ISSN: 0973-7510
E-ISSN: 2581-690X
As Jordan advances in an attempt to promote greywater reuse, it is important to investigate the composition of bacteria in these new sources. To evaluate the presence of enteric pathogens in greywater, a pilot study investigating enteric pathogens in household washing machines and kitchen sink effluents from residential premises was conducted. In the culture-dependent method, bacteria were identified after using Sanger sequencing of 16S rRNA. Bacteria in the phylum Proteobacteria have been found to be the most abundant phyla, which may indicate that they play an important environmental role and might be representative of adaptation to different environments. Klebsiella sp. and Pseudomonas sp. were the two major genera found in this study and accounted for 78.57% of the total isolates. This is the first investigation of enteric pathogens in household washing machines and kitchen sink effluents in Jordan. To my knowledge, no study has identified the microbial hazards associated with greywater reuse in Jordan yet. Additional research with more adequate methodology is needed to assist our findings.
Enteric Pathogens, Washing Machine, Kitchen Sink, Greywater, 16S rRNA, Klebsiella sp., Pseudomonas sp.
Jordan is one of the most water scarce countries of the world and greywater is the best choice for water reuse due to its large quantity and low concentration of contaminants.1 Greywater refers to wastewater produced from domestic activities such as washing machines, kitchen and wash basins, showering and bathing. It is generally defined as urban wastewater without wastewater from toilets.2 In Jordan, the automatic washing machine or dishwasher is found to consume about 50 liters in each operating case.3 Water resources per person in Jordan are far below the international water poverty line with less than 60 m3/year.4 Around 50-80% of an individual’s daily water consumption in Jordan are greywater,5,6 where washing machines account for a quarter of domestic wastewater. Thus, one of the recently undertaken strategies to enhance the water situation in Jordan is the reuse of greywater to bridge the gap and help in water balances. The reuse of greywater in Jordan is yet narrowed to rural areas where about 64% of houses are not connected to wastewater treatment plants7 therefore, can make the most use from greywater reuse for purposes of irrigation and toilet flushing.8 The current standard to control greywater reuse and reusing reclaimed greywater in Jordan (JS 1776/2013) permit the use of treated greywater for unrestricted irrigation.9 Furthermore, the climate change crisis is increasing variability in the water cycle; thus inducing reducing the predictability of water availability.10 Climate change drives increasing use of water-saving initiatives worldwide. Untreated greywater is allowed for local reuse as irrigation water in several rural areas in Jordan. Around 60 % of Amman’s households and 30% of rural in Jordan reused water within their household.11 Other countries only allow the reuse of treated greywater, while a few ban reusing it entirely due to public health concerns.12,13
As Jordan moves forward in an attempt to promote greywater reuse, it is important to investigate the composition of microbial communities and pathogens in these new sources. Microorganisms can be introduced into greywater by hand washing, baths, showers, laundry washing and food-handling in the kitchen.14,15 Contaminated food and mishandling of food in the kitchen have been recognized as sources of enteric pathogens in greywater.13,15 Enteric pathogens have also been detected in households laundry greywater during a microbial monitoring programme in Australia. The possible presence of pathogens in greywater can determine the ability to reuse it,16 which might be transmitted to humans by direct or indirect contact.15,17 Many studies reported the microbial contamination in home laundry operations and the risk of household microbial transmission.18-24 Previous studies showed that microorganisms isolated from washing machines are more tolerant to chemical surfactants when compared to control strains that covered the Gram-negative and Gram-positive bacteria in addition to the yeast.25,26 As a consequence of the self-developed matrix of extracellular polymeric substances (EPS), washing machines strains are more tolerant towards chemicals.25 Therefore, potential drug-resistant microorganisms could emerge in the domestic environment and lead to a potential health risk because of cross-contaminations throughout the washing process.21 The origins of greywater, the types of infrastructure and detergents used are all important environmental factors impacting greywater microbial communities.19 According to the Centers for Disease Control and Prevention (CDC), Gram-negative bacteria cause many infections including bloodstream infections, meningitis, pneumonia, and surgical or wound site infections in healthcare settings. However, more research is still required to determine the greywater microbial communities, including bacterial pathogens.
