Antimicrobial Resistance (AMR) has been regarded as a major public health concern as a reason of millions of deaths. Extended-spectrum β-lactamase (ESBL) is considered as a leading factor contributing to this and limiting its treatment. Thus, ESBL producing Enterobacteriaceae should be discriminated from those having other mechanism conferring resistance. Several phenotypic methods have been evaluated for this purpose. Some of these are based on conventional method (DDST, CDT, ESBL E-test, Cica-β test) while others depend on automated systems (VITEK 1, VITEK 2, Phoenix, MicroScan). All the conventional methods have been found to be more specific, sensitive and cost effective than any of the automated system though they are easy to perform and interpret. Automated system also fails to detect ESBL in presence of other interfering enzymes such as AmpC, MBL or K1 enzyme. ESBL can be detected by using third-generation cephalosporin (cefotaxime or ceftazidime) or monobactam (aztreonam) in combination with clavulanate. AmpC can be distinguished by using cloxacillin-containing agar, fourth-generation cephalosporin (cefepime) or phenylboronic acid. MBL producers remain unaffected in presence of clavulanate but gets inhibited by carbapenems (imipenem, meropenem) in combination with EDTA. Cefpodoxime-clavulanate and ceftazidime- clavulanate combinations are reliable for K1 enzyme detection but are not suitable for distinguishing blaCTX–M1.
β-Lactamase, Enterobacteriaceae, Extended-spectrum β-lactamases, AmpC, Metallo-β lactamase, K1 Enzyme, Phenotypic Detection
Antimicrobial resistance (AMR) has emerged as a serious public health concern causing millions of deaths worldwide in last 2 decades.1 The factors that mostly contribute to AMR include irrational and overuse of antibiotics.1 b-lactam antibiotics are those that contain b-lactam ring in their molecular structures are classified as penams, cephems (cephalosporins and cephamycins), monobactams, carbapenems,2 and carbacephems.3 Until 2003, in terms of selling quantity, more than half of all commercially available antibiotics in use were b-lactam compounds.4 Extended-spectrum b-lactamases (ESBLs), the enzyme produced by the microorganism like Klebsiella species, Acinetobacter, Pseudomonas aeruginosa, Citrobacter, Proteus, Enterobacter and E. coli,are known to be a potent hydrolyzer of b-lactam antibiotics.5 ESBLs producing bacteria are crucial threat for patients in the hospital, long-term care facilities and the community for their highly disseminating characteristics.6 ESBL producing E. coli strains also show resistance to other classes of antibiotic making the pathogen hard to treat.7-8
Extended-Spectrum β-Lactamase (ESBL), comes under Ambler class A and D β-lactamase enzymes,9 They mediate resistance to extended spectrum (third and fourth generation) cephalosporins (e.g. Ceftazidime, cefotaxime, ceftriaxone, cefepime etc.) and monobactams (e.g. Aztreonam) leaving antimicrobial classes like cephamycins (e.g. Cefoxitin and cefotetan) or carbapenems (e.g. meropenem or imipenem) and are inhibited by β-Lactamase inhibitors such as clavulanate, sulbactam and tazobactam.10-12 The Clinical and Laboratory Standards Institute (CLSI) in the United States issued national guidelines for laboratory detection of E. coli, Proteus mirabilis, and Klebsiella spp. with ESBL. But, the guideline for the interpretation of ESBL testing results for those species of Enterobacteriaceae that are also good AmpC producers (Ambler class C enzyme) is not provided there. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) eases this problem by recommending some tests for the detection of ESBL alone and in presence of AmpC and carbapenemase enzymes.13 Resistance, at breakpoint, is not fixed to all third- or fourth generation cephalosporins, whether based on disk-diffusion in agar, minimum inhibitory concentration (MIC) determination or automated systems, it varies with the choice of cephalosporin tested,14 and the recommended guidelines to be followed. Two detection strategies are in common use: one is the detection of ESBL using a third-generation cephalosporin, usually cefotaxime or ceftazidime, and second is the confirmation of the former by detecting the synergy between the extended spectrum cephalosporin and a β-lactamase inhibitor, usually clavulanate. Based on these strategies several phenotypic methods have been developed to detect or confirm ESBL production; some of them depend on manual handling while others on commercially available automated systems.15
This review describes the phenotypic methods for ESBL detection, their advantages, and limitations. In addition, it also describes the detection of ESBL-producing micro-organisms which are co-producers of other enzymes that interfere in ESBL detection. AmpC, Metallo-β-lactamase, and K1 enzyme act as masking agents of ESBL production. ESBL production should, therefore, be detected in the presence of these enzymes. Though phenotypic methods are out of date a bit, they are still performed in various laboratories, especially in low- and middle-income countries where the prevalence of ESBL producing Escherichia coli is high.
