ISSN: 0973-7510
E-ISSN: 2581-690X
Salmonella-related gastroenteritis and diarrheal infections pose significant health risks across all age groups in the developing countries. The high consumption of raw green leafy vegetables, particularly among health-conscious and younger populations, may further increase the risk of infection if food preparation is inadequate. In the current study, 645 vegetable samples were collected over the year, and 61 samples tested positive for bacterial contamination of Salmonella spp. The highest bacterial contamination was recorded in cabbage (Brassica oleracea var. capitata) (18.60%, positive (n)/total sample (N) = 8/43), Bathua (Chenopodium album), and fenugreek (Trigonella foenum-gracecum) (18.60%, n/N = 8/43), followed by cauliflower (Brassica oleracea var. botrytis) (13.95%, n/N = 6/43), parsley (Petroselinum crispum), spinach (Spinacia oleracea), and purslane (Portulaca oleracea) (11.62%, n/N = 5/43). The vegetable samples were collected randomly, and vegetables were subsequently assessed biochemically and microbiologically. Over the year, monthly analysis revealed peak contamination percentages in February (15.55%, n/N = 7/45), September (11.66%, n/N = 7/60), August, and January (10.66%, n/N = 8/75). The 15 samples was selected from 61 Salmonella isolates for antibiotic susceptibility profiling showed the high resistance to methicillin (93.33%, n/N = 14/15), Cefpodoxime, Clindamycin, and Teicoplanin (80%, n/N = 12/15), Linezolid, Novobiocin, Colistin, and Nitrofurantoin (53.33%, n/N = 8/15). Analysis of randomly selected vegetable samples using 16S rRNA confirmed the presence of Salmonella typhimurium subspecies as the predominant serovars. The present study is crucial for understanding the nature of bacterial flora, resistance, and transmission.
Antimicrobial Resistance, Food Safety, Salmonella spp., Prevalence, Vegetables
Salmonella spp. remain one of the most significant causes of foodborne illnesses worldwide. Globally, it is responsible for an estimated 93.8 million cases of gastroenteritis and 155,000 deaths each year.1 High-income countries, such as the United States and European Union, report consistent surveillance data for pandemic microbes in the food chain.2 In contrast, low and middle-income countries, particularly Africa and Southeast Asia, register higher incidence rates of infection due to factors such as poor sanitation, unhygienic food practices, contaminated drinking water, and limited access to healthcare.2 The rise in vegetable-associated outbreaks is often linked to contaminated irrigation water, improper handling, and cross-contamination during processing.3 India, with its largest population and diverse agricultural sector, faces significant challenges in controlling Salmonella spp. The country has documented an estimated 100,000 cases annually, with variation across different states. Outbreaks are frequently associated with poor hygiene practices, contaminated water sources, and poor food practices.4 The consumption of raw or minimally processed vegetables, often grown in contaminated environments, has emerged as a critical factor in the transmission of Salmonella spp.5 Rapid urbanization and changes in dietary patterns, with an increasing reliance on fresh produce, further exacerbate the risk of outbreaks.5 The capital city of Uttarakhand, Dehradun, reflects both national and regional trends in Salmonella spp. The city has witnessed an increase in reported cases, with approximately 2,000 cases annually, primarily linked to the consumption of contaminated vegetables.6
The combination of traditional agricultural practices, where vegetables are often irrigated with groundwater, and the growing demand for fresh produce in urban markets creates an environment conducive to the spread of Salmonella spp.7 Local studies have highlighted the presence of multiple Salmonella spp. serotypes in vegetables sold in open markets, emphasizing the need for improved food safety practices.8 Salmonella spp. transmission primarily occurs through consumption of contaminated food and water. Although poultry, eggs, and dairy products have traditionally been the main sources, there has been a significant increase in outbreaks linked to vegetables.9 Contamination of vegetables can occur at various points in the supply chain, including cultivation, harvesting, processing, and distribution. Manure contaminated with Salmonella spp. or sewage sludge can introduce bacteria into vegetables.10 Handling and processing poor hygiene practices during handling, processing, and transportation can lead to cross-contamination. Inadequate storage, particularly in warm and humid environments, can promote the growth of Salmonella spp. on vegetables.10 The emergence of antimicrobial-resistant (AMR) Salmonella spp. is a growing public health concern.
The overuse and misuse of antibiotics in agriculture, particularly chemicals (pesticides) in the cultivation of vegetables, contributes to the development of resistant strains.11 Vegetables can become reservoirs for AMR Salmonella spp. through irrigation water sources contaminated with resistant bacteria from agricultural runoff, or untreated sewage can transfer these pathogens to crops.10 During harvesting, processing, and distribution, vegetables can acquire pathogenic bacteria from contact with contaminated surfaces or other food products.12 In the present study, we characterized bacterial isolates from green leafy vegetables grown locally for a year. This study investigated seasonal variations in bacterial contamination, providing valuable insights into the factors that may contribute to the prevalence of contaminants at different times of the year. The identification of multidrug-resistant Salmonella spp. strains is a significant finding as it highlights the challenges associated with treating infections caused by these bacteria.