Little has been published on enteric pathogens in domestic washing machines and kitchen sinks effluents. Therefore, this pilot study offers a view into acquiring knowledge on the potential pathogens associated with untreated greywater reuse in Jordan through characterizing the bacterial isolates of the effluent water of the domestic washing machines and kitchen sink from residential premise using a culture-dependent method followed by Sanger sequencing of 16S rRNA gene sequences. Pathogens are generally characterized by culture-dependent methods. This study will be the first to investigate enteric pathogens in household washing machines and kitchen sink effluents in Jordan.
Sample collection and preparation
This pilot study is aimed to detect the enteric pathogens of a domestic washing machine and kitchen sink effluents that is generated at a residential premise in Aqaba City, Jordan in 2022. This study focused on the bacterial flows where the domestic washing machines and kitchen sink effluents delivered directly without treatment to storage tank through a piped distribution system. The washing machine was used twice a week. Greywater samples were collected from the storage tank during April of 2022 under aseptic condition. The tank was cleaned after each sampling to prepare for the next sampling. Samples that were collected using 1L sterile bottles were kept on ice and used within 1 hour after collection. Owner voluntarily and anonymously allowed us to use their washing machine and kitchen sink effluents. All members of the residential premises were aged between 28 and 44 years. The washing machine worked at 30°C. Washing machines were loaded with regular detergent without fabric softener (detergent name not provided by the residential premises). While kitchen sink was used on daily basis.
Microbial enumeration and identification
Bacterial numbers in greywater were estimated using surface plate techniques.27 The microorganisms from raw washing machine and kitchen sink effluents were collected by centrifugation for 10 minutes at 12,000 ×g using sterile technique. After centrifugation, the supernatant was discarded, and the cells were resuspended in nutrient broth and incubated overnight at 37°C. The broth was streaked using sterile glass spreader onto Xylose Lysine Desoxycholate agar (DIFCO, FRANCE) and incubated overnight at 37°C. XLD agar was used to enumerate enteric pathogens using the spread plate method.28 Colonies were identified based on morphological tests for initial identification that were further identified later by 16S rRNA gene sequencing. Therefore, for molecular identification, bacterial DNA was extracted using the Quick-DNA Fecal/Soil Microbe Miniprep Kit (Zymo Research /USA) following the manufacturer’s instructions. The integrity and size of genomic DNA was checked using gel electrophoresis (1% agarose), and DNA concentrations were determined using microplate spectrophotometer system (BioTek Instruments, Inc.).
The molecular-based works were carried out for microbial diversity of 16 extracts. The 16S rRNA gene for each sample was amplified using universal 16S rRNA bacterial primers 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CTACGGCTACCTTGTTACGA-3′).29 Polymerase chain reaction (PCR) mixture (25 µl) contained 2 µl of DNA template, 2U Taq DNA Polymerase, 1x PCR buffer, 1 mM Nucleotide mix, 2 mM MgCl2 and 0.2 uM of each primer. The PCR was carried out using the following protocol: one cycle at 94°C for 5 min, 30 cycles at 93°C for 1 min, 54°C for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 5 min. All PCR products were analyzed using gel electrophoresis (1% agarose).
Sequencing of amplified PCR products
A total of 16 random bacterial isolates were selected for this study and sequenced by Sanger sequencing service from Macrogen (Macrogen Inc., Korea). Sequences were edited by removing the bad quality parts from the beginning and the end of the sequences. Good quality sequences generated (n=14) were used for similarity search using BLASTn tool at the National Center for Biotechnology Information (NCBI).30 BLASTn at NCBI was used to find similarities at the species level for this study, whereas SILVA can only go down to the genus level.31 The 16S rRNA gene sequences of bacteria isolated in this study are uploaded in GenBank under the accession numbers (OP514804-OP514814) and (OP514817-OP514819).
