ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-resistant Staphylococcus aureus (MRSA) is resistant to β-lactam antibiotics owing to the presence of mecA and mecC resistance genes. Resistance genes in MRSA are carried by a genetic component named staphylococcal cassette chromosome mec (SCCmec). The mecC gene showed 63% similarity with the mecA gene. This resulted in the mecC gene not being detected by routine PCR examination, which specifically detects mecA. Data regarding the epidemiology of molecular detection of the mecC gene in Indonesia are still very limited, especially in North Sumatra Province. This study aimed to characterize MRSA resistance genes in a tertiary hospital in Medan, North Sumatra. Clinical samples of the infection were collected and identified as MRSA using the VITEK-2 compact device. A total of 80 samples from bacteremia patients in our hospital were used in this research. The detection of resistance genes is performed using conventional Polymerase Chain Reaction (PCR). Visualization of the presence of genes was performed using electrophoresis. The mec gene was detected in 79 MRSA samples (98.75%). A total of 63 samples carried two resistance genes, mecA and mecC (78.75%), 15 samples carried only mecC (18.75%), one sample carried only mecA, and only one sample carried neither mecA nor mecC. The finding of the mecC gene is a cause for concern because it cannot be detected via routine PCR. This study showed that the majority of MRSA bacteria carry a mixture of mecA and mecC genes.
Methicillin-resistant Staphylococcus aureus, Molecular Epidemiology, Polymerase Chain Reaction, Tertiary Hospital
Staphylococcus aureus is a gram-positive bacterium that is sphere-shaped with a diameter of approximately 1 µm and arranged in irregular clusters that resemble grapes. Staphylococcus spp. are naturally found on human skin and mucous membranes. However, certain Staphylococcus spp. can cause infections, with Staphylococcus aureus being potentially severe. Almost everyone has experienced an infection caused by these bacteria, and these may vary from food poisoning and minor wound infections to life-threatening infections.1
Penicillin and its derivatives are effective against Staphylococcal infections. However, shortly after the introduction of penicillin, resistant strains of S. aureus have emerged and spread globally. In 1961, methicillin-resistant S. aureus (MRSA) bacteria were discovered, posing a persistent global health problem.2 According to previous studies, the prevalence of MRSA in Indonesia is relatively high, ranging from 17% to 26.6%.3-5 This high prevalence in hospitals can lead to an increased economic burden on the healthcare sector and heightened use of specific antibiotics against MRSA. There is concern that this could also trigger the development of other resistant strains.6
MRSA carries the resistance gene mecA, which leads to the transformation of penicillin-binding protein (PBP) to PBP2a. PBP2a acts as an alternative transpeptidase with a low affinity for most β-lactam antibiotics. The resistance gene component is located in a mobile genetic element known as Staphylococcal Cassette Chromosome mec (SCCmec). The mecC resistance gene results from a mutation in the mecA resistance gene, exhibiting only 63% similarity to mecA. MRSA isolates carrying the mecC gene are known to originate from the transmission of livestock-associated MRSA or other Staphylococci to humans.2 This also results in the mecC gene not being detected through routine PCR examination which usually specifically detects mecA or through the PBP2a slide agglutination test.7
Surveillance of MRSA infections in healthcare facilities is crucial for controlling the growth of antimicrobial-resistant bacteria. Additionally, it enables the administration of efficient antibiotic therapy and more effective infection control strategies.8 Research detecting resistance genes in MRSA bacteria is still limited in Indonesia, especially in North Sumatra. Therefore, researchers aimed to determine the presence of mecA and mecC resistance genes in MRSA bacteria identified from clinical samples at Haji Adam Malik General Hospital, Medan, North Sumatra.
Bacterial isolates
Isolated DNA samples were collected from our previous study.9 A total of 80 samples were used in this research, collected from Haji Adam Malik General Hospital from September 2022 to January 2023. This study included the conventional detection of the mec gene using conventional Polymerase Chain Reaction (PCR), followed by electrophoresis and UV light visualization.
Detection method
For the detection of mecA and mecC genes, we used specific forward and reverse gene primers. The primers used in this study were previously used in another study. For primer specificity, we chose the primers by using the Basic Local Alignment Search Tool (BLAST) from NCBI. These primers were confirmed to be suitable for the detection of the mec gene. The primers used are described in Table 1.
