (p <0.001) were found between groups.
There was a higher frequency of HCMV in the age group over 51 years (20 samples; 16.7%) in malignant oral tumor than in £50 age group (10; 8.3%). HCMV- infected males comprised 21 out of 120 samples (17.5%) and this was more than females (9/120; 7.5%). The tongue area was more frequent infected with HCMV (10 cases; 8.3%) than another site of oral cavity (8 cases; 6.7%) (Table 2).
Table (2):
The Results Of HCMV-DNA-CISH and P16INK4A -IHC Among The Study Groups.
| HCMV-CISH signals | P16INK4A – IHC signals | ||||||
|---|---|---|---|---|---|---|---|
| + (%) |
|
p-value | + (%) |
|
p-value | ||
| Malignant OSCC n=40
|
17 (42.5) | 23 (57.5) | <0.001 | 21 (52.5) | 19 (47.5) | 0.004 | |
| Benign oral tumors n=40 | 10 (25) | 30 (75) | 14 (35) | 26 (65) | |||
| Control ⃰⃰ n= 40 | 3 (7.5) |
37 (92.5) | 1 (2.5) |
39 (97.5) |
|||
| Age group | ≤50 | 10 (8.3) |
54 (45) |
< 0.05 | 16 (13.3) |
35 (29.2) |
0.168 |
| 51-91 | 20 (16.7) |
36 (30) |
20 (16.6) |
49 (40.8) |
|||
| Gender | male | 21 (17.5) |
39 (32.5) |
0.698 | 19 (15.8) |
41 (34.2) | 0.289 |
| Female | 9 (7.5) |
51 (42.5) |
17 (14.2) |
43 (35.8) |
|||
| Grade/Differentiation | Poor | 2 (5) |
2 (5) |
0.286 | 2 (5) |
2 (5) |
0.343 |
| Moderate | 4 (10) |
3 (7.5) |
3 (7.5) |
4 (10) |
|||
| Well | 11 (27.5) |
18 (45) |
16 (40) |
13 (32.5) |
|||
| Site of tumor | buccal mucosa | 4 (3.3) |
2 (1.7) |
0.261 | 2 (1.7) |
4 (3.3) |
0.175 |
| Cheek | 1 (0.8) |
3 (2.5) |
2 (1.7) |
2 (1.7) |
|||
| Floor
|
1 (0.8) |
5 (4.1) |
2 (1.7) |
4 (3.3) |
|||
| Gingiva | 0 (0) |
0 (0) |
1 (0.8) |
0 (0) |
|||
| Hard palate | 0 (0) |
2 (1.6) |
1 (0.8) |
1 (0.8) |
|||
| Upper lip | 4 (3.3) |
5 (4.1) |
5 (4.1) |
4 (3.3) |
|||
| Lower lip | 1 (0.8) |
1 (0.8) |
1 (0.8) |
1 (0.8) |
|||
| Maxilla | 1 (0.8) |
2 (1.6) |
1 (0.8) |
2 (1.6) |
|||
| Oral cavity | 8 (6.7) |
50 (41.7) |
10 (8.3) |
48 (40) |
|||
| Tongue | 10 (8.3) |
30 (25) |
11 (9.2) |
22 (18.3) |
|||
Control = Healthy oral tissue
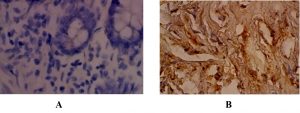
The detection of the expression of P16INK4A was carried out using IHC technology and was recognized as brownish color at nuclear localization (Fig. 2).
Fig. 2. Oral sections in OSCC: IHC staining of P16 protein ׳40. (A): Negative reaction; (B) positive reaction.
According to the results in Table 2, the expression of P16 INK4A was found in 21 tissues (52.5%) of malignant oral squamous cell tumors, 14 tissues (35%) of benign oral squamous cell tumors, and 1 tissue (2.5%) of control group.