Manual Methods of ESBL Detection Test
Double-Disc Synergy Test (DDST)
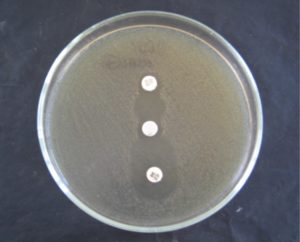
Double-disc synergy test is a kind of disc diffusion test that detects the effectiveness of an antimicrobial agent in the presence of another antimicrobial agent.16 This test utilizes two discs on the cultivated agar plate, either infused with a different antimicrobial solution.16,17 It was initially designed to differentiate between the microorganisms that over produce cephalosporinase, and those that produce ESBLs.18 In this test two discs are utilized: one containing 30µg of any third-generation cephalosporin (cefotaxime, ceftriaxone, ceftazidime) or monobactam (aztreonam) and another containing amoxicillin–clavulanate (containing 10 µg of clavulanate) usually positioned at a distance of 30 mm (centre to centre) on a cultivated agar plate (Figure 1). DDST for ESBL is considered positive when reduced susceptibility to third-generation cephalosporin or monobactam shows a prominent increase in inhibition zone of antimicrobial agent towards the clavulanate-containing disk, often resulting in a characteristic shape-zone referred to as ‘champagne-cork’ or ‘keyhole’. The DDST was first used in epidemiological studies to assess the spread of ESBL-producing Enterobacteriaceae in French hospitals.19,20 It has been generally regarded as an effective and reliable method for the detection of ESBLs in a wide range of Enterobacteriaceae species, although it is sometimes necessary to adjust the disk spacing. It has been shown that the sensitivity increased even more, to 90%, when this distance was reduced to 20 mm.21 Modified Double Disc Synergy Test (MDDST) utilizes the 4th generation cephalosporins (cefepime) and an optimum spacing of the discs infused with antimicrobial solutions and inhibitor for the detection of the synergy.22 But for this spacing modification, disks should be arranged by the help of forceps with narrower distances as several types of marketed disk-dispenser are designed for the routine spacing (i.e. 30 mm).
Figure 1. Double-disc synergy test performed on ESBL producing strain. Amoxicillin/clavulanate containing disc placed in between cefotaxime (up) and ceftazidime (down).24
Combination Disc Test (CDT)
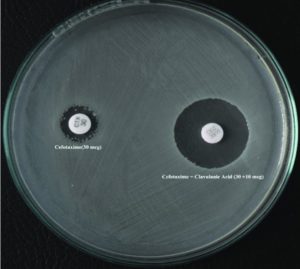
Combination Disc Test is another kind of disc diffusion test that also require the comparison of the effectiveness of two discs infused with antimicrobial solution placed on same cultured agar plate, but in this case both of the discs are having the same antimicrobial agent. For each test, 2 discs are required; One containing cephalosporin/monobactam alone (cefotaxime, ceftazidime, cefpodoxime, aztreonam) while another contains the same antimicrobial in combination with clavulanic acid. The inhibition zone around the cephalosporin/monobactam disc combined with clavulanic acid is compared with the zone around the disc with the cephalosporin/monobactam alone.23 The test is supposed be positive if the inhibition zone diameter is ≥ 5 mm larger with clavulanic acid than without (Figure 2).23 The advantage of this test is it is very easy to perform and interpret and has an upper hand over DDST as the latter one lacks sensitivity because of the problem of optimal disc spacing, the need of precision.5 The sensitivity, specificity, positive predictive value & negative predictive value of CDT were assessed as 100%, 77.9%, 83.1% & 100% respectively in one study by Chowdhury et al.6 This method is also useful in distinguishing ESBL producers from strains with AmpC enzymes and from K1 enzyme over-expressing the K. oxytoca.15
Figure 2. Combination disc test performed on ESBL producing strain. Cefotaxime and cefotaxime/clavulanate containing discs are placed apart from each other.58
ESBL E-Test
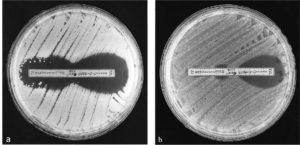
Epsilometer test or E-test is defined as the ‘exponential gradient’ method that determines the antimicrobial resistance of the microorganism.24 ESBL E-test uses commercially available rectangular plastic strips containing exponential gradient of cefotaxime, ceftazidime or cefepime alone at one end and in combination with clavulanate (4mg/L) on the other (Figure 3a). The concentration gradient is marked with numerical scale on the strip. These strips are denoted as CT/CTL, TZ/TZL, PM/PML strips respectively for cefotaxime/cefotaxime-clavulanate, ceftazidime/ceftazidime-clavulanate and cefepime/cefepime-clavulanate.25 A strain is considered ESBL positive when the inhibition ellipse made by the test cephalosporin alone and in combination with clavulanate is compared and the latter shows a reduced MIC ratio by ≥ 8.25 The test is also considered as positive when there is either a rounded zone (phantom zone) (Figure 3b) just below the lowest concentration of CTL, TZL or PML gradients, or a deformation of the CT, TZ or PM inhibition ellipse at the tapering end independent of the ratios or MICs24. A major drawback of this test is interpreting the results of the ESBL E-test strips is delicate and requires expert handling. In addition, it is also difficult to interpret when the MIC values for cephalosporins fall outside the range of MICs available on the test strip and it is also true for the multi-resistant isolates.26
Figure 3. ESBL E-test performed with cefepime and cefepime/clavulanate strip. a) Result shows ESBL negative strain. b) Result shows ESBL positive strain with clear presence of phantom zone.59