Table (1):
Primers used in this study
| Name | Primer sequence | Amplicons | Ref. |
|---|---|---|---|
| mecA Forward | AAAATCGATGGTAAAGGTTGGC- | 533 bp | 2 |
| mecA Reverse | AGTTCTGGAGTACCGGATTTGC- | ||
| mecC Forward | TCACCAGGTTCAAC[Y]CAAAA- | 356 bp | 2 |
| mecC Reverse | CCTGAATC[W]GCTAATAATATTTC- |
The identification of the gene started by preparing the master mix used. The master mix for PCR was prepared by combining GoTaq Green Master Mix 2X, primers (forward and reverse), and nuclease-free water. The PCR mix contains 12.5 µl of GoTaq Green master mix 2X, 10 µM (1 µl) each forward and reverse primer, 8.5 µl of Nuclease-free Water, and 2 µl of DNA sample. The PCR mix was placed in each PCR tube. The mix was then vortexed for homogenization and spun down to put down the sample.
The PCR process began with an initial denaturation at 95 °C for 5 min to melt the DNA strands. Subsequently, 35 cycles of denaturation at 95 °C for 30 s, hybridization at 54 °C for 30 s, and extension at 72 °C for 1 min were performed. Finally, a final extension was carried out at 72 °C for 2 min. Each cycle produced a duplication of the target DNA, and this process yielded the desired results after 35 cycles. The final PCR product then processed to electrophoresis.2,9
Electrophoresis
In the initial steps, a 1% agarose gel was prepared by combining Tris-acetate-ethylenediaminetetraacetic acid (EDTA) buffer and agarose in an Erlenmeyer flask. The solution was heated to boiling, followed by the addition of ethidium bromide and thorough mixing. Next, the solution was poured into a caster and cooled until it solidified for approximately 45 min. The prepared agar was then placed into the electrophoresis chamber, and subsequently, 1X TAE was added to the electrophoresis chamber until the agar was completely submerged.
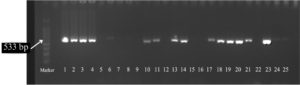
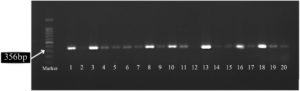
The molecular detection of mecA and mecC was conducted by loading 5 µL of a 100 bp DNA ladder into the first well as a control, followed by subsequent wells filled with 7 µl of PCR amplification products. Electrophoresis was run at a voltage of 80 V for 75 min. Subsequently, DNA bands were visualized under ultraviolet light and documented using the Gel Doc system (BioRad, California, United States of America). Based on the expected size of the amplicon from PCR results, the mecA and mecC resistance genes could be detected by observing the presence of the mecA resistance gene band (~533 bp) and the mecC resistance gene band (~356 bp).2,9,10
Figure 1 shows the mecA gene band and Figure 2 shows the mecC gene band.
In this study, there were no differences between groups that exhibited mecC gene only compared with other variants proven with the bivariate chi-square test. Both groups had relatively similar proportions in terms of sex (p > 0.05), age (p > 0.05), outcome (p > 0.05) and leukocyte count (p > 0.05) (Table 2).
Table (2):
Demographic characteristics of patients with MRSA infection
Categories |
mecC (+) only n = 15 |
Others variant n = 65 |
p-value |
|---|---|---|---|
Sex • Male • Female |
10 (66.7%) 5 (33.3%) |
42 (64.6%) 23 (35.4%) |
0.881 |
Age (years) • • 18-59 • ≥60 Mean ± standard deviation |
8 (53.3%) 0 (0%) 7 (46.7%) 27.13 ± 19.07 |
21 (32.3%) 32 (49.2%) 12 (18.5%) 36.29 ± 23.13 |
0.118 |
Outcome • Recovery • Death |
12 (80%) 3 (20%) |
47 (72.3%) 18 (27.7%) |
0.542 |
Leukocyte counts (× cell/mm3) • • 4,500-11,000 • >11,000 Mean ± standard deviation |
0 (0%) 8 (53.3%) 7 (46.7%) 25.492 ± 5.090 |
5 (7.7%) 24 (36.9%) 36 (55.4%) 15.048 ± 10.031 |
0.342 |
Of the 80 samples examined, most contained the mec gene, whereas one sample showed a negative result for the gene. Most samples (78.75%) had mixed resistance genes (mecA and mecC) (Table 3).