HCMV was statistically significantly correlated with the malignant oral squamous cell group when compared with healthy tissues (control group) (p <0.001, OR=12.7). The p16INK4A gene was significantly correlated with the malignant oral squamous cell group when compared with the healthy tissues (control group) (p<0.001, OR= 33.66 (Table 3).
Table (3):
The Association Between HCMV and P16INK4A With Malignant And Benign Oral Squamous Cell Tissues.
| Malignant (n=40) | Control (n=40) | P value | OR(95%CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| hCMV | 17 (42.5%) | 3 (7.5%) | <0.001 | 12.7 (2.6-54.1) | |||||
| p16INK4A | 21(52.5%) | 1(2.5%) | <0.001 | 33.66(2.4) | |||||
| Benign (n=40) | Control (n=40) | P value | OR(95%CI) | ||||||
| hCMV | 10(25%) | 3(7.5%) | 0.001 | 8.2(1.9-40.09) | |||||
| p16INK4A | 14(35%) | 1(2.5%) | <0.001 | 37.8(2.3) | |||||
The p16INK4A gene was significantly correlated with the benign oral squamous cell tumors group when compared with the healthy oral squamous cell tissues (p <0.001, OR 37.8).
A strong positive relationship (with highly significant correlation) was found between HCMV and p16INK4A markers in oral squamous cell tissues (r = 0.476, P = 0.003). Similarly, there is a strong positive relationship (with highly significant correlation) between p16INK4A and grade of oral squamous cell tissues (r = 0. 439; p= 0.008). However, there are no significant correlations among HCMV and other markers (and as illustrated in table 4).
Table (4):
Spearman’s Rho Statistical Testing of Age, Grade, Hcmv-CISH And P16ink4a -IHC To Evaluate The Studied Markers In Oral Squamous Cell Tissues.
| Spearman’s rho | Age groups (years) | Grade | HCMV | p16INK4A | |
|---|---|---|---|---|---|
| Grade | r | -0.139 | |||
| P | 0.465 | ||||
| HCMV | r | 0.040 | 0.133 | ||
| P | 0.867 | 0.412 | |||
| p16INK4A | r | 0.176 | 0.439 | 0.476 | |
| P | 0.366 | 0.008* | 0.003* | ||
*Correlation is highly significant (P<0.01).
OSCC is complex and multifactorial disease, and the trials of authors to investigate the possible risk factors seem to present for OSCC have proposed other factors such as genetic susceptibility, viral infections, hormonal and immunologic modulations, and other systemic diseases in initiating and promoting OSCC18.
In the present study, the mean age of patients with oral cancer whom mean age was (57.28 years) while the mean age of those who have benign oral tumor & apparently healthy tissues were 45.40 and 50.80 years, respectively. These differences could be referred to the differences in population under different studies and also since some of the researchers included either a limited numbers or only an age range of patients with OSCC. It was reported that OSCC has predominance in older male patients, while in those who were younger was almost equally distributed among smokers and/or drinkers as compared to those non-smokers and/or non-drinkers counterparts19.
In this present study, the percentage of males (60%) with oral squamous cell carcinoma was higher than their female counterparts (40%) with a ratio of male to female 1.4/1. This result is compatible with previous Iraqi study on OSCC by Al-Kawaz et al.20, who reported male to female ratio of 1.4/1.Also our results agree with studies in carried out by Andisheh-Tadbir et al.21, and Srivastava et al.22, in Saudi Arabia and Iran, respectively. The reasons for oral cancer incidence and mortality rates being higher in males than in females are not completely understood and might likely reflecting complex gender-related interactions for exposure to the genetic as well as hormonal changes, smoking, tobacco chewing, alcoholic drinking, sociodemographic and clinical as well as pathological factors23.