Collection of sample
A total of 645 vegetable samples were randomly collected from fresh produce and purchased in Dehradun, Uttarakhand, India, in a pilot study from February 2023 to January 2024; more details are shown in Table 1. The samples were obtained from fresh produce farms, vegetable cultivation fields, local markets, and supermarkets across various locations within Dehradun City. Samples were immediately transported to the laboratory in sterile containers for bacteriological analysis. The vegetables purchased and collected were Lathyrus oleraceus (green pea), Allium cepa (onion), Capsicum annuum (green chili), Brassica oleracea var. capitata (cabbage), Apium graveolens (celery), Beta vulgaris (beetroot), Trigonella foenum-graecum (fenugreek), Spinacia oleracea (spinach), Lactuca sativa L. (long lettuce), Portulaca oleracea (purslane), Chenopodium album (bathua), Brassica oleracea var. botrytis (cauliflower), Brassica oleracea var. italica (broccoli), Coriandrum sativum (cilantro), and Petroselinum crispum (parsley) all samples details in shows Table 2.
Table (1):
Monthly samples isolation
Study of month |
No of testing |
No of Contamination of samples |
Percentage of contamination in each month |
|---|---|---|---|
February |
45 |
7 |
15.55 |
March |
60 |
6 |
10 |
April |
60 |
5 |
8.33 |
May |
45 |
2 |
4.44 |
June |
45 |
3 |
6.66 |
July |
60 |
5 |
8.33 |
August |
75 |
8 |
10.66 |
September |
60 |
7 |
11.66 |
October |
30 |
2 |
6.66 |
November |
30 |
2 |
6.66 |
December |
60 |
6 |
10 |
January |
75 |
8 |
10.66 |
645 |
61 |
9.45 |
Table (2):
Contamination and occurrence of Salmonella spp. in vegetable fresh produce, Dehradun, Uttarakhand
Name of the samples |
Common name of the samples |
No. of samples |
No. of contaminated samples |
Percentage of Salmonella contamination |
|---|---|---|---|---|
P. crispum |
Parsley |
43 |
5 |
11.62 |
C. sativum |
Cilantro |
43 |
3 |
6.97 |
B. o. var. italica |
Broccoli |
43 |
3 |
6.97 |
B. o. var. botrytis |
Cauliflower |
43 |
6 |
13.95 |
C. album |
Bathua |
43 |
8 |
18.60 |
P. oleracea |
Purslane |
43 |
5 |
11.62 |
L. sativa L. |
Long lettuce |
43 |
4 |
9.30 |
S. oleracea |
Spinach |
43 |
5 |
11.62 |
T. f. graceum |
Fenugreek |
43 |
8 |
18.60 |
B. vulgaris |
Beetroot |
43 |
3 |
6.97 |
A. graveolens |
Celery |
43 |
3 |
6.97 |
B. o. var. capitata |
Cabbage |
43 |
8 |
18.60 |
C. annuum |
Green chilli |
43 |
0 |
0 |
A. cepa |
Onion |
43 |
0 |
0 |
L. oleraceus |
Green pea |
43 |
0 |
0 |
645 |
61 |
Isolation and identification of bacterial species
Each sample was washed with sterile distilled water and 1% hypochlorite to remove debris from surface. A sterile knife was used to cut a small portion of each vegetable sample and homogenize it using a sterile mortar and pestle, whereas vegetables with extraction were squeezed mechanically to extract the juice. One milliliter of the resultant homogenate was serially diluted to 10-6. Then 0.1 ml each of the 10-5 and 10-6 dilution fractions was separately spread and plated onto a nutrient agar plate (Himedia, India) and incubated at 37 °C for 24 h. The organisms were purified by successive subculturing on xylose lysine decarboxylase agar plates and incubated at 37 °C for 24 h. The isolates were further identified using the biochemical profiles of KB001 and KB002 test kits (HiMedia, India). Biochemical assays were performed, including the use of HiMedia reagents, to improve the identification of isolates (Table 3).