The colonies were counted after incubation and the total count was determined as described by Speck32 by the following equation:
cfu/ml= number of colonies × D.F/plated volume in ml
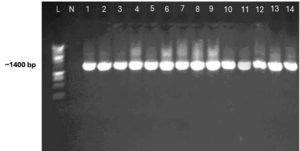
The mean total count of bacteria in greywater was 6.1×106 cfu/ml. Fourteen bacterial isolates from the storage tank of domestic washing machine and kitchen sink effluents were successfully sequenced and analyzed using a 16S rRNA gene PCR (Figure) followed by Sanger sequencing. NCBI BLASTN33 was used to find similar sequences. Identification were determined for 14 isolates identified on species level. Based on the sequencing analysis, all sequences from the storage tank of domestic washing machine and kitchen sink effluents were identified and classified as belonging to the following four genera: Klebsiella, Citrobacter, Proteus and Pseudomonas with identity percentage ranged from 95.32 to 99.93 (Table 1). The genus Klebsiella was predominant (Table 2). Identification at the species level was possible for all samples as they have a very close identity with sequences available on Genbank (Table 2).
Table (1):
BLAST analysis of PCR amplicons from bacterial isolates according the highest similarity to 16S rRNA sequences in the GenBank nucleotide sequence database
Sponge IDs |
Closet organism in GenBank |
NCBI accession No. |
Similarity (%) |
E-value |
GenBank accession No.1 |
|---|---|---|---|---|---|
GW1 |
Klebsiella grimontii |
CP091752.1 |
99.51% |
0.00 |
OP514804.1 |
GW2 |
Klebsiella oxytoca |
MT509911.1 |
99.50% |
0.00 |
OP514805.1 |
GW3 |
Klebsiella oxytoca |
MT509877.1 |
99.51% |
0.00 |
OP514806.1 |
GW4 |
Citrobacter freundii |
OQ405468.1 |
95.32% |
0.00 |
OP514807.1 |
GW5 |
Citrobacter murliniae |
NR_028688.1 |
97.54% |
0.00 |
OP514808.1 |
GW6 |
Klebsiella grimontii |
CP091752.1 |
99.58% |
0.00 |
OP514809.1 |
GW7 |
Pseudomonas aeruginosa |
FJ823152.1 |
99.93% |
0.00 |
OP514810.1 |
GW8 |
Pseudomonas aeruginosa |
KY962357.1 |
99.65% |
0.00 |
OP514811.1 |
GW9 |
Proteus vulgaris |
MN833577.1 |
99.51% |
0.00 |
OP514812.1 |
GW10 |
Klebsiella pneumoniae |
MF455200.1 |
99.36% |
0.00 |
OP514813.1 |
GW11 |
Klebsiella pneumoniae |
MZ389267.1 |
99.36% |
0.00 |
OP514814.1 |
GW12 |
Pseudomonas aeruginosa |
KC456535.1 |
96.06% |
0.00 |
OP514817.1 |
GW13 |
Pseudomonas aeruginosa |
HQ844502.1 |
98.73% |
0.00 |
OP514818.1 |
GW14 |
Klebsiella grimontii |
CP091752.1 |
99.58% |
0.00 |
OP514819.1 |
1GenBank accession number of all samples used in this study.
