ISSN: 0973-7510
E-ISSN: 2581-690X
In recent years, an increase in Escherichia coli and Klebsiella pneumoniae resistant to carbapenem was reported globally. Due to their high prevalence and extensive range of medical conditions, Escherichia coli and Klebsiella pneumoniae are both confirmed to be major public health concerns. Furthermore, carbapenem resistance restricts treatment options for individuals infected with these bacteria. Consequently, early detection of carbapenem resistance is essential for starting effective therapy, achieving successful management, and avoiding the infection from spreading further in the future. This study’s objective was to identify the phenotypic and genotypic identification of Metallo-β-lactamases (MBL) in carbapenem-resistant E. coli and K. pneumoniae in advanced healthcare facilities. Meropenem resistance was tested in E. coli and K. pneumoniae using the Kirby-Bauer disc diffusion technique. MBL was discovered using a combination of Disc diffusion testing and the Modified Hodge Test. The Polymerase Chain Reaction was used to determine the genotypes of the bla NDM-1 genes that express these enzymes. Out of 427 strains, including 223 E. coli and 204 K. pneumoniae, 35 (8.2%) consisted of carbapenem-resistant, and 29 (82.85%) showed phenotypically verified as metallo-beta-lactamase producers by using the Combined disc test and 20 (57.14%) using the Modified Hodge test. Polymerase Chain Reaction tests for genes detect those three different strains all showed the bla NDM-1 gene. Carbapenemase production and MBL can be recognized with the help of phenotypic combination disc and MHT tests in labs. Since both tests showed 100% concordance, laboratories may use the less expensive CDT instead of the MHT. The current study supports institutional antibiotic stewardship programmes to manage antibiotic use and prevent CRE worldwide.
Carbapenem-resistant, Metallo-β-lactamase, Modified Hodge Test, bla NDM-1, Gene Detection
Escherichia coli and Klebsiella pneumoniae, which are intolerant to the antibiotic carbapenem, are becoming more common and spreading. This is considered a worldwide threat to health, especially in developing countries. Carbapenems are β-lactam Antibiotics with a wide range of effects that most β-lactamases can’t break down.1 Infectious diseases caused by multidrug-resistant organisms were ultimately treatable after their debut in 1980. Unfortunately, Enterobacteriaceae and other Gram-negative organisms’ developed resistance to carbapenems is now a global health crisis.2 Class A β-lactamases, such as K. pneumoniae carbapenemase (KPC) and Guiana extended-spectrum (GES), class D β-lactamase OXA-48, and Ambler class β-lactamases MBLs such as IMP, VIM, and NDM all contribute to this resistance via plasmid-mediated clavulanic acid inhibition.3 In the year 2000, the presence of K. pneumoniae carbapenemase (KPC) was first reported in the United States. Subsequently, it has been found in many other countries and has spread all over the world.4,5
New Delhi metallo-β-lactamase (NDM) was initially found in E. coli and K. pneumoniae strains derived through a Swedish sufferer who visited India.6 There are six typical methods for demonstrating MBL emergence: the Modified Hodge test (MHT), imipenem/ceftazidime-EDTA Double Disc Synergy test, the impregnated imipenem discs, and a decrease of at least four log units in MIC values when using the imipenem-EDTA combination.7
Using straightforward methods for detecting MBL development among Gram-negative bacilli clinical strains is vital. This research aimed to identify metallo-β-lactamases to carbapenem-resistant E. coli and K. pneumoniae by analyzing their phenotypes and genomes. These strains were obtained through numerous clinical specimens at highly developed healthcare facilities.
The current investigation was conducted between February 2021 to November 2022 by the Department of Microbiology at the Adesh Medical College and Hospital in Shahabad, Kurukshetra.