Table (3):
Biochemical characterisation
Test Name |
Interpretation |
|---|---|
Triple sugar iron |
+ |
H2S production |
+ |
Nitrate reduction |
+ |
Phenolylalanine deamination |
– |
Lysine utilization |
– |
Ornithine utilization |
– |
Sucrose |
– |
Rhamnose |
+ |
Mannitol |
+ |
Sorbitol |
– |
Lactose |
+ |
Arabinose |
+ |
Adonitol |
– |
Glucose |
+ |
Citrate utilization |
+ |
Voges-Proskauer test |
– |
Methly red |
+ |
Indole |
– |
Oxidase |
– |
Catalase |
+ |
Antimicrobial Susceptibility testing (AST)
The initial preparation of the test broth was generated after culture at 37 °C for 5-6 hours, in accordance with the national clinical test operating procedure for drug susceptibility testing in vitro. A limit of 0.5 McFarland units was set for the degree of bacterial turbidity. Kirby-Bauer disc diffusion techniques were performed using Mueller-Hinton agar medium, which allowed AST to identify all 15 Salmonella spp. bacterial isolates. We prepared the media according to standard protocols. The press was prepared following the convention. To confirm the sterility of the plates, they were autoclaved and incubated for 24 hours at 37 °C.13 Using the CLSI 201514 standards, therapeutically effective antibiotics were used against each isolate, and antibacterial sensitivity testing was performed on the isolated bacterial cultures. The CLSI 201514 guidelines were followed for all minimum inhibitory doses (MIC). Forty-six (46) antibiotics were used in this investigation, including 16 of the antibiotic classes cephalosporin, lincosamide, quinoline/fluoroquinoline, penicillin, sulfonamide, macrolides, aminoglycosides, glycopeptides, chloramphenicol, oxazolidinones, tetracycline, aminocoumarin, polypeptides, monobactams, nitrofurans, and carbapenem.
Molecular analysis characterisation and identification of Salmonella isolates
Isolation of DNA
Genomic DNA was extracted from the sample by homogenizing it in 1 ml of extraction buffer in a mortar, followed by the addition of an equal volume of phenol-chloroform alcohol (25:24:1). After centrifugation at room temperature (14,000 rpm) for 15 min, the upper aqueous phase was transferred into a new tube. DNA was precipitated by adding 0.1 volume of 3 M sodium acetate (pH 7.0) and 0.7 volume of isopropanol, followed by incubation at room temperature for 15 min, and centrifugation at 4 °C (14,000 rpm) for 15 min. The DNA pellet was washed twice with 70% ethanol, briefly with 100% ethanol, and then air-dried. DNA was dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and treated with 5 µL DNase-free RNase A (10 mg/ml) to remove RNA contamination. The isolated DNA samples were used for bacterial identification via 16S rRNA sequencing to identify Salmonella spp., followed by antimicrobial susceptibility testing (AST). This version retains essential steps and details of the DNA extraction process.
16S rRNA sequencing characterization
Following DNA isolation, the sample was outsourced to CytoGene for 16S rRNA sequencing. The Sanger method was employed for 16S rRNA sequencing of the bacterial isolates. Primer details the PCR product size ~1.5 kb for forward primer is “GGATGAGCCCGCGGCCTA” and reverse primer is “CGGTGTGTACAAGGCCCGG”. F-GC content of 72.2, R-GC content of 65.0, and 30 cycles.
Phylogeny
Maximum likelihood phylogenies of all serovars compared three or more isolates from this study, in addition to the reference genomes. Completely closed genomes for each serovar downloaded from the NCBI database were calculated using core genome alignment. Six reference genomes were obtained and compared with the sequence of the isolate PQ066119. 1 Salmonella enterica strain RPGEU-1, PQ066132. 1 Salmonella enterica strain RPGEU-3, PQ066136. 1 Salmonella enterica strains RPGEU-5 and PQ135076. 1 Salmonella enterica strains RPGEU-2 and PQ066141. 1 Salmonella enterica strains RPGEU-6 and PQ066134. One Salmonella enterica strain RPGEU-4, submitted to NCBI. A phylogenetic tree was created in MEGA 11 using the neighbor-joining method and bootstrapping. The Neighbor-Joining approach was used to infer the evolutionary history.15 It is assumed that the bootstrap consensus tree, which was generated from 1000 repetitions, accurately depicts the evolutionary history of the examined species.16 branches that are collapsed correspond to partitions that are replicated in fewer than 50% of bootstrap replicates. Next to the branches are the percentage of duplicate trees, where the related taxa were grouped together in the bootstrap test (1000 repetitions).16 Evolutionary distances are shown as base substitutions per site and were calculated using the Maximum Composite Likelihood approach.17 Thirteen nucleotide sequences were used in this study. First, the locations of their codons and noncoding codons were covered. For every sequence pair, all unclear locations were eliminated (pairwise deletion). The final dataset contained 1589 locations in total. In MEGA11, evolutionary analyses were carried out.18
A total of 645 vegetable samples from 15 different vege types were analyzed for Salmonella contamination. The highest levels of contamination were found in Chenopodium album (Bathua), Trigonella foenum-graecum (Fenugreek), and Brassica oleracea var. capitata (cabbage), with 18.60% of samples testing positive for Salmonella. Petroselinum crispum (parsley), Portulaca oleracea (purslane), and Spinacia oleracea (spinach) each showed 11.62% contamination. Lower levels of contamination were observed in Brassica oleracea var. italica (broccoli), Brassica oleracea var. botrytis (cauliflower), Coriandrum sativum (Cilantro), Beta vulgaris (Beetroot), and Apium graveolens (Celery), all with contamination rates ranging between 6.97% and 13.95%. Notably, no Salmonella contamination was detected in Capsicum annuum (green chili), Allium cepa (onion), or Lathyrus oleraceus (green pea). Overall, 61 of the 645 samples tested positive for contamination, representing 9.45% of the total samples analyzed and the percentage of Salmonella in each vegetable is shown in Table 2. Monthly analysis of contamination levels across 645 samples revealed significant variation in the percentage of Salmonella-positive samples throughout the year.