Cica-β Test
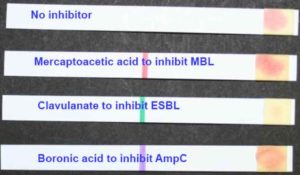
This test examines for hydrolysis of the chromogenic oxyimino-cephalosporin HMRZ-86 with and without specific inhibitors.27 This technique is simple and fast; gives result in 15 mins and it is also useful for detecting not only ESBL but also the AmpC and metallo β-lactamases (MBL). Cica-β Test kit comes with four strips; each containing chromogenic oxyimino-cephalosporin HMRZ-86 with a) no inhibitor to detect hydrolysis of extended-spectrum cephalosporins, b) clavulanic acid to detect ESBL, c) with boronic acid to detect AmpC production and d) sodium mercapto-acetic acid to detect metallo-β-lactamases.27 A colour change from yellow to red indicates positive result (Figure 4). Garrec et al. in his study, showed that Cica-β test had significantly lower sensitivity in comparison to the other phenotypic detection methods.28 Not only that, it had some other limitations such as misinterpreting the K1 enzyme and carbapenemase hyper producer as AmpC overproducer.27
Figure 4. Cica-β test perfomed on ESBL producing strain. Presented by David Livemore at ECCMID 2010 in Vienna
Three Dimensional Tests (TDTs)
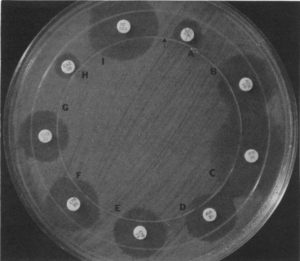
Three Dimensional susceptibility testing was first proposed by Thomson et al. in 1984.29 This test came with slight modification in disc diffusion method with a heavy inoculum load that had been thought to provide more information as some workers felt that the light inoculum of some disc techniques fails to portray the in-vivo situation.29 Two types of Three Dimensional susceptibility test are there: a) direct and b) indirect.30 In direct TDT MHA plates are inoculated with test strains matching 0.5 McFarland turbidity standards. Then this inoculated agar plate is to be stabbed vertically with a sterile scalpel blade touching the bottom of the agar at a predetermined point 3 mm inside the position at which the antibiotic discs are to be placed. A circular slit is cut in the agar concentric with the margin of the plate (Figure 5). Slit was inoculated with test strain having 109 – 1010 CFU of cells dispensed in tryptone soy broth. Then plate is to be incubated overnight at 37°C. In indirect TDT MHA plates were seeded with the inoculum of a standard sensitive strain (E. coli ATCC 25922) adjusted to McFarland 0.5 standards.31 With the exception of this, the method is the same as that described for the direct three-dimensional test. Indirect was performed when inhibition zones were small or absent. Distortion of the inhibition zone in the vicinity of its intersection with the circular three-dimensional inoculation indicated positive result. In some cases, crude enzyme extract can also be used instead of using direct bacterial isolate as Courdon et al. (2000) showed in his study.32 Thomson and Sanders compared the combination of the direct and the indirect three-dimensional tests and the DDST with 32 strains of E. coli and K. pneumoniae, 28 of which produced ESBL. 93% and 82% of the isolates were found to be ESBL positive respectively by TDTs and DDST.31 Menon et al. performed the same with 70 Enterobacteriaceae strains, of which 56 were multidrug resistant and 14 were ESBL producers.30 They reported detection rates of 86% for the three-dimensional test and 14% for DDST.30 Sahid et al. in 2004 proposed some modifications in three-dimensional susceptibility testing. He brought the modifications in both the ways stated earlier, i.e. a) using direct bacterial isolates and b) using crude enzyme extracts.33 The modifications adopted by him were that he put inoculum or enzyme extract into slits, wells or onto the surface as spots at varying distances from the discs. He showed that spot inoculation and well inoculation method gave better results than the conventional method by using whole bacterial isolates and enzyme extract respectively.33
Figure 5. Direct three-dimensional test of CAZ-2-producing E. coli MISC 234. The three-dimensional inoculation slit is represented by the arrow. Result shows minor distortions indicating antibiotic inactivation of cefamandole (D), cefoperazone (E), cefotaxime (F), and ceftriaxone (G). No distortions occur in tests with aztreonam (B), imipenem (C), or cefoxitin (I). The inhibition zones were too small to interpret result with piperacillin (A) and ceftazidime (H).31
Automated Methods of ESBL Detection Test
VITEK 1 ESBL Test
VITEK 1 analysis is an automated system that assess the antimicrobial activity of some cephalosporins with and without clavulanate simultaneously while the test interpretation is based on computerized expert system.34 VITEK 1 test utilized commercially available cards having four wells containing cefotaxime and ceftazidime alone (at 0.5 mg/ml) and in combination with clavulanic acid (at 4 mg/ml). After inoculation this card is placed inside the VITEK 1 analyzer which then indicates the isolate as either ESBL positive or negative based on the predetermined ratio of reduction in growth of the cefotaxime or ceftazidime well containing clavulanic acid with that in the well containing drug alone.34
VITEK 2 ESBL Test
VITEK 2 ESBL test is a modified version of VITEK 1 ESBL test; this test uses three antimicrobial drugs instead of two including and excluding clavulanic acid. Different sets of VITEK 2 cards are commercially available. Some cards contain cefepime (1.0 mg/L), cefotaxime and ceftazidime (0.5 mg/L) either alone or associated with 10 or 4 mg/L of clavulanate, respectively,35 while other cards contain ceftriaxone, cefepime, and aztreonam (0.5 mg/L) with and without clavulanate (4 mg/L). The latter is currently only validated for detection of ESBLs in E. coli, K. pneumoniae, and K. oxytoca.28