427 isolates of the family Enterobacteriaceae were identified in pus, blood, urine, CSF, and other body fluids collected from IPD and ICU patients to the Microbiology lab. Detection of the strains was accomplished using standard microbiological procedures. On Muller-Hinton agar plates with widely accessible discs (Hi-media), antimicrobial sensitivity was tested using the Kirby-Bauer disc diffusion procedures, and the outcomes were evaluated followed by CLSI guidelines.8,9
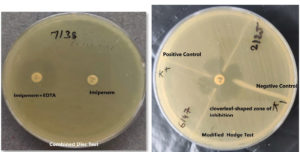
Imipenem-EDTA disc test in combination for phenotypic MBL production detection
All carbapenem-resistant strains were tested with imipenem-EDTA and appropriate positive and negative controls. Mueller Hinton agar plates were inoculated according to CLSI guidelines. On the MHA plate, a 10-mcg imipenem disc and a combination of 10-mcg imipenem-EDTA (10/750 mcg) discs were incubated at 37°C for 16-18 hours. The imipenem-EDTA disc enhanced the inhibition zone by 7 mm compared to the imipenem disc alone, indicating an MBL-positive strain.10
Modified Hodge test phenotypic detection MBL production
CLSI-recommended Modified Hodge testing was used on each strain.11 While an E. coli ATCC 25922 lawn culture was being performed at a 1:10 dilution on the Mueller Hinton’s agar plate, a disc containing ten mcg of Imipenem was placed in the middle of the test area. The test organisms were then placed in a straight line starting from the disc’s edge to the plate’s edge. Before being analyzed, plates were kept at 37°C for 24 hours. In a positive Modified Hodge test, the E. coli 25922 strain formed an indentation within the disc diffusion zone approximating a clover leaf-like structure, indicating the production of MBL. In a negative test, the Escherichia coli ATCC 25922 strain did not generate an indentation along the growth streak of the test organism within the disc diffusion zone12,13 as shown in Figure 1. Controls used in the test procedure were as follows.
- pneumoniae ATCC BAA-1705 MHT (Positive).
- pneumoniae ATCC BAA-1706 MHT (Negative).
Genotypic characterization of MBL production
Genotypic recognition was done using a marker-based method to identify the bla NDM gene at the Centre for Interdisciplinary and Biomedical Research, Adesh University, Bathinda.
DNA extraction
The DNA was extracted from bacterial strains using the Xploregen kit-based procedure. 1-2 colonies from an overnight growing culture were suspended in 100 µl ultrapure water and cooked at 90°C for twenty minutes. Five minutes were spent spinning the culture at 15,000 rpm to collect the liquid that rose to the surface. The pellets were stored at 20°C until they were needed. The manufacturer’s method was used to get the DNA out of the pellets. The DNA integrity was tested using nano-quant (260/280 ratio). The agarose gel was also used to test the DNA’s integrity.
Polymerase chain reaction and DNA sequencing
Primers with sequences 5’-GGTTTG GCGATCTGGTTTTC-3’ (NDM-Forward primer) and 5’ -CGGAATGGCTCATCACGATC-3’(NDM-Reverse primer) were used in a polymerase chain reaction to amplify the bla NDM-1, which resistance genes. We amplified under the circumstances prescribed by the kit’s (Xploregen kit) protocol, which called for a Polymerase Chain Reaction. The Ethidium Bromide loading reagent was added to the mixture of amplified PCR products and then electrophoresed at 100 volts for at least thirty-five minutes.
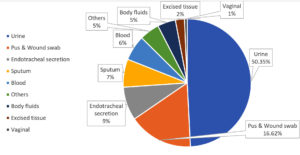
427 strains were discovered in diverse clinical specimens. As shown in Table, the most strain came from urine (215, or 50.35%), followed by pus and wound swabs (71, or 16.62%), ET secretion (37, or 8.66%), sputum (31, or 7.25%), and blood (28, or 6.55%) shown in Figure 2.