Table (2):
Number of bacterial isolates obtained from residential washing machine and kitchen sink storage tank and identified by PCR using amplification of 16S rRNA (Species level; n = 14) across the 14 different samples
| Phylum | Class | Order | Family | Genus | Species | GenBank accession number |
|---|---|---|---|---|---|---|
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Klebsiella | Klebsiella grimontii | OP514804.1, OP514809.1, OP514819.1 |
| Klebsiella oxytoca | OP514805.1, OP514806.1 | |||||
| Klebsiella pneumoniae | OP514813.1, OP514814.1 | |||||
| Citrobacter | Citrobacter freundii | OP514807.1 | ||||
| Citrobacter murliniae | OP514808.1 | |||||
| Morganellaceae | Proteus | Proteus vulgaris | OP514812.1 | |||
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | OP514810.1, OP514811.1, OP514817.1, OP514818.1 |
Applying Sanger sequencing on the full-length bacterial 16S rRNA gene of 14 isolates obtained in this study showed a clear assignment of the full-length bacterial 16S rRNA gene sequences to assign taxonomic classification down to the species level.31 Taxonomic identification of sequenced isolates was carried out by using BLAST to align sequences to the NCBI 16S BLAST database. Resulting hits in the present study were ranked by e-value, then the taxonomy of the highest scoring sequence was selected. Using the universal primers 27F and 1492R for a full-length bacterial 16S rRNA gene,34 from 14 isolates obtained in this study with an expected amplicon of ~ 1400 bp, bacteria in the phylum Proteobacteria have been found to be the most prevalent on the items laundered. Previous studies of microbial communities in washing machines have also revealed that washing machines are mostly inhabited by the phyla Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes.20,21,35 In congruence with previous molecular studies,20,35 Proteobacteria (93.75%), and Firmicutes (6.25%) also presented the most abundant phyla here.
The aim of this study was to evaluate the presence of enteric pathogens in greywater by initiating a pilot study of enteric pathogens in the effluents of domestic washing machines and kitchen sinks using the culture-dependent approach followed by using Sanger sequencing of 16S rRNA gene of all bacterial isolates. Many of the bacterial identified represent environmental bacteria, which typically found everywhere in the environment such as K. pneumoniae species.36 K. pneumoniae have been previously isolated from washing machines.37-39 K. pneumoniae is one of the species that most often involved in human infections, and frequently found in the large intestine but are also present in soil and water.40 K. pneumoniae is most pathogenic to humans among all Klebsiella sp., followed by K. oxytoca, where both K. pneumoniae and K. oxytoca cause community-acquired meningitis and brain abscesses.41 Citrobacter sp. occur in the environment and in the human colon, and can cause sepsis in immunocompromised patients. Citrobacter is predominantly spread from person-to-person,42 it normally cause urinary tract infections, intra abdominal sepsis, blood stream infections, pneumonia, and brain abscesses.43 C. freundii, can be found in feces and the environment as well as in drinking water containing relatively high concentrations of nutrients.44 In addition, some of the bacterial identified in this study are common biofilm formers, such as Pseudomonas sp., P. aeruginosa was found inside the detergent drawer.35 The major route of exposure to P. aeruginosa is direct contact with contaminated water or skin exposure.45 P. aeruginosa can also colonize on open burn wounds, causing infections, abscesses, and sepsis.46 K. oxytoca was one of the opportunistic pathogens found in the washing machines as well.37 Proteus sp., and Klebsiella sp. usually originating from food ingredients are commonly present on kitchen sponges to surfaces of stainless steel and polyethylene.47,48 Proteus sp. are commonly associated with complicated urinary tract infections (UTIs), also survive well within the environment in water, soil, and sewage.49 Where Proteus can cause uncomplicated cystitis, prostatitis, and pyelonephritis, especially in hospital-acquired cases.50
It should be noted that our study has limitations, which were expected as a pilot study. One limitation of this study was the population size (e.g., single household) and the small number of isolates that were sequenced which may have led to the identification of only some bacterial pathogens or non-pathogenic microflora of a domestic washing machine and kitchen sink effluents. It is anticipated that more samples collected and sequencing of more isolates could potentially reveal additional knowledge on the enteric pathogens might be present. In addition, due to the use of selective medium in this study, other groups of bacteria that may be present in the sample has not been shown. Related to the small sample size, we suggest that this study should be regarded more as a feasibility study of enteric pathogens in the effluents of domestic washing machines and kitchen sinks in Jordan. Another limitation of this study is that data generated by the current analysis does not include bacterial communities growth or decay during storage of the samples post-collection, as weekly samples for only one month were considered, and it may not reflect the actual contamination that may persist in greywater storage systems. However, this is more of a preliminary study, and additional research with more adequate methodology is needed to assist our findings. This is important since little has been known and published about bacterial pathogens in greywater in Jordan, and in domestic washing machines and kitchen sink effluents in particular. To my knowledge, no study has identified the microbial hazards associated with greywater reuse in Jordan yet. Overall, the culture-dependent method can address some limitations of the possibility of interaction, and the information obtained as only certain bacteria can be isolated using culture-dependent approach. Thus, using both culture-dependent and culture-independent methods for the identification of enteric pathogens will offers wider possibilities. However, it must be taken into account that the enteric pathogens of greywater in general would vary according to many factors including the background microflora, inhibitory substances, and non-culturable cells that could be present and must be taken into consideration for further future research.
