ISSN: 0973-7510
E-ISSN: 2581-690X
The Ganoderma lucidum immunomodulatory protein (FIP-glu) holds promise as a potential anti-tumor therapeutic agent. Enhancing the expression and stability of the engineered yeast strain is crucial for its industrial viability. This study focused on optimizing the fermentation parameters to produce an N-glycosylated protein, designated T36N, using the engineered Pichia pastoris GS115 strain with a mutant gene FIP-glu_T36N, a FIP-glu variant with an N-linked glycan modification. The response surface methodology (RSM) was employed to investigate the fermentation parameters that influence the yield of the N-glycosylated protein. Initially, the impact of fermentation duration on the optical density (OD600) and protein yield of P. pastoris GS115 were assessed, leading to the determination of a 96-hour fermentation period as optimal. Subsequently, the interplay among fermentation temperature, initial pH, and methanol concentration on protein yield was explored. The RSM analysis revealed the most favorable conditions for fermentation: a temperature of 26 °C, an initial pH of 6.5, and a methanol concentration of 1.00%. Verification tests confirmed that under these optimized conditions, the T36N yield peaked at 346.64 ± 3.47 mg.L-1, marking a 32.9% increase. The results underscored the capability of achieving high yields of T36N through the optimization of P. pastoris GS115 fermentation conditions. This research laid the groundwork for the industrial-scale production of N-glycosylated immunomodulatory proteins, offering a platform for the development of novel therapeutic agents.
N-glycosylated, Fungal Immunomodulatory Protein, Fermentation Conditions, Optimization
Fungal immunomodulatory proteins (FIPs) are a class of small proteins that exhibit immunomodulatory activity like plant hemagglutinins and immunoglobulins. These proteins are primarily derived from macrofungi that are both edible and medicinal.1,2 To date, over 30 types of FIPs have been isolated and identified, with the majority originating from the Ganoderma genus.3 These proteins have been shown to possess a range of biological activities, including anti-cancer, immunoregulation, and anti-allergy properties.4
Glycosylation is a crucial post-translational modification of proteins that plays a significant role in various biological processes, such as cell proliferation, differentiation, immune inflammation, and protein stability.5,6 Glycosylated proteins have a stable structure and hold potential for industrial applications.7-9 However, the development and application of FIPs in disease prevention and treatment have been limited, and there is a lack of research on glycoprotein production.
FIP-glu, first isolated from G. lucidum,10,11 was one fungal immunomodulatory protein. With the advancement of biotechnology, extensive studies have been conducted on the production of recombinant FIPs (r-FIP) using various microorganisms through recombination technology.12-15 These studies have laid a solid foundation for the industrial application of FIPs and their potential clinical use.14 Yeasts, in particular, have been widely used in the industrial production of medicinal proteins due to their numerous advantages.16,17
Fermentation conditions, such as temperature, pH and fermentation time significantly impact the yield of fermentation products. For engineered yeast, the inducer is also a critical factor for protein expression. Response surface methodology (RSM) is an effective technique for identifying the optimal combination of variables to produce the desired outcome in experimental design.18,19
In a previous study, based on bioinformatics analysis, we have successfully constructed an N-glycosylated mutant FIP-glu_T36N gene by mutating the threonine (Thr, T, T36) in the FIP-glu sequence to the asparagine (Asn, N), resulting in the production of the fungal immunomodulatory protein FIP-glu_T36N (referred to as T36N), which demonstrated enhanced anti-inflammatory activity in vitro.20 In the current study, RSM was employed to optimize the fermentation conditions of the engineered P. pastoris GS115/FIP-glu_T36N (referred to as P. pastoris GS115) to improve the yield of T36N. The goal of this study was to provide a reference for the industrial production of glycosylated proteins.