Table:
MBL-positive strains determined by phenotypic testing
Organism |
No. of isolates |
Carbapenem-resistant isolates |
MBL positive by Combined disc test |
MBL positive by Modified Hodge test |
|---|---|---|---|---|
E. coli |
223 |
12 (5.38%) |
9/12(75%0 |
5/12 (42%) |
K. pneumoniae |
204 |
23(11.27%) |
20/23 (87.1%) |
15/23 (65.2%) |
Total |
427 |
35 (8.2%) |
29/35 (82.8%) |
20/35 (57.2%) |
Carbapenem resistance was found in 35 of these 427 strains (223 E. coli and 204 K. pneumoniae). Out of 35 carbapenem-resistant strains, CDT identified a greater proportion of MBL producers than MHT producers. In contrast to the modified Hodge test, which found five E. coli and fifteen K. pneumoniae specimens obtaining MBL, the Combined disc test revealed nine E. coli and twenty K. pneumoniae strains as resulting MBL.
Analysis of genotypes (bla NDM-1)
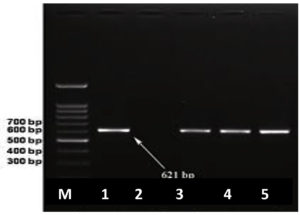
As confirmed by the detection of the bla NDM-1 gene in two K. pneumoniae and one E. coli, shown in Figure 3.
This investigation aimed to determine if MBL-producing E. coli and K. pneumoniae were obtained through various patient specimens. Carbapenem-resistant Escherichia coli (E. coli) and Klebsiella pneumoniae (KP) strains derived through numerous clinical specimens in advanced Health care facilities were resistant to carbapenem. This study discovered Carbapenem-resistant bacteria in 23 (11.27%) K. pneumoniae strains and 12 (5.38%) E. coli strains. In one E. coli and two K. pneumoniae specimens, the bla NDM-1 gene was discovered using a polymerase chain reaction.
There were 35 (8.19%) carbapenem-resistant strains out of 427.
Hedge et al. also reported 16% resistance towards Imipenem, concordant with this study.14 According to Agarwal et al., 14.27% of Enterobacteriaceae are resistant to the antibiotic meropenem.9 This value is comparable to Balan et al.15 (15%) and Behera et al.16 (19.50%) reported in this study.
35 out of 427 (8.19%) of the strains in our study made metallo-β-lactamases. Other studies also show a lower prevalence of MBL production. As an example, a study that was carried out by Kaur et al.2 revealed a prevalence of MBL of 6.4%, and a study by Deshmukh DG et al.17 reported a prevalence of MBL of 2.9%.
In a similar investigation, Meel et al. discovered that the prevalence of MBL producers 10 (8.84%) of Klebsiella pneumoniae.18
The polymerase chain reaction was used in the current work to amplify the bla NDM-1 gene. Only 8.33% (1/12) and 8.69% (2/23), respectively, of the 35 carbapenem-resistant specimens of K. pneumoniae and E. coli were determined. According to the investigation, K. pneumoniae and E. coli strains that produce carbapenem are responsible for 8% of all illnesses.
Similar results were reported by Bora et al. in 2016; 8.67% (19/219) of the K. pneumoniae strain tested positive for bla NDM-1.19
In a study by Mohasen et al., bla NDM 1 was determined in three strains of K. pneumoniae, but the carbapenemase gene was not discovered in any of the E. coli specimens.11 The three NDM-1-positive strains were susceptible to polymyxin-B, but almost every other drug tested, except carbapenems, did not work against them. In a study by Sahin et al., Metallo-β-lactamases were found with the MBL antibiotic gradient test. Two of the 43 strains had MBL enzyme, but only one of these two strains was bla NDM-1 positive.20
Thus, the phenotypic technique, the combined disc test and the MHT test can be used to identify carbapenemase production and MBL in laboratories. Since both tests showed complete concordance, laboratories may use the CDT instead of the MHT, which is expensive. When genotypic assays are unavailable, employ phenotypic ones. Drug-resistant microorganisms can be reduced by following antimicrobial stewardship programs and antibiotic policies, including carbapenem antimicrobials.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046-5054.
Crossref - Kaur J, Khanna P, Chand K. Phenotypic Detection of Carbapenem Resistance among Escherichia coli and Klebsiella Isolates Obtained from Various Clinical Samples. Int J Curr Microbiol Appl Sci. 2017;6(10):1566-1573.
Crossref - Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-458.