The highest number of Salmonella-positive samples was observed in February (15.55%), followed by January and August (both 10.66%), suggesting the role of possible seasonal or environmental factors that influence bacterial load during these months. Conversely, fewer Salmonella-positive samples were observed in May. Other months, such as March, September, and December, showed moderate contamination levels ranging from 10% to 11.66%, while contamination rates for April, June, July, October, and November were relatively low, between 6.66% and 8.33%. These fluctuations may be attributed to variations in temperature, humidity, and farming practices, which could affect the survival and proliferation of the contaminants. Understanding these patterns is crucial for optimizing testing and control measures, particularly during months with higher contamination risks. Overall, 9.45% of the total samples were found to be contaminated, highlighting the need for continuous monitoring and improved food safety measures.
Antimicrobial susceptibility profiling of Salmonella isolates from 15 green leafy vegetables provided valuable insights into the resistance patterns of this pathogen (Table 4). The data revealed a trend of increasing resistance to multiple antibiotics across different classes, highlighting the urgent need for effective strategies to combat antimicrobial resistance. According to Peacock and Paterson,19 methicillin is frequently used to treat Staphylococcus aureus infections. Our investigation showed that the penicillin category had a considerably enhanced methicillin-resistance of 93% in Salmonella isolates. This is a unique observation, where methicillin-resistance was observed against Salmonella spp., whereas it was primarily reported in Staphylococcus infection.
Table (4):
Antibiotic resistance pattern of Salmonella spp.
| Class of Antibiotics | Name of Antibiotics | Code | % of resistance isolates n = 15 | ||
|---|---|---|---|---|---|
| R | I | S | |||
| Cephalosporin | Cepholexime (30 mcg) | CEP | 0 (0) | 33.33 (5) | 66.66 (10) |
| Cefuroxime (30 mcg) | CXM | 0 (0) | 40 (6) | 60 (9) | |
| Cefoperazone (75 mcg) | CPZ | 0 (0) | 26.66 (4) | 73.33 (11) | |
| Ceftazidime (30 mcg) | CAZ | 0 (0) | 33.33 (5) | 66.66 (10) | |
| Cefaclor (30 mcg) | CF | 6.66 (1) | 33.33 (5) | 60 (9) | |
| Cefotaxime (Cepholexime) (30 mcg) | CTX | 6.66 (1) | 13.33 (2) | 80 (12) | |
| Ceftriaxone (30 mcg) | CTR | 6.66 (1) | 20 (3) | 73.33 (11) | |
| Cefoxitin (Cephoxitin) (30 mcg) | CX | 13.33 (2) | 40 (6) | 46.66 (7) | |
| Cefpodoxime (10 mcg) | CPD | 80 (12) | 6.66 (1) | 13.33 (2) | |
| Quinoline/Fluoroquinolone | Sparfloxacin (5 mcg) | SPX | 0 (0) | 6.66 (1) | 93.33 (14) |
| Norfloxacin (10 mcg) | NX | 0 (0) | 33.33 (5) | 66.66 (10) | |
| Ciprofloxacin (5 mcg) | CIP | 0 (0) | 46.66 (7) | 53.33 (8) | |
| Levofloxacin (5 mcg) | LE | 6.66 (1) | 66.66 (10) | 26.66 | |
| Ofloxacin (5 mcg) | OF | 6.66 (1) | 46.66 (7) | 46.66 (7) | |
| Gatifloxacin (5 mcg) | GAT | 6.66 (1) | 33.33 (5) | 60 (9) | |
| Moxifloxacin (5mcg) | MO | 26.66 | 40 (6) | 33.33 (5) | |
| Nalidixic Acid (30 mcg) | NA | 33.33 (5) | 40 (6) | 26.66 (4) | |
| Penicillin | Ampicillin/ Sulbactam (10 mcg) | A/S | 6.66 (1) | 13.33 (2) | 80 (12) |
| Ampicillin (10 mcg) | AMP | 6.66 (1) | 13.33 (2) | 80 (12) | |
| Augmentin (30 mcg) | AMC | 6.66 (1) | 60 (9) | 33.33 (5) | |
| Cloxacillin (1 mcg) | COX | 6.66 (1) | 26.66 (4) | 66.66 (10) | |
| Amoxicillin/Clavulanic acid (30 mcg) | AMC | 20 (3) | 60 (9) | 20 (3) | |
| Ampicillin/ Cloxacillin (10 mcg) | AX | 26.66 (4) | 40 (6) | 33.33 (5) | |
| Penicillin-G (10 units) | P | 33.33 (5) | 40 (6) | 26.66 (4) | |
| Oxacillin (1 mcg) | OX | 40 (6) | 40 (6) | 20 (3) | |
| Methicillin (5 mcg) | MET | 93.33 (14) | 6.66 (1) | 0 (0) | |