Strain preservation and media preparation
The genetically modified P. pastoris GS115/FIP-glu_T36N strain was stored at -80 °C within the Plant Biotechnology Research Center at Shanghai Jiao Tong University’s School of Agriculture and Biology. A Bicinchoninic acid (BCA) protein quantification kit was sourced from Sangon Biotech (C503021, Shanghai, China). The Buffered Glycerol-complex Medium (B540130, BMGY) was formulated with 10 g.L-1 yeast extract, 20 g.L-1 peptone, 10 mL.L-1 glycerin, 13.4 g.L-1 Yeast nitrogen base, potassium phosphate buffer at pH 6.0, and 0.4 mg.L-1 biotin. The Buffered Methanol-complex Medium (BMMY) included 10 g.L-1 yeast extract, 20 g.L-1 peptone, 13.4 g.L-1 yeast nitrogen base, potassium phosphate buffer at pH 6.0, 0.4 mg.L-1 biotin, and 5 mL.L-1 methanol, all procured from Sangon Biotech (B540131, Shanghai, China). Sterilization of all media was conducted at 121°C for 20 min.
Fermentation time impact on P. pastoris GS115 growth and protein production
P. pastoris GS115 was cultivated in BMGY until the optical density (OD600) value reached between 2.0 and 6.0. Subsequently, the culture was transferred into 250 mL Erlenmeyer flasks containing 60 mL of BMMY, with the OD600 adjusted to 1.0, and incubated at 28 °C with agitation at 220 rpm. Methanol at a concentration of 1.00% (v/v) was added daily for a period of 120 hours. To assess the impact of fermentation duration on the growth of P. pastoris GS115 and protein yield, the OD600 was measured every 24 hours using a microplate reader (BioTek, Winoosik, VT, USA). Post-centrifugation at 4000 × g for 20 min, the supernatant was collected for protein quantification using the BCA assay kit, following the manufacturer’s protocol.
Fermentation condition optimization via RSM
Recognized for its high protein production capabilities, P. pastoris requires fermentation condition optimization to address strain-specific and product-related challenges such as temperature, methanol consumption, and initial pH.21 RSM was utilized to identify optimal conditions for protein yield in P. pastoris GS115 fermentation, following the Box-Behken central composite design. Based on preliminary findings regarding the influence of methanol concentration, temperature, and pH on protein yield (data not presented), the independent variables and their levels are detailed in Table 1. A total of 17 randomized trials were executed using Design-Expert 13.0 software (Stat-Ease Inc., Minnesota, USA), examining the interactive effects of temperature (X1), initial pH (X2), and methanol concentration (X3) on the protein yield outcome (Table 2).
Table (1):
Factors and levels of Box-Behken central design
| Independent variables | Coded variables Levels | ||
|---|---|---|---|
| -1 | 0 | 1 | |
| Temperature (℃), X1 | 26 | 28 | 30 |
| Initial pH, X2 | 5.5 | 6.0 | 6.5 |
| Methanol content (%), X3 | 0.75 | 1.00 | 1.25 |
Table (2):
Response surface methodology experimental design and results
| Run | Independent variables | Protein yield (Y), mg.L-1 | |||
|---|---|---|---|---|---|
| Temperature (℃) X1 | Initial pH X2 | Methanol content (%) X3 | Experimental | Predicted | |
| 1 | -1 | 0 | 1 | 268.16 | 266.07 |
| 2 | 0 | 0 | -1 | 250.42 | 257.43 |
| 3 | 0 | 0 | 0 | 261.84 | 260.85 |
| 4 | 0 | 0 | 0 | 262.43 | 260.85 |
| 5 | 0 | 0 | 0 | 259.97 | 260.85 |
| 6 | 0 | -1 | 1 | 285.08 | 285.85 |
| 7 | -1 | -1 | 0 | 322.53 | 323.85 |
| 8 | 0 | 1 | -1 | 289.16 | 288.39 |
| 9 | 0 | 1 | 1 | 292.15 | 300.48 |
| 10 | 0 | -1 | -1 | 287.06 | 278.73 |
| 11 | 0 | 0 | 0 | 263.04 | 260.85 |
| 12 | 1 | -1 | 0 | 310.93 | 317.17 |
| 13 | -1 | 1 | 0 | 352.37 | 346.13 |
| 14 | 1 | 0 | 1 | 257.25 | 250.24 |
| 15 | 1 | 0 | -1 | 237.57 | 239.66 |
| 16 | 1 | 1 | 0 | 320.51 | 319.19 |
| 17 | 0 | 0 | 0 | 256.98 | 260.85 |
Model and peak validation
To ascertain the accuracy of the RSM model, five combinations were selected for validation with peak values confirmed under the optimal fermentation conditions. Protein yield was then determined under these conditions with five replicates.