In this study, 72.5% of OSCC have well differentiated grade, whereas moderately and poorly differentiated OSCC grade (III) constituted 17.5% and 10%, respectively. As reported in other studies, the majority of the patients presented with well or moderately differentiated tumors, most frequently located in the mobile tongue, floor of mouth and alveolar rim35. Alshawi (2010)24 studied OSCC grading and revealed likewise percentages to our results (71.42% in grade I, 17.14% in grade II and 11.41 % in grade III). Kashif et al.25, found 60.9% OSCC grading in grade I, 32.6% in grade II, and 6.5% in grade III. Furthermore, Sangle et al.38, found that 50 % grade I, 33.3% grade II and 16.6% grade III Consistently to our results. However, Minhas et al.26, revealed that moderate (II) grade OSCC cases were more common than well differentiated (I) grade. These results might related both to the poor early detection as well as health education programs, and to the non-specific symptoms of oral cancers so as the patients being presented with a high grade tumors.
The most affected site in the group of malignant oral tumors was tongue (35%); followed by oral cavity (25%), (7.5%) in both maxilla and cheek, tumors in buccal mucosa, lower lip of mouth, hard palate and floor of mouth each constituted (5% ) and lastly, tumors in both lips & gingiva (2.5%).Tumors arising in the floor of mouth has been explained by the accumulation of carcinogens in the floor of the mouth27.
Human Cytomegalovirus (HCMV) encodes various proteins that increase stimulation of cellular proliferation and migration, inhibition of apoptosis, and release stimulating factors28. It have been reported that the overall prevalence of HCMV in OSCC in respecting different areas was ranging from 0 to 91.5%29-32.
In our study, the percentage of HCMV in OSCC was higher (42.5%) than in the benign (25%) & control oral tissues (7.5%). The presence of such HCMV infection in the examined OSCC tissues implies that this virus could either increase the possibility of oncogenesis or opportunistically infected these cancerous tissues. Similarly33, was found the positive rates of HCMV DNA sequence were 0%, 14.3%, 28.1% in normal oral tissues & OSCC, respectively. In infected hosts, the cellular tropism of the virus is extensive where HCMV is particularly present in monocytes and lymphocytes, the main focus sites for CMV reactivation34.
Detection of viral DNA in tissue biopsies via immunohistochemistry or in situ hybridization have been used for diagnosis; however, if only few cells are affected, their results can be ambiguous .Both transient and established CMV infections are likely to involve multiplicity of infection of a few cells, indicating that a very small number of infectious viruses reached a susceptible target cell and/or an inefficient initial infection of those cells.
The p16INK4A gene is located on chromosome 9p21, and as a tumor suppressor gene, playing a key role in cell cycle regulation. It codes for a protein which binds and inhibits cyclin-dependent kinases (CdK-4and -6)35. It also inhibits phosphorylation of Rb and promotes the formation of an Rb-E2F repressive transcriptional complex, so as to blocks the progression of cell cycle at the G1-S point. The p16INK4A gene plays a significant role to inhibit malignancies. Altered and / or loss of p16 expression in oral premalignant and tumors lesions of the oral cavity have been reported36.
In our study, p16 expression was immunohistochemical analyzed in malignant oral squamous cell tumors (52.5%), while (35%) in benign oral squamous cell tumors, and 2.5% of control group .These present results are in accordance with studies conducted by Muirhead et al.37 and Shah et al.38 who demonstrated a decreased immunohistochemical expression of p16 in OSCC as compared with dysplasia and normal oral epithelia . Studies on p16 expression have reported that the percentage varied from 13%–94% in oral cancers (57-59). These wide range of variations and discrepancies could be attributed to the differences in sample size as well as the scoring scheme used for p16 assessment. It was observed that high frequency of OSCC has revealed a decreased expression of p16 which could happened by p16 gene inactivation via homozygous p16 gene deletion, mutation and hyper-methylation of its promoter36.
Dauren Adilbay et al.39, revealed p16-IHC positive signals in 25.7% of cases. The p16INK4Amethylation of promoter region has been reported to be from 23% to 67% in primary oral squamous cell carcinomas40.
It was observed that high frequency of OSCC has revealed a decreased expression of p16 which could happen by p16 gene inactivation via homozygous p16 gene deletion, mutation and hyper-methylation of its promoter36. p16INK4A promoter methylation in premalignant oral lesions tend to transformed these lesions into OSCC (41). Considering the cancer grade (from well differentiated grades towards the worse one, Kresty, Mallery42 also found that p16INK4A methylation is predominant high-grade OSCC patients.