| Macrolides | Azithromycin (15 mcg) | AZM | 13.33 (2) | 53.33 (8) | 33.33 (5) |
| Clarithromycin (15 mcg) | CLR | 20 (3) | 66.66 (10) | 13.33 (2) | |
| Roxithromycin (30 mcg) | RO | 33.33 (5) | 53.33 (8) | 13.33 (2) | |
| Erythromycin (15 mcg) | E | 40 (6) | 33.33 (5) | 26.66 (4) | |
| Aminoglycosides | Gentamicin (10 mcg) | GEN | 6.66 (1) | 20 (3) | 46.66 (7) |
| Amikacin (30 mcg) | AK | 13.33 (2) | 66.66 (10) | 20 (3) | |
| Netillin (Netilmicin Sulphate) (30 mcg) | NET | 13.33 (2) | 40 (6) | 46.66 (7) | |
| Tobramycin (10 mcg) | TOB | 13.33 (2) | 40 (6) | 46.66 (7) | |
| Glycopeptides | Vancomycin (30 mcg) | VA | 26.66 (4) | 53.33 (8) | 20 (3) |
| Teicoplanin (10 mcg) | TEI | 80 (12) | 13.33 (2) | 6.66 (1) | |
| Sulphonamide | Co-Trimoxazole (Sulpha/Trimethoprim) (25 mcg) | COT | 0 (0) | 20 (3) | 80 (12) |
| Carbapenem | Imipenem (10 mcg) | IPM | 20 (3) | 40 (6) | 40 (6) |
| Tetracycline | Tetracycline (30 mcg) | TE | 26.66 (4) | 53.33 (8) | 20 (3) |
| Monobactams | Aztreonam (30 mvg) | AT | 33.33 (5) | 53.33 (8) | 13.33 (2) |
| Chloramphenicol | Chloramphenicol (30 mcg) | C | 46.66 (7) | 40 (6) | 13.33 (2) |
| Oxazolidinones | Linezolid (30 mcg) | LZ | 53.33 (8) | 33.33 (5) | 13.33 (2) |
| Aminocoumarin | Novobiocin (5 mcg) | NV | 53.33 (8) | 26.66 (4) | 20 (3) |
| Polypeptides | Colistin (Methane Sulphonate) (10 mcg) | CL | 53.33 (8) | 26.66 (4) | 20 (3) |
| Nitrofurans | Nitrofurantoin (300 mcg) | NIT | 53.33 (8) | 13.33 (2) | 33.33 (5) |
| Lincosamide | Clindamycin (2 mcg) | CD | 80 (12) | 13.33 (2) | 6.66 (1) |
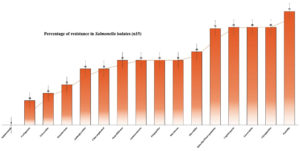
The current study raised serious concerns by showing that Salmonella spp. isolates from green leafy vegetables had a high resistance rate of 80% to cefpodoxime, teicoplanin, and clindamycin. This finding is consistent with previous research by Fatima et al.,20 indicating a persistent and rising trend of resistance among these antibiotics. The growing prevalence of multidrug-resistant Salmonella spp. poses a major threat to public health as limited treatment options may lead to high morbidity and mortality. Contamination of green leafy vegetables with resistant Salmonella spp. is particularly worrying, as these foods are widely consumed, and this can serve as a reservoir for the transmission of resistant bacteria. Our results revealed a considerable increase in the frequency of multidrug-resistance in Salmonella spp. isolates (Figure 1), with resistance to colistin, nitrofurantoin, novobiocin, and linezolid (53.33% each), exceeding those reported by Castro-Vargas et al.21 In chloramphenicol category, 46.66% resistance in vegetable isolates was notably higher than the rates reported by Castro-Vargas et al.,21 in which 40.5% resistance observed in chicken meet, and eggs and egg-laying hens, on average.
Figure 1. Percentage of multidrug-resistance in Salmonella isolates in present study (samples n = 15)
In the previous study, Shoaib et al.22 reported the highest resistance to azithromycin (50%) against Salmonella spp. for azithromycin. However, our study found a lower resistance trend of 40% to azithromycin compared with their findings. The current study recorded a higher resistance of 33.33% to aztreonam in comparison to 5% resistance reported by Lu et al.,23 highlighting the increasing occurrence of bacterial isolates that are resistant to aztreonam. Compared to the results of Yin et al.,24 our investigation revealed a noteworthy 33.33% resistance to nalidixic acid, underscoring the growing incidence of resistance to quinolone/fluoroquinolone medicines. Salmonella spp. resistance in food samples was reported by Gargano et al.25 to be 33.3%, greater than the 26.66% resistance against tetracycline, we observed in isolates from green leafy vegetables.