Phoenix ESBL Test
The automated Phoenix ESBL test relies on the growth response to selected extended-spectrum cephalosporins, with or without clavulanic acid.36 Phoenix ESBL panel is composed of five wells containing cefpodoxime, ceftazidime, ceftazidime with clavulanic acid, cefotaxime with clavulanic acid and ceftriaxone with clavulanic acid respectively.36 After inoculation, the panel is placed in the instrument for continuous growth monitoring and result is interpreted through BDXpert system. This BDXpert system depends on three rules: a) Rule no. 1505: “Enterobacteriaceae with ESBLs are resistant to all β-lactam drugs, except carbapenems.”;b) Rule no. 1502: “Enterobacteriaceae that are susceptible to a carbapenem and resistant to ureidopenicillins and 3rd generation cephems or cefpodoxime or aztreonam are also resistant to all β-lactam, except carbapenems.”; c) Rule no. 106 may be provided: “Screening test suggests a possible ESBL producer; confirmatory testing is recommended.” No rule is supplied if test result is negative.26
MicroScan Analysis
The Siemens MicroScan technology was introducedasthe first advanced technology that was based on true minimum inhibitory concentration.37 The MicroScan WalkAway-96 SI System comes with automated features including automated maintenance and result interpretation. Dehydrated inoculation panels are provided by the company to be inoculated overnight, placed inside the WalkAway-96 SI instrument for result and interpretation. The following antimicrobial agents (concentration ranges in mg/L) are used in the MicroScan ESBL plus panel: aztreonam (0.5–16); cefepime (1–32); cefotetan (1–32); cefpodoxime (0.5–64); cefotaxime(0.5–128); cefotaxime/clavulanate (0.125/4–16/4); cefoxitin (2–32); ceftazidime (0.5–128); ceftazidime/clavulanate (0.125/4–16/4); ceftriaxone (1–64); imipenem (0.5–16); meropenem (0.5–16); piperacillin (16–64)38 and results are interpreted according to the CLSI guidelines.37
There are many studies that have evaluated the ability of automated systems to detect ESBL-producing Enterobacteriaceae and some of these studies are summarized below: Leverstein-van hall et al., in his study, worked with the 74 multi-resistant Enterobacteriaceae isolates (34 E. coli, 26 K. pneumoniae, and 14 K. oxytoca isolates) and 17 controls to evaluate the efficacy of the VITEK 1, VITEK 2, and Phoenix automated system. The three automated systems detected ESBL production with a sensitivity ranging from 78% (VITEK 2) to 83% (VITEK 1) to 89% (Phoenix).26 Concerning multi-resistant isolates the VITEK 2 performed worse than the Phoenix as it gave highest percentage of indeterminate test results.26 Wiegand et al. took three automated system into his consideration i.e. VITEK 2, Phoenix and MicroScan automated system. He performed his experiment using 147 Enterobacteriaceae isolates (E. coli, 62 isolates; K. pneumoniae, 29 isolates; E.cloacae, 17 isolates; K. oxytoca, 16 isolates; Enterobacter aerogenes,6 isolates; Proteus mirabilis, 6 isolates; Morganella morganii, 5 isolates; C. freundii, 4 isolates; Proteus vulgaris, 1 isolate; Serratia marcescens, 1 isolate) of which 85 were ESBL positive. The Phoenix automated system showed highest sensitivity for the detection of ESBLs with 99%, followed by the VITEK 2 (86%) and the MicroScan (84%); while in the case of specificity assessment, result was more variable. VITEK 2 showed highest specificity (78%) followed by a specificity of 72% for MicroScan and 52% for Phoenix automated system.37 Thomson et al., in their study, they used 76 ESBL producing and 26 ESBL non-producing Enterobacteriaceae strains to compare Phoenix automated system with VITEK 2. They found that the Phoenix ESBL confirmatory test and expert system exhibited 96% sensitivity and 81% specificity for ESBL detection initially. Activation of the two additional rules i.e. rules 325 and 1437, increased sensitivity to 99% but unfortunately reduced the specificity to 58%. The VITEK 2 ESBL confirmatory test exhibited 91% sensitivity, which was reduced to 89% sensitivity by its expert system, while its specificity was ranging from 85% to 88%.39
Detection of ESBL in the Presence of Other Interfering Enzymes
Combination of Several Tests for Detection of ESBL in Strains Overproducing AmpC
The production of AmpC enzyme in gram-negative bacteria is controlled by either chromosomal gene (inducible) or by a plasmid (stable production).40 Several bacterial species (Enterobacter spp., K. aerogenes, C. freundii, M. morganii, P. stuartii, and S. marcescens) have inducible chromosomally encoded AmpC cephalosporinase and are considered in serious infections.41 Expression of inducible AmpC is regulated by AmpR. E coli lacks this AmpR. Thus, expresses this enzyme at a low level consecutively.42 Other bacterial species, such as K. pneumoniae, which lack chromosomal genes, can acquire plasmid genes. E coli may harbor plasmid carrying AmpC.42 AmpC enzymes confer resistance to oxyimino-cephalosporins, a-methoxy- β-lactams or cephamycins (cefoxitin and cefotetan), and monobactams and are poorly inhibited by clavulanic acid.40 Thus, AmpC productions can mask the detection of ESBL.