This study demonstrated the presence of enteric pathogens in the untreated greywater from residential premises. The bacteria in the phylum Proteobacteria have been found to be the most abundant phyla in this study, which may indicate that they play an important environmental role and might be representative of adaptation to different environments. The search for identification of the enteric pathogens in domestic washing machines and kitchen sink effluents as described in this current study will be an important aspect considering the health risk assessment and the potential microbial hazards associated with greywater reuse in Jordan. This is the first study in Jordan to investigate enteric pathogens in household washing machines and kitchen sink effluents.
ACKNOWLEDGMENTS
The author would like to thank the Molecular Biology Laboratory, Marine Science Station, University of Jordan-Aqaba, for laboratory assistance.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by the author.
- DeOreo WB, Mayer PW, Dziegielewski B, Kiefer J. Residential End Uses of Water. Version 2: Executive Report. Water Research Foundation. 2016. https://www.circleofblue.org/wp-content/uploads/2016/04/WRF_REU2016.pdf. Accessed November 9, 2022.
- De Gisi S, Casella P, Notarnicola M, Farina, R. Grey water in buildings: a mini review of guidelines, technologies and case studies. Civ Eng Environ Syst. 2016;33:35-54.
Crossref - Al-Zoubi M, El-Amad W, Sultan N, et al. Sanitation for Millions – WASH in Islam – Guide on Water, Sanitation and Hygiene (WASH) from an Islamic Perspective. 2020. Accessed November 9, 2022.
- Deutsche Gesellschaft fur Technische Zusammenarbeit (GTZ). Managing water scarcity in Jordan. 2021. Accessed November 9, 2022.
- Ammari TG, Al-Zu’bi Y, Al-Balawneh A, et al. An evaluation of the re circulated vertical flow bioreactor to recycle rural greywater for irrigation under arid Mediterranean bioclimate. Ecol Eng. 2014;70:16-24.
Crossref - Albalawneh A, Chang TK. Review of the greywater and proposed greywater recycling scheme for agricultural irrigation reuses. Int J Res Granthaalayah. 2015;3(12):16-35.
Crossref - Ulimat AA. Wastewater Production, Treatment, and Use in Jordan. In Second Regional Workshop ‘Safe Use of Wastewater in Agriculture’. New Delhi, India. 2012. https://www.ais.unwater.org/ais/pluginfile.php/356/mod_page/content/111/
Jordan_ULIMAT_ICID_AHMA_2012-1.pdf. Accessed February 18, 2023. - Al Arni S, Elwaheidi M, Salih A, Ghernaout D, Matouq M. Greywater reuse: an assessment of the Jordanian experience in rural communities. Water Sci Technol. 2022;85(6):1952-1963.