The 20% imipenem resistance found in our study was less than 38% carbapenem resistance found in Kanaan et al.26 The 13.33% resistance to aminoglycosides was lower than the highest (32%) resistance reported by Lauteri et al.27 Unlike the significant prevalence of co-trimoxazole resistance reported by Lauteri et al.,27 our study did not detect any resistance in samples among Salmonella isolates. This discrepancy may be due to differences in the number of study populations, geographic regions, or time periods. However, it is important to remain cautious, as resistance patterns can evolve quickly. Continued monitoring of antibiotic resistance trends is essential for effective Salmonella spp. infections. The high resistance rates observed in this study underscore the importance of implementing strict food safety measures to prevent contamination of green leafy vegetables with resistant Salmonella spp. Additionally, prudent antibiotic stewardship practices are essential to minimize the development and spread of antibiotic resistance.
Molecular phylogeny extends our knowledge of organism relationships and provides the foundation for conventional identification techniques.28 Based on 16S rRNA gene sequence analysis, and submitted to NCBI ID, RPGEU-1, RPGEU-2, RPGEU-3, RPGEU-4, RPGEU-5, and RPGEU-6 were identified as Salmonella enterica subsp. Similarly, strains NR 119108.1, MF804992.1, NR 074910.1 and CP053585.1 appear in the same cluster and were close to other members of Salmonella enterica spp. Comparative sequence analysis of 16S rRNA is currently the most widely used approach for the reconstruction of microbial phylogenies. In our study, we found that the isolates PQ066119.1, PQ135076.1, PQ066132.1, PQ066134.1, PQ066136.1, and PQ066141.1, belonged to Salmonella enterica spp. In our study we found that in our study similarity when control to PQ066119.1 (99.29%) with NR 119108.1, PQ135076.1 (81.37%) with MF804992.1, PQ066132.1 (98.77%), PQ066134.1 (98.77%), PQ066136.1 (97.96%) with NR 0749101, and PQ066141.1 (96.42%) CP053585.1 showed sequence similarity in Table 5.
Table (5):
The source and location of isolated strains, identification based on 16S rRNA gene sequence and their accession numbers
Accession no |
Strain ID in NCBI |
Strain name genus |
Source of isolation |
Location of Isolation |
Similarity % |
Number of nucleotides of 16S rRNA gene |
Closely related taxa identified by using Database |
|---|---|---|---|---|---|---|---|
PQ066119.1 |
Salmonella enterica strain RPGEU-1 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
99.29% |
1536 |
NR_119108.1. |
PQ135076.1 |
Salmonella enterica strain RPGEU-2 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
81.37 % |
1447 |
MF804992.1 |
PQ066132.1 |
Salmonella enterica strain RPGEU-3 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
98.77 % |
1538 |
NR_074910.1 |
PQ066134.1 |
Salmonella enterica strain RPGEU-4 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
98.77 % |
1449 |
NR_074910.1 |
PQ066136.1 |
Salmonella enterica strain RPGEU-5 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
97.96 % |
1464 |
NR_074910.1 |
PQ066141.1 |
Salmonella enterica strain RPGEU-6 |
Salmonella enterica subsp. |
Green Leafy Vegetables |
Dehradun, Uttarakhand, India |
96.42 % |
1483 |
CP053585.1 |
The similarity of the isolates in the phylogenetic tree was demonstrated using different control sequences (Figure 2). Upon harvest, many farmers have poor control over the management of their produce when sold as intermediaries. This can result in cross-contamination, particularly in informal market environments, where raw meat and vegetables are often sold in proximity and under unsanitary conditions.29 Additionally, both vendors and consumers frequently neglect fundamental food hygiene practices.10 Among the vegetables analyzed in this study, vegetables was found to be particularly susceptible to contamination by Salmonella, E. coli, and Coliform bacteria.30 This finding is consistent with previous reports of bacterial contamination of lettuce in various foodborne outbreaks, including those involving Salmonella in different Indian states.31
Figure 2. Phylogenetic tree showing inter-relationship of Salmonella isolates (RPGEU-1, RPGEU-2, RPGEU-3, RPGEU-4, RPGEU-5, and RPGEU-6) with closely related species of the genus Salmonella enterica subsp.