Several modifications in DDST and CDT have been introduced to detect AmpC production in strains in which ESBL is masked by the overproduction of AmpC cephalosporinases. Those modifications can be summarized as follows:
- Cefepime, a fourth-generation cephalosporin, is less rapidly hydrolysed by AmpC enzyme than ESBL. Tzelepi et al. reported a sensitivity of 16% only when placing cefotaxime, ceftriaxone, ceftazidime and aztreonam discs adjacent (30mm) to amoxicillin–clavulanate containing disc but an increased sensitivity by 61% and 90% when used cefepime disc and placed this disc at a distance of 20mm respectively.21
- Clavulanic acid, sulbactam, and tazobactam have an effect in inhibiting ESBLs but AmpC β-lactamases are poorly inhibited by these inhibitors.43 Cloxacillin, oxacillins however are proved to be good inhibitors of AmpC β-lactamases.44 Performing the DDST on cloxacillin containing agar (200 mg/L) has been shown to enhance the ability of the test to detect ESBL in Acinetobacter baumannii45 and Pseudomonas aeruginosa.46
- A small set of boronic acids act as low nanomolar inhibitors of AmpC β-lactamase but do not affect ESBL.47 Nagarathnamma et al. reported identification of AmpC β-lactamase and ESBL co-producers correctly by CDT method using cefotaxime (CTX), cefotaxime plus clavulanic acid (CTX/CA), cefotaxime plus boronic acid (CTX/BA) and cefotaxime plus clavulanic acid with boronic acid (CTX/CA/BA) discs.48 But this method fails to detect inducible AmpC (iAmpC). Nagarathnamma overcame this limitation by placing imipenem disc adjacent to the earlier mentioned discs as imipenem had been known to be a potent inducer of iAmpC.48
- Drieux et al. recommended the use of the ESBL E-test on cloxacillin-containing agar when the MIC values are higher than those measurable on the strips.35 He had shown a significant reduction in MIC value when using cloxacillin-containing agar rather than using Mueller–Hinton agar (MHA)and a further reduction in MIC in presence of clavulanate.35
Modification of Some Phenotypic Tests to Distinguish Metallo-β-Lactamases Producers from Carbapenemase Producing Enterobacteriaceae and Detection of ESBL in It
Carbapenem-resistant Enterobacteriaceae (CRE) can be defined by their resistance to any carbapenems such as imipenem, meropenem, or ertapenem. Carbapenemase class A such as KPC enzyme49 hydrolyzes a broad range of β-lactam antibiotics including pencillins, cephalosporins and carbapenems. This enzyme also efficiently hydrolyzes cefotaxime but only poorly hydrolyzes ceftazidime50 and can be inhibited by enzyme inhibitors such as clavulanic acid and tazobactam. Carbapenemase class B enzymes (IMP, VIM, GIM and SPM-1) or metallo-β-lactamases (MBL) generally hydrolyze third generation cephalosporins as well as carbapenems, but not aztreonam35 and can be inhibited by ethylene diamine tetra-acetic acid (EDTA) and mercaptopropionic acid.51 Several modifications were adapted for detecting MBL from other carbapenemase producers in these manners:
- Liao et al. used imipenem discs with and without EDTA for the detection of MBL.51 A difference of 5mm in the zone of inhibition created by those two discs clearly indicated the production of MBL. But in his study, he inoculated the test strain onto the imipenem disc instead of inoculating the disc in test strain suspension indicating that the strains were inoculated onto the MHA plate following the routine disc diffusion procedure (simple EDTA synergistic carbapenem inactivation method or esCIM). He reported that esCIM performs better than eCIM in detecting MBL with 91% sensitivity and 100% specificity.50
- Wei et al. developed rapid EDTA-modified carbapenem inactivation method (reCIM) combined with modified rapid carbapenem inactivation method (mrCIM) that might give result within 4 hours.52 For this purpose, he used meropenem discs incubated with test strain for 45 mins. He placed these discs inside the Eppendorf tube containing indicator E. coli ATCC 25922 already incubated for 2.5 hours. The growth of indicator E. coli with OD > 2.0 indicated carbapenemase production, whereas OD < 1.0 could be considered as carbapenemase negative. Interpretation reCIM was same as mrCIM but in this case OD>2.0 indicated presence of MBL.52
- If an isolate shows resistance to imipenem and ceftazidime simultaneously then there are four possibilities of being either class A or class A carbapenemase and ESBL positive or metallo-β-lactamase plus ESBL positive or only class B carbapenemase positive. If this same isolate shows resistance to aztreonam too, it can be considered as class A or KPC enzyme producer or co producer of class A carbpenemase and ESBL or producer of MBL with ESBL as only MBL is susceptible to aztreonam.35 Now if this same strain confers resistance to EDTA, it can be considered as co-producer KPC enzyme and ESBL or the producer of KPC enzyme alone. The presence of ESBL in MBL producing strain can be demonstrated by detecting synergy between clavulanate & EDTA containing disc and third generation cephalosporin or cefepime.35 KPC enzyme production can be detected by a combined disc test using aminophenylboronic acid (400µg/disc) with and without imipenem.53