Crossref - JISM. Water-Reclaimed Grey Water (JS 1776:2013); Jordan Standards and Metrology Organization: Amman, Jordan. 2013:1-12.
- UN-Water. Climate Change and Water – UN-Water Policy Brief. Geneva: United Nations for Water. 2019. https://www.unwater.org/sites/default/files/app/uploads/2019/10/UN_Water
_PolicyBrief_ClimateChange_Water.pdf. Accessed November 9, 2022. - Iskandarani M. Analysis of Demand, Access and Usage of Water by Poor Households in Jordan. Bonn: Center for Development Research (ZEF). 1999.
- Finley S, Barrington S, Lyew D. Reuse of domestic greywater for the irrigation of food crops. Water Air Soil Pollut. 2009;199:235-245.
Crossref - Maimon A, Tal A, Friedler E, Gross A. Safe on-site reuse of greywater for irrigation – a critical review of current guidelines. Environ Sci Technol. 2010;44(9):3213-3220.
Crossref - Eriksson E, Auffarth K, Henze M, Ledin A. Characteristics of grey wastewater. Urban Water J. 2002;4(1):85-104.
Crossref - Ottoson J, Stenstrom TA. Faecal Contamination of Greywater and Associated Microbial Risks. Water Res. 2003;37(3):645-655.
Crossref - Benami M, Herzberg M, Gillor O, Gross A, Vonshak A, Orlofsky E. Assessment of pathogenic bacteria in graywater systems and irrigated soils. Sci Total Environ. 2013;458-460:298-302.
Crossref - Toze S. Reuse of effluent water-benefits and risks. Agric Water Manag. 2006;80(1-3):147-159.
Crossref - Babic MN, Gostincar C, Gunde-Cimerman N. Microorganisms populating the water-related indoor biome. Appl Microbiol Biotechnol. 2020;104(15):6443-6462.
Crossref - Nagarkar M, Keely SP, Brinkman NE, Garland JL. Human- and infrastructure-associated bacteria in greywater. J Appl Microbiol. 2021;131(15):2178-2192.
Crossref - Callewaert C, Van Nevel S, Kerckhof FM, Granitsiotis MS, Boon N. Bacterial Exchange in Household Washing Machines. Front Microbiol. 2015;6:1381.
Crossref - Nix ID, Frontzek A, Bockmühl DP. Characterization of Microbial Communities in Household Washing Machines. Tenside Surfact Det. 2015;5(6):432-440.
Crossref - Belkin NL. Home laundering of soiled surgical scrubs: surgical site infections and the home environment. Am J Infect Control. 2001;29(1):58-64.
Crossref - Kagan LJ, Aiello AE, Larson E. The role of the home environment in the transmission of infectious diseases. J Community Health. 2002;27(4):247-267.
Crossref - Larson E, Duarte CG. Home hygiene practices and infectious disease symptoms among household members. Public Health Nurs. 2001;18(2):116-127.
Crossref - Gattlen J, Amberg C, Zinn M, Mauclaire L. Biofilms isolated from washing machines from three continents and their tolerance to a standard detergent. Biofouling. 2010;26(8):873-882.
Crossref - Gattlen J, Zinn M, Guimond S, Korner E, Amberg C, Mauclaire L. Biofilm formation by the yeast Rhodotorula mucilaginosa: process, repeatability and cell attachment in a continuous biofilm reactor. Biofouling. 2011;27(9):979-991.
Crossref - APHA. Standard Methods for the Examination of Water and Wastewater. 20th ed. American Public Health Association, American Water Works Association and Water Environmental Federation, Washington DC; 1998.
- Friedler E, Gilboa Y. Performance of UV disinfection and the microbial quality of greywater effluent along a reuse system for toilet flushing. Sci Total Environ. 2010;408(9):2109-2117.
Crossref - Chadwick P, Delisle GJ, Byer M. Biochemical identification of hospital enterobacteria by replica agar plating. Can J Microbiol. 1974;20(12):1653-1664.