Before fresh herbs are delivered to the store and eventually to the consumer, they go through a number of processing stages and are exposed to many sources of contamination, which might contaminate the edible parts of the herbs. The antimicrobial-resistant bacterial load might also be explained by this microbial contamination. Notably, 61 Salmonella-positive isolates were subjected to 16 different classes of antibiotics, and resistance observed in many isolates, and the pathogen was recovered from around 9.45% of fresh produce samples. The presence of enterobacteria in the food chain that carry AMR determinants, such as ESBL or AmpC, or that display both phenotypes and can be encoded in plasmids with transferable capability, poses a relatively high risk for both sporadic human pathogenicity and the spread of these resistance genes to neighboring bacteria, according to the most recent reports from EFSA (2023). Green leafy vegetables become contaminated because Salmonella spp. are transmitted with fecal debris, particularly raw manure, and with inadequate food handler cleanliness, they are frequently associated with outbreaks related to produce.31
Disease transmission to vegetables has been caused by biological soil additive-contaminated waste pathogens and insufficiently composted manure.32 The likelihood of Salmonella spp. and other microbes of fecal origin contaminating food may rise due to India’s widespread use of animal waste as fertilizer and non-compliance with international guidelines for treating manure.31 In the current study, estimates of the frequency of Salmonella in vegetables were greater than those found in Chinese lettuce samples.33 Compared to the dry season, there were more Salmonella spp. in Dehradun vegetables than in the wet and winter seasons. As shown in the present study, bacterial growth was more conducive during the rainy season, which was marked by warmer temperatures, higher humidity, and more rainfall.
These findings are in line with research assessing the seasonal impact on bacterial development in soil treated with manure and irrigation water.34 Green leafy vegetables may be subjected to further cross-contamination throughout the import process, such as handling and international shipping, which might account for the higher bacterial levels observed in the winter and wet seasons. This study examined the effects of seasonal fluctuations on Salmonella spp. in 15 green leafy vegetable varieties. The investigation found that these markets had considerable levels of enteric bacterial contamination caused by insufficient handling during shipping, subpar farming methods, and unhygienic market environments.
Implementing effective preventive measures is further hampered by several sociocultural issues, such as poor literacy among farmers and inadequate infrastructure for monitoring and training. The purpose of this study was to characterize the isolated microbial strains from fresh produce and commercially used green leafy vegetables marketed in Dehradun, India, in terms of their susceptibility to antimicrobial-resistant pathogens. Periodic studies are required for the monitoring and surveillance of food products, such as dairy, meat, and vegetables.
The prevalence of diarrheal disease poses a significant health risk in developing countries. Poor hygiene and low accessibility to clean drinking water contribute to many infections annually. The present study confirms the occurrence of pathogenic bacteria such as Salmonella spp. in green leafy vegetables. Furthermore, the presence of drug-resistant Salmonella typhimurium in green vegetables, as confirmed by drug susceptibility testing further complicates the disease management. The study emphasizes the role of green leafy vegetables in disease transmission, especially in health-conscious individuals and young people who consume raw, uncooked green vegetables. The study also necessitates the importance of periodic surveillance of vegetables for pathogenic microorganisms to alert consumers.
ACKNOWLEDGMENTS
The authors would like to thank Graphic Era (Deemed to be University) for the support and help for the current study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PG conceptualized and supervised the study and applied methodology. RK performed investigation and formal analysis. VK and PG performed validation. RK wrote the original draft. NKA and VK wrote the manuscript. NKU, PG, VK and SK reviewed the manuscript. NKA, NKU, PG and SK edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by Graphic Era (Deemed to be University), Dehradun, India.
DATA AVAILABILITY
All datasets generated or analysed during the study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Galan-Relano A, Diaz AV, Lorenzo BH, et al. Salmonella and Salmonellosis: An Update on Public Health Implications and Control Strategies. Animals. 2023;13(23):3666.
Crossref - Faour-Klingbeil D, Todd ECD. Prevention and control of foodborne diseases in Middle-East North African countries: Review of national control systems. Int J Environ Res Public Health. 2019;17(1):70.
Crossref - Dyda A, Nguyen PY, Chughtai AA, MacIntyre CR. Changing epidemiology of Salmonella outbreaks associated with cucumbers and other fruits and vegetables. Global Biosecurity. 2020;2(1).
Crossref - Narayan KG, Sinha DK, Singh DK. Food-borne infections and intoxications. In: Veterinary Public Health & Epidemiology, Springer, Singapore. 2023:185-200.
Crossref - Santos MI, Gracio M, Silva MC, Pedroso L, Lima A. One Health perspectives on food safety in minimally processed vegetables and fruits: From farm to fork. Microorganisms. 2023;11(12):2990.
Crossref - Maurya S, Kalra C, Mahto RK, Singh S, Sharma N, Semwal S. Epidemiological Study of Salmonella typhi and Its Month-Wise Effect on Different Age Groups in Dehradun. J Adv Med Med Res. 2020;32(24):256-260.
Crossref - Lenzi A, Marvasi M, Baldi A. Agronomic practices to limit pre- and post-harvest contamination and proliferation of human pathogenic Enterobacteriaceae in vegetable produce. Food Control. 2021;119(107486):107486.