Statistical analysis
Experiments were conducted in triplicate, with results expressed as mean ± standard deviation (SD). Analysis of variance (ANOVA) was applied to the independent variables to determine significance levels. Significance was denoted as ns (not significant), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001), and **** (P ≤ 0.0001).
Influence of fermentation time on P. pastoris GS115 growth and protein output
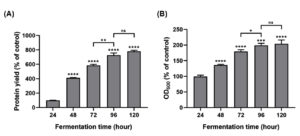
The optical density at 600 nm (OD600) and the protein yield served as indicators to evaluate the impact of fermentation duration on the growth of P. pastoris GS115 and its protein production. The findings indicated a positive correlation between fermentation time and protein yield, with a plateau observed after 96 hours of fermentation. A significant increase was noted when comparing the 96-hour mark to the 72-hour mark (Figure 1; P ≤ 0.0001). At 96 and 120 hours, the protein yield was 726% and 780% higher than at 24 hours, respectively, with no significant difference between these two time points (Figure 1A). This suggested that the engineered strain achieved a stable protein yield by the 96-hour fermentation mark. Secreted protein is linked to the cell density, as mirrored by the OD600 values, which is indicative of P. pastoris GS115 growth. Compared to the 24 hours, the OD600 values were 176.33% and 195.61% higher after 72 and 96 hours of fermentation, respectively, with a significant difference between these two time points (Figure 1B; P ≤ 0.05). However, at 120 hours, the OD600 value reached 202.15%, with no significant difference observed between the 96 hour and 120 hour marks. Mao et al.22 found that the concentration of rFIP-glu expressed by P. pastoris GS115 increased from 2-5 days and the OD600 value increased from 3-5 days. Externally expressed protein might vary due to different strains and incubation time.23 Appropriate incubation time was one of the important factors for obtaining high proteins expression.
Figure 1. Effects of fermentation time on the protein concentration and the growth of engineered Pichia pastoris GS115. Data was expressed as mean ± SD (n = 3). ns (not significant), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001), and **** (P ≤ 0.0001)
Optimization of fermentation conditions via RSM
Employing the Box-Behnken design principle, a three-factor, three-level RSM experiment was devised, encompassing 17 test points. These tests were categorized to assess experimental error, with fermentation temperature, initial pH, and methanol concentration as independent variables, and protein yield as the dependent response. The experimental and model-predicted values from the response surface analysis are detailed in Table 2. Design-Expert 13.0 software was utilized to fit the experimental data through a quadratic polynomial regression model. The relationship between the response value Y (protein yield) and the independent variables could be described by the following equation:
Y = 260.85 – 8.40 X1 + 6.07 X2 + 4.80 X3 – 5.07 X1X2 – 0.49 X1X3 – 1.24 X2X3 + 15.36 X12 + 50.37 X22 – 22.86 X32
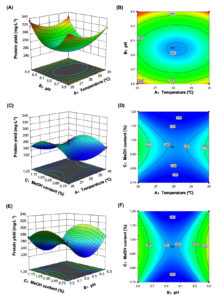
In this equation, Y, X1, X2, and X3 represented the predicted T36N yield, temperature, initial pH, and methanol content, respectively. The effects of independent variables and their interaction on the production yield of T36N were analyzed and the results are shown in Figure 2. Further analysis found that the higher the initial pH tending to 6.5 and the lower the temperature tending to 26 °C, accordingly resulted in the 3D surface higher (Figure 2A) and the contoured lines denser (Figure 2B). At this point, the yield of T36N revealed a greater increase. The initial pH played a relatively dominant role in the interaction between temperature and initial pH, but it was not obvious. On the other hand, at the level of interaction between temperature and methanol content, when the methanol content was inclined to 1.00%, the 3D surface was relatively high (Figure 2C) and the contours were relatively dense (Figure 2D), in the direction of the lower temperature. At this point, the yield of T36N showed a greater increase. The fermentation temperature obviously played a dominant role in the interaction between temperature and methanol content. In a similar analysis, initial pH played a more dominant role than methanol content, and when methanol content was 1.00%, the higher initial pH was, the greater yield of T36N reached (Figures 2E-F).
