ISSN: 0973-7510
E-ISSN: 2581-690X
The expected rise in world population and variability of climate change cause biotic and abiotic stress conditions that add uncertainty and complexity to food security and agro-industries. Plants are physiologically, biochemically, and molecularly affected when exposed to stressful conditions. Endophytic microbes that inhabit internal plant tissues without causing tissue damage or disease symptoms play a prominent role in the growth and development of host plants under both normal and abnormal conditions. In the current study, a pot experiment was conducted to verify that the same bacteria with multiple plant growth-promoting traits and osmotolerance were inoculated onto surface-sterilized maize seeds sown in sterile soil, re-isolated from these seedlings, and tested for their endophytic colonization to fulfill Koch’s postulate, proving their endophytic competence and persistence. The bacterial isolates were found to colonize plants at levels ranging from 4.30 to 5.26 Log10 CFU g-1, and the maximum colonization of inoculated isolates was observed in roots, followed by stems, and least in leaves. The re-isolated bacteria were compared with inoculated isolates in terms of their carbon source utilization, antibiotic sensitivity, and 16S rRNA gene sequences, thus determining which endophytic bacteria had the ability to colonize and persist at high levels in plant hosts by experimentally inoculating plants.
Plant Growth Promotion, Endophytic Colonization, Re-isolation
Few authors have proposed a functional definition of endophytic behavior, considering any bacterium as an endophyte if it can be isolated from surface-disinfested plant tissue or extracted from inside the plant, and if it does not visibly harm the plant. Endophytes are defined as fungi or bacteria, which for all or part of their life cycle invade the tissues of living plants and cause unapparent and asymptomatic infections entirely within plant tissues, but no symptom of disease.1
Plants are greatly tentative on microorganisms capable of improving their metabolic activity to withstand stress conditions.2 Under adverse conditions microbes foresight the cell metabolism of plants, enabling microbe-treated plants to respond more swiftly than their untreated counter plants.3 Thereupon employing beneficial microbial inoculants for enhancing plant growth and to combat adverse conditions could provide a pathway for economical and environmentally sustainable agriculture.
Endophytic bacteria which are inhabitants of plant tissues are advantageous over rhizospheric bacteria because of its direct contact and have no competition for nutrition or habitat with rhizospheric microorganisms.4 Moreover, the aptitude of plant growth and stress alleviation by endophytic bacteria was well studied.5-7 The plant and endophytic bacterial interaction improve the drought stress tolerance8 and have been proved have different defense mechanisms against plant pathogens.9
Microbial colonization in endophytic tissues of plant is a complex process that requires the capacity of bacteria to compete in the rhizosphere soil to find a place to communicate and interact with the plant roots. Root exudates are chemically diverse and include those molecules involved in attracting microorganisms to the root, or in the case of endophytes, to be able to colonize the internal plant tissues.
Endophytes are present in different plant compartments such as apoplast, and vascular elements, and less frequently on the inner parts of cells.10 To achieve a successful colonization, endophytes require a compatible host plant.11 The endophytic colonization and movement of microorganisms may be greatly influenced by the micro-environment, as well as developmental and environmental factors.12,13
Bacterial strains
Potential plant growth promoting bacterial isolates Kosakonia radicincitans strain GAD1813 (MW822555), Priestia megaterium strain GAD181_8 (MW880714), Bacillus licheniformis strain GAD181_11 (MW880717), Pseudomonas aeruginosa, Klebsiella pneumoniae strain DSM 30,104 (MW812251) and Methylorubrum populi strain GAD18_16 (MZ484406) isolated from maize plant tissues14 were used to study their colonization ability inside the plant to confirm their endophytic nature.
Inoculation of endophytic bacteria into maize plants
Inoculum was prepared by pelleting the bacteria grown to the mid-log phase by centrifugation (Centrifuge REMI R-248M). Washed the pellet twice and suspended in phosphate buffer to maintain inoculation density of 108 per mL. Surface sterilization of maize seeds were done with 70% ethanol for 40 seconds and 2.5% NaOCl for 20 minutes followed by four washings with sterile distilled water. Surface sterilized seeds were primed with bacterial isolates by soaking in individual bacterial suspensions for about 2 hrs. Treated seeds were air dried and sown in clay pots having 12 inch diameter and 12 inch depth with three seeds in each pot filled with sterilized loamy soil to avoid indigenous soil microorganisms. Each treatment was maintained in triplicates.