Crossref - Mohanapriya R, Paranidharan V, Karthikeyan S, Balachandar D. Surveillance and source tracking of foodborne pathogens in the vegetable production systems of India. Food Control. 2024;162(110427):110427.
Crossref - Ehuwa O, Jaiswal AK, Jaiswal S. Salmonella, Food Safety and Food Handling Practices. Foods. 2021;10(5):907.
Crossref - Yang Q, Chen J, Dai J, et al. Total coliforms, microbial diversity and multiple characteristics of Salmonella in soil-irrigation water-fresh vegetable system in Shaanxi, China. Sci Total Environ. 2024;924:171657.
Crossref - Miller SA, Ferreira JP, LeJeune JT. Antimicrobial use and resistance in plant agriculture: A one health perspective. Agriculture. 2022;12(2):289.
Crossref - Possas A, Perez-Rodriguez F. New insights into cross-contamination of fresh-produce. Curr Opin Food Sci. 2023;49:100954.
Crossref - Nassar MSM, Hazzah WA, Bakr WMK. Evaluation of antibiotic susceptibility test results: how guilty a laboratory could be? J Egypt Public Health Assoc. 2019;94(1):4.
Crossref - CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard Twelfth Edition. CLSI document M02-A12. Clinical and Laboratory Standards Institute, Wayne, PA. 2015.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030-11035.
Crossref - Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022-3027.
Crossref - Peacock SJ, Paterson GK. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84:577-601.
Crossref - Fatima A, Saleem M, Nawaz S, Khalid L, Riaz S, Sajid I. Prevalence and antibiotics resistance status of Salmonella in raw meat consumed in various areas of Lahore, Pakistan. Sci Rep. 2023;13(1):22205.
Crossref - Castro-Vargas RE, Herrera-Sanchez MP, Rodriguez-Hernandez R, Rondon-Barragan IS. Antibiotic resistance in Salmonella spp. isolated from poultry: A global overview. Vet World. 2020;13(10):2070-2084.
Crossref - Shoaib M, Satti L, Hussain A, Khursheed N, Sarwar S, Shah AH. Disc Diffusion Testing of Azithromycin Against Clinical Isolates of Typhoidal Salmonellae: A Diagnostic Conundrum. Cureus. 2021;13(7):e16777.
Crossref - Lu Y, Sun P, Shao W, et al. Detection and Molecular Identification of Salmonella Pathogenic Islands and Virulence Plasmid Genes of Salmonella in Xuzhou Raw Meat Products. J Food Prot. 2022;85(12):1790-1796.
Crossref - Yin X, Dudley EG, Pinto CN, M’ikanatha NM. Fluoroquinolone sales in food animals and quinolone resistance in non-typhoidal Salmonella from retail meats: United States, 2009-2018. J Glob Antimicrob Resist. 2022;29:163-167.
Crossref - Gargano V, Sciortino S, Gambino D, et al. Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics. 2021;10(7):809.
Crossref - Kanaan MHG, Khalil ZK, Khashan HT, Ghasemian A. Occurrence of virulence factors and carbapenemase genes in Salmonella enterica serovar Enteritidis isolated from chicken meat and egg samples in Iraq. BMC Microbiol. 2022;22(1):279.
Crossref - Lauteri C, Festino AR, Conter M, Vergara A. Prevalence and antimicrobial resistance profile in Salmonella spp. isolates from swine food chain. Ital J Food Saf. 2022;11(2):9980.
Crossref - Avise JC. Molecular Markers, Natural History and Evolution. 1994th ed. Springer; 2012.
Crossref - Albert V, Ramamurthy T, Das S, et al. Comprehending the risk of foodborne and waterborne disease outbreaks: Current situation and control measures with Special reference to the Indian Scenario. Heliyon. 2024;10(16):e36344.
Crossref - Osafo R, Balali GI, Amissah-Reynolds PK, et al. Microbial and parasitic contamination of vegetables in developing countries and their food safety guidelines. J Food Qual. 2022;2022:1-24.
Crossref - Verma P, Saharan VV, Nimesh S, Singh AP. Phenotypic and virulence traits of Escherichia coli and Salmonella strains isolated from vegetables and fruits from India. Journal of applied microbiology. 2018;125(1):270-281.
- Akanmu AO, Babalola OO, Venturi V, et al. Plant Disease Management: Leveraging on the Plant-Microbe-Soil Interface in the Biorational Use of Organic Amendments. Front Plant Sci. 2021;12:700507.
Crossref - Yang X, Wu Q, Huang J, et al. Prevalence and characterization of Salmonella isolated from raw vegetables in China. Food Control. 2020;109(106915):106915.
Crossref - Li H, Wang H, Jia B, Li D, Fang Q, Li R. Irrigation has a higher impact on soil bacterial abundance, diversity and composition than nitrogen fertilization. Sci Rep. 2021;11(1):16901.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.