In summary, the degree of influence of three factors on protein yield was ranked as B > A > C, that is, initial pH> temperature > methanol content. As a result, the optimal combination conditions of the model for protein yield were fermentation temperature of 26 °C, initial pH 6.5, methanol content of 1.00%.
Figure 2. Surface plot and contour plot of the effects of protein concentration by interaction of fermentation temperature, initial pH and methanol content (T36N)
Model variance analysis
The model’s variance analysis is presented in Table 3. The regression model was subjected to variance analysis, revealing an overall P-value of ≤0.0001, signifying the model’s high statistical significance. The determination coefficient R2 equaled 0.9767, indicating that 97.67% of the response variability was attributed to the model’s nine factors, suggesting a robust fit. Additionally, the adjusted determination coefficient R2Adj equaled 0.9467, further validating the model’s predictive accuracy.
Table (3):
ANOVA of regression model
Source |
Sum of Squares |
Df |
Mean Square |
F-Value |
P-Value |
|
|---|---|---|---|---|---|---|
Model |
14770.86 |
9 |
1641.21 |
32.58 |
significant |
|
X1 |
564.82 |
1 |
564.82 |
11.21 |
0.0123 |
significant |
X2 |
295.12 |
1 |
295.12 |
5.86 |
0.0461 |
significant |
X3 |
184.61 |
1 |
184.61 |
3.66 |
0.0971 |
not significant |
X1X2 |
102.62 |
1 |
102.62 |
2.04 |
0.1966 |
not significant |
X1X3 |
0.9409 |
1 |
0.9409 |
0.0187 |
0.8951 |
not significant |
X2X3 |
6.18 |
1 |
6.18 |
0.1226 |
0.7365 |
not significant |
X12 |
993.42 |
1 |
993.42 |
19.72 |
0.0030 |
significant |
X22 |
10683.85 |
1 |
10683.85 |
212.08 |
significant |
|
X32 |
2200.77 |
1 |
2200.77 |
43.69 |
0.0003 |
significant |
Residual |
352.63 |
7 |
||||
Lack of fit |
328.61 |
3 |
||||
Pure Error |
24.02 |
4 |
||||
Cor Total |
15123.49 |
16 |
||||
R2 |
0.9767 |
|||||
R2Adj |
0.9467 |
|||||
R2Pred |
0.6499 |
|||||
C.V. % |
2.53 |
To test the reliability of the regression fitting model, 9 sets of validation experiments were conducted (Table 4). The results showed that the correlation coefficient between measured and predicted protein yield was 0.9356 > 0.9, which demonstrated that the regression fitting model was valid. According to the regression fitting model of response surface optimization, the predicted yield of T36N was 346.13 mg.L-1 under the optimal combination conditions (temperature 26 °C, initial pH 6.5, and methanol content 1.00%) for inducing expression of T36N by P. pastoris GS115. At the same time, the P. pastoris GS115 fermentation experiment was conducted under the above optimal combination conditions for 5 replicates. The results showed that the actual expression yield of T36N was 346.64 ± 3.47 mg.L-1 under the optimal condition, while the original T36N yield was 260.85 ± 2.45 mg.L-1, obtaining an increase of 32.89% (Supplementary Table S1). These data indicated that the optimal fermentation combination conditions obtained by RSM were consistent with the actual conditions. Lin et al.24 obtained the expression of rFIP-fve produced by P. pastoris GS115, with the optimal yield of 258.2 mg.L-1. The same host, under different fermentation conditions or expressing different heterologous proteins, will lead to different yields.