It could be concluded from the main findings of this research paper that the contributions of HCMV along with co-expression of P16 gene in a respective rate of OSCC patients (42.5% and.52.5%, respectively) could point for or supporting the possible hypothesized role for HCMV infection along with mutated and /or defected p16 gene in either OSCC pathogenesis or carcinogenesis.
ACKNOWLEDGMENTS
We Ought to Acknowledge the Department of Biology, College of Science, University of Babylon and Clinical Communicable Diseases Research Unit (CCDRU), College of Medicine University of Baghdad for their Permission to perform the Practical Part of this Research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SHMA and SHMAA designed the study. SHMA collected data. SHMA, SHMAA, HGS, TFA and ZMMA analyzed the data. SHMAA, HGS, TFA and ZMMA interpreted the data. SHMA, HGS, TFA and ZMMA wrote the manuscript. HGS, TFA and ZMMA critically revised the manuscript. All authors read and approved the final version of the manuscript.
FUNDING
None.
ETHICS STATEMENT
This project has formal ethics approval by the college of medicine, University of Baghdad and college of medicine, University of Babylon, Iraq.
Ethics approvals for the follow up of individuals who donated normal oral tissues was given by the college of dentistry, University of Baghdad, Iraq.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Cesar R and bernardo V. Histological and molecular aspects of oral squamous cell carcinoma (Review). Oncol Lett., 2014; 8(1): 7–11.
Crossref - Malay Kumar, Ronak Nanavati, Tapan G Modi, Chintan Dobariya. Oral cancer: Etiology and risk factors: A review. J. Can. Res. Ther., 2016; 12(2): 458-463.
- Gondivkar SM, Parikh RV, Gadbail AR, et al. Involvement of viral factors with head and neck cancers. Oral Oncology, 2012; 48(3): 195–199.
Crossref - Greer RO, Jr., Douglas JM, Jr., Breese P, Crosby LK. Evaluation of oral and laryngeal specimens for human papillomavirus (HPV) DNA by dot blot hybridization. Journal of Oral Pathology and Medicine, 1990; 19(1): 35–38.
Crossref - Greer RO, Shroyer K, Crosby L. NIH Publication No. 92-3461. Bethesda, Md, USA: US Department of Health and Human Services, Public Health Service, National Institutes of Health; Identification of human papillomavirus DNA in smokeless tobacco keratoses and premalignant and malignant oral lesions by PCR amplification with consensus sequence primers, 1992.
- Park NH, Byung MM, Sheng LL. NIH Publication No. 92-3461. Bethesda, Md, USA: US Department of Health and Human Services, Public Health Service, National Institutes of Health; 1992. Role of viruses in oral carcinogenesis.
- Johnson NW. Risk Markers for Oral Disease, Oral Cancer Detection of Patients and Lesions at Risk.Cambridge, UK: Cambridge University Press; 1991.
- Nagao Y, Sata M, Tanikawa K, Itoh K, Kameyama T. High prevalence of hepatitis C virus antibody and RNA in patients with oral cancer. Journal of Oral Pathology and Medicine, 1995; 24(8): 354–360.
Crossref - Porter SR, Lodi G, Chandler K, Kumar N. Development of squamous cell carcinoma in hepatitis C virus-associated lichen planus. Oral Oncology, 1997; 33(1): 58–59.
Crossref - Shirin Saravani; Hamideh Kadeh; Ebrahim Miri-Moghaddam; Ali Zekri; Nima Sanadgol; Aliye Gholami. Human Cytomegalovirus in Oral Squamous Cell Carcinoma in Southeast of Iran. Jundishapur J. Microbiol., 2015; 8(8): e21838.
Crossref - Mario Dioguardi, Giuseppe Troiano, Lucio Lo Russo, Giovanni Giannatempo, Corrado Rubini, Dario Bertossi, Roberto Cocchi and Lorenzo Lo Muzio. Occult co-infection in the oral cavity with cytomegalovirus during immuno-suppression. J. Transl. Sci., 2015; 1(2): 26-28.