Crossref - Kuo J, Yang YT, Lu MC, Wong TY, Sung PJ, Huang YS. Antimicrobial activity and diversity of bacteria associated with Taiwanese marine sponge Theonella swinhoei. Ann Microbiol. 2019;69(3):253-265.
Crossref - Balvociute M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT – how do these taxonomies compare? BMC Genomics. 2017;18 (Suppl 2):114.
Crossref - Speck ML. Compendium of Methods for the Microbiological Examination of Foods. 2nd ed. Washington (D.C.): American public health association; 1984. https://lib.ugent.be/catalog/rug01:001211748
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-3402.
Crossref - Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63(8):3233-3241.
Crossref - Jacksch S, Kaiser D, Weis S, et al. Influence of Sampling Site and other Environmental Factors on the Bacterial Community Composition of Domestic Washing Machines. Microorganisms. 2019;8(1):30.
Crossref - Cabral JP. Water microbiology. Bacterial pathogens and water. Int J Environ Res Public Health. 2010;7(10):3657-3703.
Crossref - Schmithausen RM, Sib E, Exner M, et al. The Washing Machine as a Reservoir for Transmission of Extended-Spectrum-Beta-Lactamase (CTX-M-15)-Producing Klebsiella oxytoca ST201 to Newborns. Appl Environ Microbiol. 2019;85(22):e01435-19.
Crossref - Boonstra MB, Spijkerman D, Voor In’t Holt AF, et al. An outbreak of ST307 extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in a rehabilitation center: An unusual source and route of transmission. Infect. Control Hosp Epidemiol. 2020;41(1):31-36.
Crossref - Jacksch S, Zohra H, Weide M, Schnell S, Egert M. Cultivation-Based Quantification and Identification of Bacteria at Two Hygienic Key Sides of Domestic Washing Machines. Microorganisms. 2021;9(5):905.
Crossref - Levinson W. Review of Medical Microbiology and Immunology. 9th Ed. Lang medical books, McGraw-Hill. 2006.
- Janda JM, Abbott SL. The Genera Klebsiella and Raoultella. The Enterobacteria. 2nd Ed.Washington, USA: ASM Press. 2006:115-129.
- Doran TI. The role of Citrobacter in clinical disease of children: Review. Clin Infect Dis. 1999;28(2):384-394.
Crossref - Pepperell C, Kus JV, Gardam MA, Humar A, Burrows LL. Low-Virulence Citrobacter Species Encode Resistance to Multiple Antimicrobials. Antimicrob. Agents Chemother. 2002;46(11):3555-3560.
Crossref - World Health Organization. WHO. Guidelines for Drinking water Quality. Addendum: Microbiological agents in drinking water. World Health Organization, Geneva. 2001.
- Mena KD, Gerba CP. Risk assessment of Pseudomonas aeruginosa in water. Rev Environ Contam Toxicol. 2009;201:71-115.
Crossref - Pruitt BA, Jr McManus AT, Kim SH, Goodwin CW. Burn wound infections: current status. World J Surg. 1998;22(2):135-145.
Crossref - Rossi EM, Scapin D, Tondo EC. Survival and transfer of microorganisms from kitchen sponges to surfaces of stainless steel and polyethylene. J Infect Dev Ctries. 2013;7(3):229-234.
Crossref - Cardinale M, Kaiser D, Lueders T, Schnell S, Egert M. Microbiome analysis and confocal microscopy of used kitchen sponges reveal massive colonization by Acinetobacter, Moraxella and Chryseobacterium species. Sci Rep. 2017;7(1):5791.
Crossref - Abbott SL. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and Other Enterobacteriaceae. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, ed. Manual of Clinical Microbiology. 9th ed. Washington, USA: ASM Press; 2007:698-711.
Crossref - O’Hara CM, Brenner FW, Miller JM. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev. 2000;13(4):534-546.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.