False –Positive ESBL Detection with K1 Enzyme Hyperproducers
K1 enzyme is also called KOXY enzyme, named as it is frequently encountered in some isolates of K. oxytoca.54 Hyperproduction of this chromosomal β-lactamase results in false positive detection of ESBL as it also shows resistance to some extended spectrum cephalosporins and monobactams. KOXY enzyme is characterized by resistance to aztreonam and cefuroxime, moderate resistance to ceftriaxone, cefotaxime and cefepime and susceptibility to ceftazidime.36,55 This resistance pattern is little bit different from ESBL. For this reason, ceftazidime must be included in the panel of antibiotics for routine susceptibility testing56,57 for ESBL. However, CTX-M ESBL also shows susceptibility to ceftazidime making it difficult to differentiate from K1 enzyme.55 Potz et al., reported only one false positive ESBL result among the 25 K. oxytoca isolates hyperproducing K1 β-lactamase but lacking ESBLs when performed agar dilution method with ceftazidime-clavulanate.55 In addition, cefpodoxime discs with and without clavulanate were consistently give negative results for the K1 hyperproducers proving that Cefpodoxime–clavulanate combination disc tests were reliable in distinguishing ESBL producers from hyperproducers of K1 enzyme.
ESBLs are becoming more complex, diverse and widespread day by day that a single susceptibility testing approach for detection must be diminished. A wide genetic diversity including the presence of other enzymes that confer broad spectrum drug resistance makes it more difficult to be diagnosed. Conventional phenotypic methods sometimes remain difficult in practice. Therefore, some modifications and modulations have been adapted. Nowadays, some kits are commercially available in the market for the detection of ESBL alone and in combination with other enzymes by CDT. All the above-mentioned studies unanimously evidenced that though these automated methods were easy to perform as all the panel card were commercially available and analyzed by a previously set computerized system but were less effective in detecting simultaneous expression of the different β-lactamases, possibly in combination with outer membrane porin changes or overproduction of AmpC or K1 enzyme that might mask the ESBL production. Performance of the DDST, CDT and ESBL E-Test showed better result than those of the automated systems. Use of single cephalosporin as an indicator has been known for the outcome with low sensitivity while the use of two or more antimicrobials of this category enhances both sensitivity and specificity. Although, it is difficult to attain 100% sensitivity with any of these conventional methods. Molecular assay is always regarded as the gold standard method. PCR can detect accurately the presence of resistance-conferring genes (blaTEM, blaSHV, blaCTX–M1, blaCTX–M 2, blaCTX–M 9 etc.) though it is time consuming whereas Microarray can detect simultaneously several enzyme-encoding genes from a single strain. The approach of gene sequencing remains helpful in detecting mutation that makes it more resistant to newer antibiotics. However, phenotypic methods are regarded as more convenient, easy to perform and cost-effective methods for screening in most of the clinical laboratories.
ACKNOWLEDGMENTS
The authors would like to thank Mr. K. Shaw, Mr. T. Ghorai, Mr. T. Chatterjee of Rammohan College, University of Calcutta, for their continuous support and encouragement. Authors are also thankful to Dr. T. J. Chowdhury and Dr. S. Dasgupta of Sinha Institute of Medical Science and Technology for their valuable suggestions and guidance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This project was supported by the Council of Industrial and Scientific Research (CSIR) with grant number 08/0749(0001)/2019-EMR-I.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Dixit A, Kumar N, Kumar S, Trigun V. Antimicrobial Resistance: Progress in the Decade since Emergence of New Delhi Metallo-b-Lactamase in India. Indian J Commun Med. 2019;44(1):4-8.
Crossref - Holten KB, Onusko EM. Appropriate prescribing of oral beta-lactam antibiotics. American Family Physician. 2020;62(3):611-620.
- Yao JDC, Moellering RC. Antibacterial agents. In Murray P, Baron E, Pfaller M, Tenover F, Yolken R. Manual of Clinical Microbiology. 9th ed. Washington DC: ASM press; 2007.
- Elander RP. Industrial production of beta-lactam antibiotics. Appl Microbiol Biotechnol. 2003;61(5-6):385-392.
Crossref - Tantry BA, Mohammed AH, Shaik ,Tantry MN. Detection of Extended-Spectrum -lactamases Production by Escherichia coli: A Phenotypic Comparative Study. J Pure Appl Microbiol. 2018;12(4):2245-2252.
Crossref - Chowdhury AHMSK, Nandi R, Karim AA, et al. Comparison Between Phenotypic Confirmatory Test & Double Disc Synergy Test in Detection of Extended Spectrum b-Lactamases Producers Among Gram-Negative Bacilli. Chattagram Maa-O-Shishu Hospital Medical College Journal. 2016;15(2):3-8.