- Georges Herbein and Amit Kumar. The oncogenic potential of human cytomegalovirus and breast cancer. Frontiers in Oncology. Molecular and Cellular Oncology, 2014; 4: Article 230.
Crossref - Witkiewicz, A.K., Knudsen, K.E., Dicker, A.P., and Knudsen, E.S. The meaning of p16 (ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle, 2011; 10: 2497–2503.
Crossref - Doxtader EE, Katzenstein ALA. The relationship between p16 expression and high-risk human papillomavirus infection in squamous cell carcinomas from sites other than uterine cervix: a study of 137 cases. Hum Pathol., 2012; 43: 327–332.
Crossref - Eleftherios Vairaktaris, Christos Yapijakis, Amanda Psyrri, Sofia Spyridonidou, Athanasios Yannopoulos et al. Loss of Tumour Suppressor p16 Expression in Initial Stages of Oral Oncogenesis. Anticancer Research, 2007; 27: 979-984.
- Shah NG, Trivedi TI, Tankshali RA, Goswami JV, Jetly DH, Shukla SN, Shah PM, Verma RJ. Prognostic significance of molecular markers in oral squamous cell carcinoma: a multivariate analysis. Head Neck, 2009; 31: 1544–1556.
Crossref - Harris SL, Thorne LB, Seaman WT, Hayes DN, Couch ME, Kimple RJ. Association of p16INK4a overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head and Neck, 2011; 33: 1622–1627.
Crossref - Beena, SS, Binisree, T Ayswarya, Ismayil Paikkadan, SK Padmakumar, R Sivakumar. Oral Squamous Cell Carcinoma in Patients Younger than 40 Years: A 10 Year Retrospective Study. International Journal of Scientific Study, 2016; 4(4): 150-153.
- Vera Panzarella, Giuseppe Pizzo, Francesco Calvino, Domenico Compilato, Giuseppe Colella, and Giuseppina Campisi. Diagnostic delay in oral squamous cell carcinoma: the role of cognitive and psychological variables. International Journal of Oral Science, 2013; 6: 39–45.
Crossref - Al-Kawaz AB. Oral squamous cell carcinoma in Iraq: clinical analysis. MDJ, 2010; 7(1): 100-5.
- Andisheh-Tadbir A, Mehrabani D, Heydari ST. Epidemiology of squamous cell carcinoma of the oral cavity in Iran. J. Craniofac. Surg., 2008; 19: 1699–1702.
Crossref - Srivastava S., Roy R., Gupta V., Tiwari A., Srivastava A.N., Sonkar A.A. Proton HR-MAS MR spectroscopy of oral squamous cell carcinoma tissues: an ex vivostudy to identify malignancy induced metabolic fingerprints. Metabolomics, 2011; 7: 278–288.
Crossref - Vera Panzarella, Giuseppe Pizzo, Francesco Calvino, Domenico Compilato, Giuseppe Colella, and Giuseppina Campisi. Diagnostic delay in oral squamous cell carcinoma: the role of cognitive and psychological variables. International Journal of Oral Science, 2013; 6: 39–45.
Crossref - Ali Abbas Alshawi. Age And Biopsy As Predictive Factors In The Management Of Oral Squamous Cell Carcinoma. Bas. J. Surg., 2010; 16.
Crossref - Muhammad Kashif, Muhammad Ishfaq, and A.H. Nagi. Expression of Prostanoid EP3 Receptors in Oral Squamous Epithelium and Oral Squamous Cell Carcinoma. Pathology Research International, 2015.
Crossref - Minhas, S., Kashif, M., Altaf, W. & Nagi, A.H. Oral Squamous Cell Carcinoma: Epidemiological, clinical and histological features. RMJ, 2016; 41: 81-85.
- Liviu F, Johan L. Oral squamous cell carcinoma: epidemiology, clinical presentation and treatment. J. Cancer Therapy, 2012; 3: 263-8.