After 7 to 14 days of seed germination, when plants reach height of 7.6 cm and diameter around 0.5 cm, bacterium was inoculated in triplicates. Around 1 cm over the crown of the plant bacterial suspension was injected horizontally through the stem of the seedling with the help of a 26-gauge needle and a syringe. Inoculation method was replicated by passing inoculum with needle perpendicular to the first made channel by rotating plant to 90°. After 15 days of germination bacterial isolates were applied to soil at the rate of 1 mL (108) of suspension near the root zone. After 20-25 DAS bacterial inoculation was done through foliar spay at the rate of 1 mL (108) of inoculum per plant.
Re-isolation of endophytic bacteria from inoculated plants
The endophytic bacteria were re-isolated from inoculated plants at 40-45 days after sowing to confirm their endophytic nature and survivability. Respective plant samples were ramified into leaves, stem and root separately for surface sterilization. To preclude passive diffusion of chemical disinfectants used in surface sterilization into the plant tissues osmotic pressure was maintained by soaking phosphate buffered saline (PBS NaCl 8 g, KCl 0.2 g, Na2HPO4 1.44 g, KH2PO4 0.245 g, Distilled water 1 L) at pH 7.0 for 15 minutes.15 Each plant tissue weighing about 0.5 g, cut with sterile knife into small pieces of around 1 cm, and roots are surface sterilized with 70% ethanol and 2.5% sodium hypochlorite for 40 seconds and 20 minutes, respectively, while leaves and stem were surface sterilized with same concentration of disinfectants but with 20 seconds and 10 minutes of duration respectively. To remove chemical residues the tissue pieces were washed with sterile distilled water for 4 times later with help of sterile motor and pestle the tissues were ground in 1 ml of 0.9% sodium chloride. The last washed water (100 µl) was spread plated onto nutrient agar as a sterile control. Tissue extracts were diluted up to 10-4 and 100 µl of each dilution was spread plated onto nutrient agar plates (Peptone 5 g, NaCl 5 g, Yeast extract 3 g, Agar Agar 20 g, Distilled water 1 L) and incubated at 28 °C for 2-3 days. Attained morphologically similar bacterial isolates as of inoculated ones were used for further screening.16
Qualitative comparison of recovered isolates with inoculated isolates
The re-isolated morphologically similar bacterial colonies were referred with the original cultures qualitatively by characterizing phenotypically, antibiotic resistance profiles and carbohydrate utilization to confirm their identity with inoculated isolates. Firstly phenotypic attributes viz., color, texture, elevation, margin, opacity, surface of re-isolated bacterial colonies were identified. Secondly, bacterial isolates were evaluated for antibiotic resistance by placing antibiotic discs of different concentration on already spread plated agar plates. The antibiotics evaluated are sulphatriad, streptomycin, ampicillin, penicillin G, chloramphenicol and tetracycline (Himedia, India).7 And then the carbon source utilization of recovered isolated were evaluated using peptone broth supplemented with different carbohydrates viz., glucose, sucrose, dextrose and lactose individually.17
Comparision of 16S rRNA sequences of recovered isolates with inoculated isolates
The 10 re-isolated bacteria which were confirmed as inoculated ones by the above tested qualitative parameters, further confirmed by sequencing of 16S rRNA gene and blasting with NCBI gene bank. The isolation of DNA from bacterial isolates was done and amplified using universal primers18 16S rRNA-27F (AGAGTTTGATCCTGGCTCAG) and 16S rRNA-1492R (TACGGTTACCTTGTTACGACTT) using BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer. The quality of high molecular weight DNA was evaluated on 1.0% agarose gel. By Sanger’s chain termination method19 the amplified genes were sequenced in both directions using aligner software (Barcode Biosciences, Banglore, India). The consensus sequences obtained from forward and reverse sequences of 16S rRNA gene were BLAST checked in NCBI GenBank database and identified at species level based on similarity index. The obtained sequences of re-isolated bacteria were compared with the inoculated bacterial sequences constructing phylogenetic tree by using multiple alignment software program in MEGA X 10.0 version.20
The effective osmotolerant and plant growth promoting endophytic isolates which were previously screened21 for their biochemical characters, osmotolerant activity, antagonistic activity and seed vigor index for development of microbial consortium and selected 7 isolates namely Kosakonia radicincitans (NL3E3), Priestia megaterium (JC3E2), Priestia aryabhattai (PL3E2) Bacillus licheniformis (VaR3E1), Pseudomonas aeruginosa (LS3E1), Klebsiella pneumoniae (LS3E3) and Methylorubrum populi (LL3E1) for compatibility and endophytic colonization studies.