Table (3):
Distribution of mec gene in this study
Variable |
N (%) |
|---|---|
mecA and mecC |
63 (78.75) |
mecA |
1 (1.25) |
mecC |
15 (18.75) |
None |
1 (1.25) |
MRSA infections are often found both as community infections and as infections acquired from health services. The presence of mecC increases the number of antibiotic-resistant S. aureus bacteria. In this study, most MRSA isolates had a mixture of mec genes (78.75%). This is different when compared with research by Idrees et al., in a Pakistani hospital, the majority of MRSA bacteria (57.1%) had a mixture of mecA and mecC genes, and around 7.9% of isolates only had the mecC gene.2 In a meta-analysis study by Diaz et al, (using data from studies in Europe, The United States, and Jordan) the prevalence of MRSA infections carrying mecC reached 0.004%.11 Another study in a tertiary hospital in China using 212 S. aureus samples did not find the presence of the mecC gene.12
Our studies found no difference in demographic and laboratory data between S. aureus that expressed mecC gene only and other phenotypes of S. aureus. This is a novel analysis that we presented. It must be emphasized that mecC has no role as a virulence factor. The mecC gene is a resistance gene that is primarily known to function as a mecA mutation, which encodes a trans-peptidase against β-lactam antibiotics.2
The presence of the mecC gene results in it not being detected through routine PCR examination. The PBP2c protein formed has a higher affinity for oxacillin than cefoxitin; therefore, it can be said that PBP2c has a higher affinity for penicillins than cephalosporins.13 mecC (previously named mecALGA251) is found in the mobile genetic element (MGE) called SCCmec, which is inserted at the 3’end of the orfX locus. The mecC gene is also known to be possessed by other Staphylococci bacteria such as S. xylosus and S. saphrophyticus.14
The origin of mecC is still debated. There is suspicion that the mecC gene comes from livestock due to the high number of cases occurring in people who have contact with livestock. MRSA with the mecC gene is also found in wild animals, such as red foxes, wild rabbits, wild boars, hedgehogs, and fallow deer. Another study reported that MRSA mecC was found in river water, where some wild animals (such as fallow deer) also carried MRSA mecC. This indicates that water can be a means of spreading MRSA bacteria into the environment that originate from various animal species.15
The high incidence of MRSA mecC findings has an impact on reducing the effectiveness of antibiotic therapy, and this will certainly affect patient recovery. MRSA mecC also carries other resistance genes along with regulators of their expression. In previous studies, MRSA was found to be resistant to many antibiotics. Antibiotics such as vancomycin, linezolid, teicoplanin, chloramphenicol, and clindamycin can be used because the resistance rate is lower in MRSA.2 Therefore, routine molecular epidemiological examination of resistance genes such as mec needs to be carried out not only on MRSA bacteria, but also on other bacteria. This is necessary to avoid transmission of the mecC gene from one bacterium to another. These changes in MRSA molecular data will influence local antibiotic therapy guidelines and MRSA infection control.
The limitation of this study is that we did not examine the antibiotic resistance profiles of the isolates. Apart from that, we also did not carry out clonal complex analysis, so we do not know the origin of the MRSA mecC bacteria circulating in our tertiary hospital. However, this research can be used as the main basis for carrying out further molecular examinations not only in North Sumatra but also in other regions in Indonesia.
In this study, we concluded that mecC is highly prevalent in tertiary hospitals in the city of Medan, Indonesia. Our finding can be said to be very high compared to other studies. This study enforces the need for genetic surveillance in hospitals with high patient traffic, especially in urban regions. Multiple genotypic examinations may be necessary to ensure that no resistant strain can evade detection. Further research is needed on this particular topic.