Crossref - Johnsen JI, Baryawno N, Soderberg-Naucler C. Is human cytomegalovirus a target in cancer therapy? Oncotarget., 2011; 2(12): 1329–38.
Crossref - Delavarian Z, Pakfetrat A, Falaki F, Pazouki M, Pazouki N. The Role of Viruses in Oral Squamous Cell Carcinoma in Young Patients in Khorasan (Northeast of Iran). J. Appl. Sci., 2010; 10(11): 981–5.
Crossref - Yang YY, Koh LW, Tsai JH, Tsai CH, Wong EF, Lin SJ, et al. Involvement of viral and chemical factors with oral cancer in Taiwan. Jpn. J. Clin. Oncol., 2004; 34(4): 176–83.
Crossref - Sugiura H, Yamawaki T, Toyoshima K, Kimura M, Yamaguchi A, Shibasaki K. Association between oral squamous cell carcinoma and virus infection, 2006.
- Saad Hasan Mohammed A, Majed Mohammed Mahmood Al J, Noor Al Huda Ali AHS. Localization of human cytomegalovirus- late gene DNA, expression of P53 gene and CD8-tumor infiltrating lymphocytes in oral squamous cell carcinoma. Iraqi Postgrad Med J., 12(2): 296–305.
- Wei H, Xiaoming SQX. Primary study on human papillomavirus type 16 and human cytomegalo-virus infections in oral squamous cell carcinoma. J. ComprStomatol., 1996; 2(001): 001.
- Griffiths P, Baraniak I, Reeves M. The pathogenesis of human cytomegalovirus. J. Pathol., 2015; 235: 288–297.
Crossref - Sushma PS, Jamil K, Kumar PU, Satyanarayana U, Ramakrishna M, Triveni B . PTEN and p16 genes as epigenetic biomarkers in oral squamous cell carcinoma (OSCC): a study on south Indian population. Tumour Biol., 2016; 37(6): 7625-32.
Crossref - Vairaktaris E, Yapijakis C, Psyrri A, Spyridonidou S, Yannopoulos A, Lazaris A, et al. Loss of tumour suppressor p16 expression in initial stages of oral oncogenesis. Anticancer Res., 2007; 27: 979–84.
- Muirhead DM, Hoffman HT, Robinson RA. Correlation of clinicopathological features with immunohistochemical expression of cell cycle regulatory proteins p16 and retinoblastoma: distinct association with keratinization and differentiation in oral cavity squamous cell carcinoma. J. Clin. Pathol., 2006; 59: 711–5.
Crossref - Shah NG, Trivedi TI, Tankshali RA, Goswami JV, Jetly DH, Shukla SN, et al. Prognostic significance of molecular markers in oral squamous cell carcinoma: A multivariate analysis. Head Neck, 2009; 31: 1544–56.
Crossref - Dauren Adilbay, Galim Adilbayev, Gulzhan Kidirbayeva, Viktoria Shipilova, Zhanat Sadyk, Gulsum Koyanbekova, Ekaterina Sokolenko and Jan Kloza. HPV infection and P16 expression in oral and oropharyngeal cancer in Kazakhstan. Nfectious Agents and Cancer, 2018; 13: 2.
Crossref - Soria JC, Felip E, Cobo M, Lu S, Syrigos K, Lee KH, Goker E, Georgoulias V, Li W, Isla D, et al. Afatinib versus erlotinib as second-line treatment of patients with advanced squamous cell carcinoma of the lung (LUX-Lung 8): an open-label randomised controlled phase 3 trial. Lancet Oncol., 2015; 16: 897–907.
Crossref - Hall GL, Shaw RJ, Field EA, Rogers SN, Sutton DN, Woolgar JA, Lowe D, Liloglou T, Field JK, Risk JM, 2008. p16 promoter methylation is a potenial predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiology, Biomarkers & Prevention, 17: 2174-2179.
Crossref - Kresty LA, Mallery SR, Knobloch TJ, et al. Alterations of p16INK4Aand p14ARF in patients with severe oral epithelial dysplasia. Cancer Research, 2002; 62(18): 5295–5300.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.