Crossref - Amira ME, Areej ME, Hemat KA, Ramadan HI, Heba IA. Phenotypic and genotypic detection of-lactams resistance in Klebsiella species from Egyptian hospitals revealed carbapenem resistance by OXA and NDM genes. Afr J Microbiol Res. 2016;10(10):339-347.
Crossref - Gupta V, Singh M, Datta P, Goel A, Singh S, Prasad K, Chander J. Detection of various beta-Lactamases in Escherichia coli and Klebsiella sp.: A study from Tertiary Care Centre of North India. Indian J Med Microbiol. 2020;38(3-4):3906.
Crossref - Collee JC, Fraser AG, Marmion BP, Simmons A. Mackie and MaCartney practical medical microbiology 14th edition, 131-140. Church hill. Livingstone, London. 2006.
- Fazlul MKK, Deepthi S, Farzana Y, Najnin A, Rashid Ma, Munira B, Srikumar C, Nazmul. Detection of Metallo-\b{eta}-Lactamases-Encoding Genes Among Clinical Isolates of Escherichia Coli in a Tertiary Care Hospital, Malaysia. arXiv preprint arXiv:1904.05198;2019.
Crossref - Kamel NA, El-Tayeb WN, El-Ansary MR, Mansour MT, Aboshanab KM. Phenotypic screening and molecular characterization of carbapenemase-producing Gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PloS one. 2018;13(8):e0202119.
Crossref - Agarwal N, Chauhan S, Singh P, Sharma VK. Phenotypic detection of carbapenem resistance and metallo-beta-lactamase among Enterobacteriaceae from clinical samples in tertiary care hospital. Int J Res Med Sci. 2019;7(6):2373-2376.
Crossref - Dortet L, Poirel L, Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother. 2012;56(12):6437-6440.
Crossref - Mohsen SM, Hamzah HA, Al-Deen Mustafa MI. Phenotypic and molecular study of carbapenem-resistant Enterobacteriaceae in a referral hospital in the East coast Malaysia. Int J Health Allied Sci. 2018;7(1):17-22.
Crossref - Srivastava P, Bisht D, Kumar A, Tripathi A. Prevalence of Carbapenem-Resistant Escherichia coli and Klebsiella pneumoniae in Rural Uttar Pradesh. Journal of Datta Meghe Institute of Medical Sciences University. 2022;17(3):584-588.
Crossref - Nagaraj S, Chandran SP, Shamanna P, Macaden R. Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in south India. Indian J Med Microbiol. 2012;30(1):93-95.
Crossref - Archana Hegde M, Tejashree A. Genotyping of Carbapenem-Resistant Klebsiella pneumoniae from Clinical Isolates. Int J Curr Microbiol Appl Sci. 2019;8(05):825-835.
Crossref - Balan K, Sireesha P, Setty CR. Study to detect incidence of carbapenemase among Gram-negative clinical isolates from tertiary care hospital. J Dent Med Sci. 2012;1(6):08-12.
Crossref - Behera B, Mathur P, Das A, Kapil A. Ertapenem susceptibility of extended spectrum beta-lactamase-producing Enterobacteriaceae at a tertiary care centre in India. Singapore Med J. 2009;50(6):628-632.
- Deshmukh DG, Damle AS, Bajaj JK, Bhakre JB, Patwardhan NS. Metallo-b-lactamase-producing clinical isolates from patients of a tertiary care hospital. J Lab Physicians. 2011;3(2):93-97.
Crossref - Meel P, Maheshwari M, Arora B, Malhotra V. Phenotypic detection of Metallo-b-lactamase mediated carbapenem resistance in gram-negative bacilli of family Enterobacteriaceae in a tertiary care hospital in north India. J Crit Rev. 2019;7(3):2020.
Crossref - Bora A, Ahmed G. Detection of NDM-1 in clinical isolates of Klebsiella pneumoniae from Northeast India. J Clin Diagn Res. 2012;6(5):794-800.
- Sahin K, Tekin A, Ozdas S, et al. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann Clin Microbiol Antimicrob. 2015;14(1):1-6.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.