ACKNOWLEDGMENTS
The authors would like to thank the staff of the Clinical Microbiology installation at Adam Malik Hospital and Dr. Rina Yunita, M.D. (Head of the Microbiology Installation Subunit) for assisting with sample collection.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Riedel S, Morse SA, Mietzner TA, Miller S. Jawetz, Melnick & Adelberg’s Medical Microbiology. 28th ed. New York: McGraw-Hill Education; 2019
- Idrees MM, Saeed K, Shahid MA, et al. Prevalence of mecA– and mecC-Associated Methicillin-Resistant Staphylococcus aureus in Clinical Specimens, Punjab, Pakistan. Biomedicines. 2023;11(3):878.
Crossref - Thirafi SZT, Sarassari R, Bramantono, Kuntaman. Susceptibility Patten of Methicillin-Resistant Staphylococcus aureus Bacteria in Dr. Soetomo General Academic Hospital Surabaya. Jurnal Berkala Epidemiologi. 2022;10(3): 331-340.
Crossref - Johan MP, Arden F, Usman MA, et al. Correlation between carriers of Methicillin-resistant Staphylococcus aureus and the incidence of MRSA surgical site infections in orthopedic surgery patients. Eur Rev Med Pharmacol Sci. 2024;28(10):3503-3512.
Crossref - Turbawaty DK, Legito V, Tjandrawati A. Methicillin-Resistant Staphylococcus aureus (MRSA) Patterns and Antibiotic Susceptibility in Surgical and Non-Surgical Patients in a Tertiary Hospital in Indonesia. MKB. 2021; 53(3):148-54.
Crossref - Fitranda M, Salasia SIO, Sianipar O, et al. Methicillin-resistant Staphylococcus aureus isolates derived from humans and animals in Yogyakarta, Indonesia. Vet World. 2023;16(1):239-245.
Crossref - Paterson GK, Morgan FJ, Harrison EM, et al. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother. 2014;69(4):907-910.
Crossref - Venugopal N, Mitra S, Tewari R, et al. Molecular detection and typing of methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci isolated from cattle, animal handlers, and their environment from Karnataka, Southern Province of India. Vet World. 2019;12(11):1760-1768.
Crossref - Amelia S, Kusumawati RL, Hasibuan M, Winda L, Balatif R, Ivander A. Prevalence of Panton-Valentine leucocidin (pvl) and exfoliative toxin A ( eta) gene within methicillin resistant and susceptible Staphylococcus aureus in an urban tertiary hospital: A molecular epidemiology pilot study. F1000 Res. 2024;12:1002.
Crossref - Amelia S, Wahyuni DD, Yunita R, Rozi MF. The active surveillance of Staphylococcus aureus using polymerase chain reaction-based identification method among hospitalized-patient of haji adam malik general hospital, medan, indonesia. Open Access Maced J Med Sci. 2021;9:622-625.
Crossref - Diaz R, Ramalheira E, Afreixo V, Gago B. Methicillin-resistant Staphylococcus aureus carrying the new mecC gene-a meta-analysis. Diagn Microbiol Infect Dis. 2016;84(2):135-140.
Crossref - Hou Z, Xu B, Liu L, et al. Prevalence, drug resistance, molecular typing and comparative genomics analysis of MRSA strains from a tertiary A hospital in Shanxi Province, China. Front Microbiol. 2023;14:1273397.
Crossref - Ba X, Harrison EM, Lovering AL, et al. Old Drugs To Treat Resistant Bugs: Methicillin-Resistant Staphylococcus aureus Isolates with mecC Are Susceptible to a Combination of Penicillin and Clavulanic Acid. Antimicrob Agents Chemother. 2015;59(12):7396-7404.
Crossref - Abdullahi IN, Latorre-Fernandez J, Reuben RC, et al. Beyond the Wild MRSA: Genetic Features and Phylogenomic Review of mecC-Mediated Methicillin Resistance in Non-aureus Staphylococci and Mammaliicocci. Microorganisms. 2023;12(1):66.
Crossref - Sahin-Toth J, Albert E, Juhasz A, et al. Prevalence of Staphylococcus aureus in wild hedgehogs (Erinaceus europaeus) and first report of mecC-MRSA in Hungary. Sci Total Environ. 2022;815:152858.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.