Compatibility studies
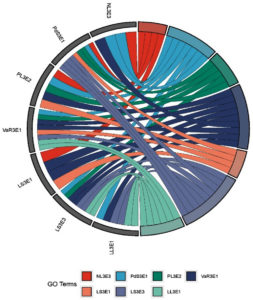
For checking the compatibility among the selected efficient endophytic bacteria cross streaking method was followed on nutrient agar medium. Kosakonia radicincitans (NL3E3) was found compatible with the isolates Priestia megaterium, Priestia aryabhattai and Pseudomonas aeruginosa while the growth was inhibited by the isolates Bacillus licheniformis, Klebsiella pneumoniae and Methylorubrum populi. The endophytic bacterial isolate Priestia megaterium (JC3E2) was found compatible with all the tested isolates except Pseudomonas aeruginosa as an inhibition zone was observed around P. aeruginosa when streaked on a mat of P. megaterium on solid nutrient agar medium. An inhibition zone was observed around P. aeruginosa and M. populi when streaked on Priestia aryabhattai (PL3E3) bacterial lawn. Bacillus licheniformis (VaR3E1) was found compatible with all the tested isolates except for K. radicintans. The endophytic bacterial isolate Pseudomonas aeruginosa (LS3E1) was found compatible with only B. licheniformis and K. pneumoniae while inhibiting the growth of all other tested isolates. Klebsiella pneumoniae (LS3E3) was found compatible with P. megaterium, B. licheniformis and M. populi. The pink pigmented methylotrophic bacteria Methylorubrum populi (LL3E1) was found compatible with P. megaterium, B. licheniformis and K. pneumoniae (Table 1). A graphical image (Figure 1) was created by R version 4.1.1.9 for clear understanding of compatibility among the bacterial isolates. The same colored lines which are drawn towards different isolates are proved as compatible with each other.
Table (1):
Evaluation of compatibility among the effective endophytic bacterial isolates
NL3E3 |
PdS3E1 |
PL3E2 |
VaR3E1 |
LS3E1 |
LS3E3 |
LL3E1 |
|
|---|---|---|---|---|---|---|---|
Kosakonia radicincitans (NL3E3) |
+ |
+ |
+ |
– |
+ |
– |
– |
Priestia megaterium (JC3E2) |
+ |
+ |
+ |
++ |
– |
++ |
++ |
Priestia aryabhattai (PL3E2) |
+ |
+ |
+ |
+ |
– |
+ |
– |
Bacillus licheniformis (VaR3E1) |
– |
+ |
+ |
+ |
+ |
++ |
++ |
Pseudomonas aeruginosa (LS3E1) |
+ |
– |
– |
+ |
+ |
+ |
– |
Klebsiella pneumoniae (LS3E3) |
– |
++ |
+ |
+ + |
+ |
+ |
+ |
Methylorubrum populi (LL3E1) |
– |
+ |
– |
++ |
– |
++ |
+ |
– Inhibition zone formed, + Low bacterial growth, + +More bacterial growth
Inoculation of maize plants with bacterial endophytes
The effective bacterial cultures were inoculated to maize individually and in combinations as consortium based on their compatibility with each other. Four methods of inoculation including seed bacterization, soil drenching, stem injection and foliar application were followed to ensure the infection of applied bacterial culture into plants because a single method may not be equally effective for all bacterial strains.
Endophytic bacterial re-isolation from treated maize plants
A pot experiment was conducted to fulfill Koch’s postulate by verifying that bacteria inoculated to surface-sterilized maize seeds sown in sterile soil could be re-isolated from the seedlings. The study also examined the endophytic competence of the bacteria by determining their ability to colonize and persist at high levels in experimentally inoculated plant hosts.
Seven bacterial isolates were inoculated in maize plants as individual isolates and in consortium. Based on morphological characterization of re-isolated bacterial colonies by comparing with inoculated isolates, microbial load in each tissue (root, stem and leaf) were taken at 40-45 days after sowing (DAS). The plants were not treated with any other pesticide which may affect the establishment of inoculum in plants and there is no incidence of pest or establishment of disease was observed within 40-45 DAS. The population count of each inoculum in different plant niche were given in Table 2.