Crossref - Jacoby GA, Munoz-Price LS. The new b-lactamases. N Eng J Med. 2005;352(4):380-391.
Crossref - Livermore DM, Canton R, Gniadkowski M, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;52(2):165-174.
Crossref - Shahandeh Z, Sadighian F, Rekabpou KB. Phenotypic study of Extended-spectrum beta-lactamase, AmpC and Carbapenemase among E.coli clinical isolates in affiliated hospitals of Babol University of Medical Sciences. Int J Health Syst Disaster Manag. 2015;3(2):74-78.
Crossref - Metri BC, Jyothi P, Basavaraj VP. The Prevalence of ESBL among Enterobacteriaceae in a Tertiary Care Hospital of North Karnataka, India. J Clin Diagn Res. 2011;5(3):470-475.
- Al-JasserAM. Extended-Spectrum Beta-Lactamases (ESBLs): A Global Problem. Kuwait Medical Journal. 2006;38:171-185.
- Livermore DM. b-Lactamases in Laboratory and Clinical Resistance. Clin Microbiol Rev. 1995;8(4):557-584.
Crossref - EUCAST. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. 2017(2).
- Bradford PA. Extended-Spectrum b-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev. 2001;14(4):933-951.
Crossref - Carter MW, Oakton KJ, Warner M, Livermore DM. Detection of Extended-Spectrum b-Lactamases in Klebsiellae with the Oxoid Combination Disk Method. J Clin Microbiol. 2000;38(11):4228-4232.
Crossref - Fong I, Shlaes D, Drlica K, editors. Antimicrobial Resistance and Implications for the 21st Century. 2nd ed.: Springer Science & Business Media. 2007.
- Wormser GP, Tang YW. Antibiotics in Laboratory Medicine. In Wilkins VLPLW&, editor. Clin Infect Dis. 2005:577.
- Jarlier V, Nicolas M, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867-878.
Crossref - Legrand P, Fournier G, Bure A, et al. Detection of extended broad-spectrum beta-lactamases in Enterobacteriaceae in four French hospitals. Eur J Clin Microbiol Infect Dis. 1989;8(6):527-529.
Crossref - Bure A, Legrand P, Arlet G, Jarlier V, Paul G, Philippon A. Dissemination in five French hospitals of Klebsiella pneumoniae serotype K25 harbouring a new transferable enzymatic resistance to third generation cephalosporins and aztreonam. Eur J Clin Microbiol Infect Dis. 1988;7(6):780-782.
Crossref - Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of Extended-Spectrum b-Lactamases in Clinical Isolates of Enterobacter cloacae and Enterobactera erogenes. J Clin Microbiol. 2000;38(2):542-546.
Crossref - Kaur J, Chopra S, Sheevani, Mahajan G. Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella pneumoniae. J Clin Diagn Res. 2013;7(2):229-233.
Crossref - CLSI. Performance Standards for Antimicrobial Susciptibility Testing. In CLSI Supplement M100. 30th ed.: Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- Dalela G. Prevalence of Extended Spectrum Beta Lactamase (ESBL) Producers among Gram Negative Bacilli from Various Clinical Isolates in a Tertiary Care Hospital at Jhalawar, Rajasthan, India. J Clin Diagn Res. 2012;6(2):182-187.
- Cormican MG, Marshall SA, Jones RN. Detection of extended-spectrum beta-lactamase (ESBL)-producing strains by the Etest ESBL screen. J Clin Microbiol. 1996;34(8):1880-1884. doi: 10.1128/jcm.34.8.1880-1884.1996.
Crossref - Hall MAL-v, Fluit AC, Paauw A, Box ATA, Brisse S, Verhoef J. Evaluation of the Etest ESBL and the BD Phoenix, VITEK 1, and VITEK 2 Automated Instruments for Detection of Extended-Spectrum Beta-Lactamases in Multiresistant Escherichia coli and Klebsiella spp. J Clin Microbiol. 2002;40(10):3703-3711.
Crossref - Livermore DM, Warner M, Mushtaq S. Evaluation of the chromogenic Cica-b-Test for detecting extended-spectrum, AmpC and metallo-b-lactamases. J Antimicrob Chemother. 2007;60(6):1375-1379.
Crossref - Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum beta-lactamase production by Enterobacteriaceae. J Clin Microbiol. 2011;49(3):1048-1057.
Crossref - Thomson KS, Mejglo ZA, Pearce GN, Regan TJ. 3-Dimensional susceptibility testing of beta-lactam antibiotics. J Antimicrob Chemother. 1984;13(1):45-54.
Crossref - Menon T, Bindu D, Kumar CP, Nalini S, Thirunarayan MA. Comparison of double disc and three dimensional methods to screen for ESBL producers in a tertiary care hospital. Indian J Med Microbiol. 2006;24(2):117-120.
Crossref - Thomson KS, Sanders CC. Detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992;36(9):1877-1882.
Crossref - Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38(5):1791-1796.
Crossref - Shahid M, Malik A, Agrawal M, Singhal S. Phenotypic detection of extended-spectrum and AmpC beta-lactamases by a new spot-inoculation method and modified three-dimensional extract test: comparison with the conventional three-dimensional extract test. J Antimicrob Chemother. 2004;54(3):684-687.