Table (4):
Validation results of the model
| Run | Independent variables | Protein yield (Y), mg.L-1 | |||
|---|---|---|---|---|---|
| Temperature (℃) | Initial pH | Methanol content (%) | Experimental | Predicted | |
| 1 | -1 | -1 | -1 | 329.74 | 297.91 |
| 2 | -1 | 0 | 1 | 268.16 | 266.07 |
| 3 | -1 | 1 | 0 | 352.37 | 346.13 |
| 4 | 0 | 0 | 0 | 261.84 | 260.85 |
| 5 | 0 | 1 | -1 | 285.08 | 288.39 |
| 6 | 0 | -1 | 1 | 289.16 | 285.85 |
| 7 | 1 | 1 | 1 | 294.69 | 302.86 |
| 8 | 1 | -1 | 0 | 310.93 | 317.17 |
| 9 | 1 | 0 | -1 | 247.57 | 239.66 |
| Correlation coefficient | 0.9356 | ||||
P. pastoris expression system is one of the most popular and standard tools to produce recombinant protein in the biomedical industry. However, the fermentation conditions of engineered P. pastoris are different from those of wild strains. In addition to temperature and pH, it is also necessary to consider the induction time of foreign protein expression and the concentration of inducer.23,25 For example, methanol is the inducer of gene expression in most P. pastoris expression systems. However, high concentration of methanol is toxic to the cells, while low concentration of methanol is not conducive to improving the productivity of heterologous proteins.23 To obtain a lot of FIP-glu, the FIP-glu had successfully expressed in various host cells, and the yield of rFIP-glu in P. pastoris ranged from 191.2 mg.L-1 to 270 mg.L-1, which was not enough for a pharmaceutical use. After optimization of the fermentation parameters, Mao et al. 22 improved the yield of rFIP-glu up to 368.71 mg.L-1 by P. pastoris in the shake-flask, under the condition of 1.0% methanol, initial pH 6.5 and 26 °C at 280 rpm for 4 days.22 In the present study, the gene FIP-glu_T36N was a gene FIP-glu variants with a N-linked glycan modification. It was still unknown whether the engineered yeast had the potential to produce protein T36N on a large scale after the gene FIP-glu_T36N was constructed into P. pastoris. This experiment demonstrated that T36N yield under these optimal fermentation conditions reached up to 346.64 ± 3.47 mg.L-1, which was increased by 32.9% comparing with not optimized.