Table (2):
Endophytic bacterial re-isolation from treated maize plants
| Treatments | Log10 CFU/g (fresh weight) | |||
|---|---|---|---|---|
| Root | Stem | Leaf | ||
| T1 (Kosakonia radicincitans NL3E3) | 4.60 | 4.30 | – | |
| T2 (Priestia megaterium JC3E2) | 5.20 | 5.11 | 4.60 | |
| T3 (Priestia aryabhattai PL3E2) | 4.90 | 4.60 | 4.90 | |
| T4 (Bacillus licheniformis VaR3E1) | 5.26 | 4.85 | – | |
| T5 (Pseudomonas aeruginosa LS3E1 | 4.30 | 4.60 | – | |
| T6 (Klebsiella pneumoniae LS3E3) | 5.08 | 5.00 | 4.78 | |
| T7 (Methylorubrum populi LL3E1) | 4.78 | – | 5.08 | |
| T8 (No inoculum- indegenious bacteria) | 4.48 | – | 4.60 | |
| MC1 (T1+T3+T6) | NL3E3 | 4.90 | – | – |
| PL3E2 | – | – | 4.48 | |
| LS3E3 | 4.90 | 5.00 | – | |
| MC2 (T1+T5+T6) | NL3E3 | 4.60 | 4.30 | 4.60 |
| LS3E1 | 4.78 | 4.48 | – | |
| LS3E3 | 5.08 | 4.90 | 4.30 | |
| MC3 (T1+T2+T4+T6) | NL3E3 | – | – | 4.48 |
| JC3E2 | 5.11 | 4.90 | – | |
| VaR3E1 | 5.23 | 4.78 | – | |
| LS3E3 | 4.90 | 5.04 | 4.60 | |
| MC4 (T2+T4+T6+T7) | JC3E2 | 5.04 | 4.60 | 4.00 |
| VaR3E1 | 5.20 | 5.00 | – | |
| LS3E3 | 5.08 | 4.90 | 4.60 | |
| LL3E1 | 4.60 | – | 4.85 | |
In the present investigation some population of indigenous bacteria was also reported in T8 regardless of using sterilized soil as potting media which might be the seed born or seed transmitted microbes (Table 2). Similarly in early growth stage of maize rhizosphere were found have dominant 16S rDNA sequences in both sterile and non-sterile potting media considering as seed transmitted bacterial cells22 and it was reported that maize rhizosphere contains wide range of bacteria from soil and seed dominated by species of Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes.
Qualitative examination of recovered isolates with inoculated isolates
The highly similar colony morphologies of re-isolated endophytic bacteria when compared with inoculated bacterial colonies were selected for further confirmation of their identity. About 15 re-isolated strains RI1T1R, RI7T2S, RI4T3R, RI5T12R, RI6T10S, RI8T7L, RI10T12L, RI9T5S, RI11T9S, RI2T9L, RI3T11R, RI12T4R, RI13T10L, RI14T9S and RI15T12R were further examined for antibiotic sensitivity and carbon source utilization.
Profiling of Antibiotic sensitivity
Based on colony morphology RI1T1R was identified as Pseudomonas and their antibiotic sensititivity has also shown similar pattern as of inoculated P. aeruginosa. The re-isolated strains RI7T2S and RI3T11R were identified as Priestia megaterium and their antibiotic sensitivity assay has shown that all the tested antibiotics were in similar pattern with inoculated P. megaterium except for Streptomycin (10 mcg) against RI7T2S which has shown resistant. RI4T3R and RI14T9S were identified as P. aryabhattai morphologically but they differed in sensitivity against Streptomycin (10 mcg) and Tetracyclin (25 mcg) respectively when compared with the inoculated isolate. The isolates RI5T12R, RI12T4R and RI15T12R were phenotypically identified as Bacillus licheniformis and the trend of antibiotic sensitivity of all the three isolates were found similar with the inoculated isolate B. licheniformis. The isolates RI6T10S, RI2T9L and RI11T9S were identified as Klebsiella pneumoniae former two isolate was found similar with the inoculated isolate in antibiotic assay while the later isolate was varied with the sensitivity against Ampicillin (10 mcg). The re-isolated strains RI8T7L and RI10T12L were phenotypically identical with Methylorubrum populi and the results of antibiotic sensitivity assay has also obtained in similar with the inoculated isolate (Table 3).