Crossref - Sanders C, Barry A, Washington J, et al. Detection of Extended-Spectrum-b-Lactamase-Producing Members of the Family Enterobacteriaceae with the Vitek ESBL Test. J Clin Microbiol. 1996;34(12):2997-3001.
Crossref - Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14(1):90-103.
Crossref - Sanguinetti M, Posteraro B, Spanu T, et al. Characterization of clinical isolates of Enterobacteriaceae from Italy by the BD Phoenix extended-spectrum beta-lactamase detection method. J Clin Microbiol. 2003;41(4):1463-1468.
Crossref - Wiegand I, Geiss HK, Mack D, Sturenburg E, Seifert H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J Clin Microbiol. 2007;45(4):1167-1174.
Crossref - Sturenburg E, Lang M, Horstkotte MA, Laufs R, Mack D. Evaluation of the MicroScan ESBL plus confirmation panel for detection of extended-spectrum beta-lactamases in clinical isolates of oxyimino-cephalosporin-resistant Gram-negative bacteria. J Antimicrob Chemother. 2004;54(5):870-875.
Crossref - Thomson KS, Cornish NE, Hong SG, Hemrick K, Herdt C, Moland ES. Comparison of Phoenix and VITEK 2 extended-spectrum-beta-lactamase detection tests for analysis of Escherichia coli and Klebsiella isolates with well-characterized beta-lactamases. J Clin Microbiol. 2007;45(8):2380-2384.
Crossref - Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22(1):161-182.
Crossref - Manoharan A, Sugumar M, Kumar A, et al. Phenotypic & molecular characterization of AmpC b-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian Medical Centers. Indian J Med Res. 2012;135(3):359-364.
- Peter-Getzlaff S, Polsfuss S, Poledica M, et al. Detection of AmpC Beta-Lactamase in Escherichia coli: Comparison of Three Phenotypic Confirmation Assays and Genetic Analysis. J Clin Microbiol. 2011;49(8):2924-2932.
Crossref - Bush K, Macalintal C, Rasmussen BA, Lee VJ, Yang Y. Kinetic interactions of tazobactam with beta-lactamases from all major structural classes. Antimicrob Agents Chemother. 1993;37(4):851-858.
Crossref - Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;33(6):1211-1233.
Crossref - Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol. 2003;41(8):3542-3547.
Crossref - Aubert D, Girlich D, Naas T, Nagarajan S, Nordmann P. Functional and structural characterization of the genetic environment of an extended-spectrum beta-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob Agents Chemother. 2004;48(9):3284-3290.
Crossref - Tondi D, Calע S, Shoichet BK, Costi MP. Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors. Bioorg Med Chem Lett. 2010;20(11):3416-3419.
Crossref - Shoorashetty RM, Nagarathnamma T, Prathibha J. Comparison of the boronic acid disk potentiation test and cefepime-clavulanic acid method for the detection of ESBL among AmpC-producing Enterobacteriaceae. Indian J Med Microbiol. 2011;29(3):297-301.
Crossref - Walther-Rasmussen J, Hרiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60(3):470-482.
Crossref - Palzkill T. Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC b-lactamases. Front Mol Biosci. 2018;5(16).
Crossref - Liao Q, Xie Y, Wang C, et al. Development and evaluation of the method for detecting metallo-carbapenemases among carbapenemase-producing Enterobacteriaceae. J Microbiol Methods. 2019;163(105652).
Crossref - Wei Q, Sun J, Wang Z, Yan L, Zhang C, Xu X. Evaluation of Modified Rapid Carbapenem Inactivation Method (mrCIM) Combined with Rapid EDTA-Modified Carbapenem Inactivation Method (reCIM) to Detect Carbapenemase and Distinguish Metallo-Carbapenemase in Enterobacteriaceae Within Four Hours. Infect Drug Resist. 2020;23(13):1919-1927.
Crossref - Pournaras S, Poulou A, Tsakris A. Inhibitor-based methods for the detection of KPC carbapenemase producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J Antimicrob Chemother. 2010;65:1319-1321.
Crossref - Essack SY. Laboratory detection of extended-spectrum beta-lactamases (ESBLs)—the need for a reliable, reproducible method. Diagn Microbiol Infect Dis. 2000;37(4):293-295.
Crossref - Potz NA, Colman M, Warner M, Reynolds R, Livermore DM. False-positive extended-spectrum beta-lactamase tests for Klebsiella oxytoca strains hyperproducing K1 beta-lactamase. J Antimicrob Chemother. 2004;53(3):545-547.
Crossref - Livermore DM. Beta-lactamase-mediated resistance and opportunities for its control. J Antimicrob Chemother. 1998;41(Suppl):D25-D41.
Crossref - Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48(Suppl 1):59-64.
Crossref - Parajuli NP, Maharjan P, Joshi G, Khanal PR. Emerging Perils of Extended Spectrum -Lactamase Producing Enterobacteriaceae Clinical Isolates in a Teaching Hospital of Nepal. BioMed Res Int. 2016;3:1-7.
Crossref - MacKenzie FM, Miller CA, Gould IM. Comparison of screening methods for TEM- and SHV-derived extended-spectrum b-lactamase detection. Clin Microbiol Infect. 2002;8(11):715-724.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.