In this study, three factors including temperature, initial pH and methanol content were taken into consideration. It is well-known that the suitable fermentation temperature is conducive to the growth of P. pastoris and the synthesis or hydrolysis degradation of protein.26,27 Some studies have suggested that glycosylation could affect the thermostability of proteins to some extent.28,29 In our study, the optimal fermentation temperature of the engineered P. pastoris was 26 °C that generally consistent with the previous results.22 The glycosylation modification does not seem to have much effect on the yield and thermal stability of the protein, which need further experiments confirmed. Secondly, initial pH of media also played an important role in cell growth, protein formation, and protein stability. In our study, initial pH was the most important of the three variables during P. pastoris GS115 fermentation for T36N glycoprotein. Under the condition of initial pH 6.5, the T36N yield reached up to the highest, which was basically consistent with the results of similar studies in previously.22 In Mao et al. study, the initial pH had a better effect on the rFIP-glu production of P. pastoris GS115 culture than fermentation temperature and methanol concentration, and under the optimum fermentation parameters (fermentation temperature 26 °C, initial pH 6.5, methanol concentration 1%), rFIP-glu concentration reached 368.71 ± 10.34 μg·mL-1.22 Generally, the optimal initial pH was obtained by running a series of fermentations at different pH. The yield of recombinant porcine lactoferrin expressed in P. pastoris increased to 12 mg·L-1 through increasing initial pH from 6.0 to 7.0.30 Therefore, it is necessary to control initial pH in the media and the best pH is related to protein properties, especially stability. Besides, methanol contributed to cell growth and was an inducer for the expression of foreign proteins in methanotrophic P. pastoris. It was reported that in the P. pastoris expression system at least 0.5% concentration of methanol was necessary for recombinant protein production, and methanol concentration should not exceed 2-2.5% for cell toxic effect of high level of methanol.23
This research successfully demonstrated the optimization of fermentation conditions for the production of the glycoprotein FIP-glu_T36N using the engineered strain P. pastoris GS115, achieved through the application of response surface methodology (RSM). The expression of various bioactive proteins by P. pastoris often requires tailored medium compositions and fermentation parameters. The current study revealed that the optimal conditions for T36N production were a fermentation temperature of 26 °C, an initial pH of 6.5, and a methanol concentration of 1.00%. Under these conditions, the T36N yield was maximized, reaching 346.64 mg.L-1. It is important to note that the experiments were conducted at the shake flask level, highlighting the utility of these methods for fine-tuning fermentation processes. The optimization of such processes in submerged culture offered a viable and efficient alternative to traditional culture techniques, particularly for engineered yeast strains like P. pastoris GS115. The findings of this study not only provided a valuable reference for the industrial-scale production of glycoprotein T36N but also underscored its potential for further development and application. The optimized parameters laid a solid foundation for future research and commercialization efforts in the field of bioactive protein production.
Additional file: Additional Table S1.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Dhama K, Saminathan M, Jacob SS, et al. Effect of immunomodulation and immunomodulatory agents on health with some bioactive principles, modes of action and potent biomedical applications. Int J Pharmacol. 2015;11(4):253-290.
Crossref - Liu Y, Bastiaan-Net S, Wichers HJ. Current understanding of the structure and function of fungal immunomodulatory proteins. Front Nut. 2020;7:132.
Crossref - Lin J, Chen H, Bai Y, et al. Ganoderma immunomodulatory proteins: mushrooming functional FIPs. Appl Microbiol Biotechnol. 2022;106(7):2367-2380.
Crossref - Li QZ, Chang YZ, He ZM, et al. Immunomodulatory activity of Ganoderma lucidum immunomodulatory protein via PI3K/Akt and MAPK signaling pathways in RAW264. 7 cells. J Cell Physiol. 2019;234(12):23337-23348.
Crossref - Kaji H, Shikanai T, Sasaki-Sawa A, et al. Large-scale identification of N-glycosylated proteins of mouse tissues and construction of a glycoprotein database, glycoProtDB. J Proteome Res. 2012;11(9):4553-4566.
Crossref - Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448-462.
Crossref - Wang X, Su K, Bao T, et al. Immunomodulatory effects of fungal proteins. Curr Top Nutraceut Res. 2012;10(1):1-12.
- Zhou S, Guan S, Duan Z, et al. Molecular cloning, codon-optimized gene expression, and bioactivity assessment of two novel fungal immunomodulatory proteins from Ganoderma applanatum in Pichia. Appl Microbiol Biotechnol. 2018;102(13):5483-5494.
Crossref - Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21(5):576-582.
Crossref - Li QZ, Wang XF, Zhou XW. Recent status and prospects of the fungal immunomodulatory protein family. Crit Rev Biotechnol. 2011;31(4):365-375.
Crossref - Kino K, Yamashita A, Yamaoka K, et al. Isolation and characterization of a new immunomodulatory protein, Ling Zhi-8 (LZ-8), from Ganoderma lucidium. J Biol Chem. 1989;264(1):472-478.