Table (3):
Assay of re-isolated endophytic bacteria against antibiotic sensitivity
No. |
Isolate code |
Expected isolate |
Ampicillin (10 mcg) |
Chloramphenicol (25 mcg) |
Penicillin-G (0.6 mcg) |
Streptomycin (10 mcg) |
Sulphatriad (300 mcg) |
Tetracyclin (25 mcg) |
|---|---|---|---|---|---|---|---|---|
1 |
RI1T1R |
Pseudomonas aeruginosa |
R |
S |
R |
R |
R |
S |
2 |
RI7T2S |
Priestia megaterium |
R |
S |
S |
S |
S |
S |
3 |
RI4T3R |
Priestia aryabhattai |
R |
S |
S |
R |
S |
S |
4 |
RI5T12R |
Bacillus licheniformis |
R |
S |
R |
S |
S |
S |
5 |
RI6T10S |
Klebsiella pneumoniae |
R |
S |
R |
S |
S |
S |
6 |
RI8T7L |
Methylorubrum populi |
S |
S |
S |
R |
S |
R |
7 |
RI10T12L |
Methylorubrum populi |
S |
S |
S |
R |
S |
R |
8 |
RI9T5S |
Pseudomonas aeruginosa |
S |
R |
S |
R |
S |
R |
9 |
RI11T9S |
Klebsiella pneumoniae |
S |
S |
S |
R |
S |
R |
10 |
RI2T9L |
Klebsiella pneumoniae |
R |
S |
R |
S |
S |
S |
11 |
RI3T11R |
Priestia megaterium |
R |
S |
S |
S |
S |
S |
12 |
RI12T4R |
Bacillus licheniformis |
R |
S |
R |
S |
S |
S |
13 |
RI13T10L |
Kosakonia radicincitus |
R |
S |
R |
S |
S |
S |
14 |
RI14T9S |
Priestia aryabhattai |
S |
S |
S |
R |
S |
R |
15 |
RI15T12R |
Bacillus licheniformis |
R |
S |
R |
S |
S |
S |
Bold font– Recovered isolates showing dissimilarity in antibiotic sensitivity with inoculated isolates
Profiling of carbon source utilization
All the re-isolated bacterial strains have shown similar pattern of carbon source utilization as of their inoculated bacterial isolates except for four including RI11T9S, RI12T4R, RI13T10L and RI14T9S which are expected as the isolates Klebsiella pneumoniae, Bacillus licheniformis, Kosakonia radicincitus and Priestia aryabhattai respectively (Table 4).
Table (4):
Assay of re-isolated endophytic bacteria against various carbon source utilization
No. |
Isolate code |
Expected isolate |
Dextrose |
Sucrose |
Maltose |
Lactose |
|---|---|---|---|---|---|---|
1 |
RI1T1R |
Pseudomonas aeruginosa |
+ |
+ |
+ |
– |
2 |
RI7T2S |
Priestia megaterium |
+ |
+ |
+ |
+ |
3 |
RI4T3R |
Priestia aryabhattai |
+ |
+ |
+ |
– |
4 |
RI5T12R |
Bacillus licheniformis |
+ |
+ |
+ |
– |
5 |
RI6T10S |
Klebsiella pneumoniae |
+ |
+ |
+ |
+ |
6 |
RI8T7L |
Methylorubrum populi |
+ |
+ |
+ |
– |
7 |
RI10T12L |
Methylorubrum populi |
+ |
+ |
+ |
– |
8 |
RI9T5S |
Pseudomonas aeruginosa |
+ |
+ |
– |
– |
9 |
RI11T9S |
Klebsiella pneumoniae |
+ |
+ |
+ |
– |
10 |
RI2T9L |
Klebsiella pneumoniae |
+ |
+ |
+ |
+ |
11 |
RI3T11R |
Priestia megaterium |
+ |
+ |
+ |
+ |
12 |
RI12T4R |
Bacillus licheniformis |
+ |
+ |
+ |
– |
13 |
RI13T10L |
Kosakonia radicincitus |
+ |
+ |
+ |
– |
14 |
RI14T9S |
Priestia aryabhattai |
+ |
+ |
– |
– |
15 |
RI15T12R |
Bacillus licheniformis |
+ |
+ |
+ |
– |
Bold font– Recovered isolates showing dissimilarity in carbon source utilization with inoculated isolate
The re-isolated bacteria displaying variations in antibiotic resistance and carbohydrate utilization compared to the original strains were excluded from further studies such as sequencing. Although they were comparable in colony morphology and certain other traits, they underwent additional screening focused on antibiotic resistance and carbohydrate utilization to ensure accurate confirmation of their similarity with the originally inoculated strains. Only those isolates that demonstrated a high degree of similarity across all characteristics were considered as the inoculated ones and were subsequently verified through 16S rRNA sequencing.