Crossref - Frelet-Barrand A. Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins. Biomolecules. 2022;12(2):180.
Crossref - Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol. 2015;32:9-17.
Crossref - Li QZ, Zheng YZ, Zhou XW. Fungal immunomodulatory proteins: characteristic, potential antitumor activities and their molecular mechanisms. Drug Discov Today. 2019;24(1):307-314.
Crossref - Popa CM, Tabuchi M, Valls M. Modification of bacterial effector proteins inside eukaryotic host cells. Front Cell Infect Microbiol. 2016;6:73-73.
Crossref - Berends E, Scholtmeijer K, Wosten HAB, et al. The use of mushroom-forming fungi for the production of N-glycosylated therapeutic proteins. Trends Microbiol. 2009;17(10):439-443.
Crossref - Wosten HAB, Scholtmeijer K, de Vries RP. Hyperproduction of enzymes by fungi. Food Mycology: CRC Press; 2007:197-210.
Crossref - Firatligil-Durmus E, Evranuz O. Response surface methodology for protein extraction optimization of red pepper seed (Capsicum frutescens). LWT – Food Sci Technol. 2010;43(2):226-231.
Crossref - Shao Y, Bai Y, Cai Z, et al. Optimization of stationary liquid fermentation conditions for N-methylsansalvamide production by the endophytic strain Fusarium sp. R1. Fermentation. 2024;10(3):140.
Crossref - Li QZ, Chen X, Mao PW, et al. N-Glycosylated Ganoderma lucidum immunomodulatory protein improved anti-inflammatory activity via inhibition of the p38 MAPK pathway. Food Funct. 2021;12(8):3393-3404.
Crossref - Yang Z, Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: A review. Biotechnol Adv. 2018;36(1):182-195.
Crossref - Mao PW, Li LD, Wang Yl, et al. Optimization of the fermentation parameters for the production of Ganoderma lucidum immunomodulatory protein by Pichia pastoris. Prep Biochem Biotechnol. 2020;50(4):357-364.
Crossref - Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020;235(9):5867-5881.
Crossref - Lin JW, Jia J, Shen YH, et al. Functional expression of FIP-fve, a fungal immunomodulatory protein from the edible mushroom Flammulina velutipes in Pichia pastoris GS115. J Biotechnol. 2013;168(4):527-533.
Crossref - Lin J, Liao Y, Yang S, et al. Identification a novel Ganoderma FIP gene from Ganoderma capense and its functional expression in Pichia pastoris. World J Microbiol Biotechnol. 2024;40(2):69.
Crossref - Curvers S, Linnemann J, Klauser T, et al. Recombinant protein production with Pichia pastoris in continuous fermentation-kinetic analysis of growth and product formation. Eng Life Sci. 2002;2(8):229-235.
https://doi.org/10.1002/1618-2863(20020806)2:8<229::AID-ELSC229>3.0.CO;2-9 - Curvers S, Brixius P, Klauser T, et al. Human chymotrypsinogen B production with Pichia pastoris by integrated development of fermentation and downstream processing. Part 1. fermentation. Biotechnol Prog. 2001;17(3):495-502.
Crossref - Li P, Anumanthan A, Gao XG, et al. Expression of recombinant proteins in Pichia pastoris. Appl Biochem Biotechnol. 2007;142(2):105-124.
Crossref - Tull D, Gottschalk TE, Svendsen I, et al. Extensive N-glycosylation reduces the thermal stability of a recombinant Alkalophilic Bacillus a-amylase produced in Pichia pastoris. Protein Expr Purif. 2001;21(1):13-23.
Crossref - Wang SH, Yang TS, Lin SM, et al. Expression, Characterization, and Purification of Recombinant Porcine Lactoferrin in Pichia pastoris. Protein Expr Purif. 2002;25(1):41-49.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.