Quantitative examination of recovered isolates with inoculated isolates
The recovered isolates which were confirmed to be inoculated ones based on morphology, antibiotic sensitive assay and carbon source utilization assay (RI1T1R, RI7T2S, RI4T3R, RI5T12R, RI6T10S, RI8T7L, RI10T12L, RI2T9L and RI3T11R) were further examined for their similarity with 16S rRNA gene sequences as of inoculated bacterial isolates.
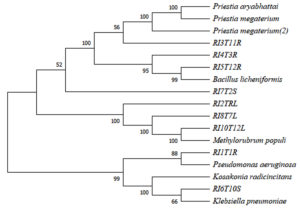
The 16S rRNA gene of re-isolated strain RI5T12R was 99.82% homologous with the 16S rRNA gene of the Bacillus licheniformis. The recovered isolate RI6T10S has exhibited 98.92% of similarity of 16S rRNA gene with Klebsiella pneumoniae gene. The 16S rRNA gene of re-isolated strain RI10T12L was found 99.40% of homologous with 16S rRNA gene of Methylorubrum populi. The 16S rRNA gene of the recovered isolate RI3T11R has shown 97.06% and 97.07% of similarity with the 16S rRNA gene of the strains Priestia megaterium and Priestia aryabhattai (Table 5). In phylogentic tree analysis RI3T11R was found to have similarity with the cluster of P. megaterium and P. aryabhattai, re-isolates RI4T3R and RI5T12R were found to have similarity with B. licheniformis with 95% and 99% respectively. The recovered strains RI8T7L, RI10T12L and RI2T9L were found to have 100% similarity with the inoculated isolate M. populi. RI1T1R have shown 88% similarity index with P. aeruginosa (Figure 2).
Table (5):
Comparison of recovered isolates with inoculated endophytic bacterial isolates based on 16S rRNA gene sequence
| No. | Isolate code | Expected bacterial strain | Accession number of inoculated endophytic bacteria | Identified bacteria (homologous with sequence run in NCBI database) | Similarity | NCBI Accession number |
|---|---|---|---|---|---|---|
| 1 | RI1T1R | Pseudomonas aeruginosa | — | Pseudomonas hibiscicola | 99.72% | NR_024709.1 |
| 2 | RI7T2S | Priestia megaterium | MW880711 | Arthrobacter pascens | 98.13% | NR_02619.1 |
| 3 | RI4T3R | Priestia aryabhattai | MW880712 | Bacillus subtilis | 100.0% | NR_118383.1 |
| 4 | RI5T12R | Bacillus licheniformis | MW880717 | Bacillus licheniformis | 99.82% | NR_118996.1 |
| 5 | RI6T10S | Klebsiella pneumoniae | MW812251 | Klebsiella pneumoniae | 98.92% | NR_113702.1 |
| 6 | RI8T7L | Methylorubrum populi | MZ484406 | Methylobacterium radiotolerance | 98.98% | NR_074244.1 |
| 7 | RI10T12L | Methylorubrum populi | MZ484406 | Methylorubrum populi | 99.40% | NR_074257.1 |
| 8 | RI2T9L | Klebsiella pneumoniae | MW880713 | Ochrobactrumgrignonense | 99.41% | NR_028901.1 |
| 9 | RI3T11R | Priestia megaterium | MW880714 | Priestia megaterium | 97.06% | NR_117473.1 |
| MW880711 | Priestia aryabhattai | 97.07% | NR_115953.1 |
Endophytic bacterial re-isolation from treated maize plants
The present investigation shows that from the root tissues recovery of inoculated bacteria was found to have higher magnitude while the recovery from the arial plant parts (stem and leaves) has shown lower magnitude of endophytic bacterial population. Almost all the inoculated bacteria in single or in combinations as consortium were found to recovered from roots while few isolates were not been observed in arial plant parts. In correlation with our results it has been proved that lower root tissues with more number of CFU/g fresh weight and leaves and stem were found with lesser magnitude of endophytic bacterial CFU g-1.23
Results have demonstrated that nonpathogenic endophytes can be re-isolated from maize plants following different inoculation techniques. Furthermore, the success of process depends on method of inoculation and endophytic bacterial isolate. The unique traits of each isolate seem to play crucial role in its ability to inhabit plants internal tissues. The results suggest a method will have to be tested specifically for the endophytic bacterial isolate to be delivered. Therefore, practical delivery of different isolates to maize plants may require different methods and the relationship of the biological control properties to the ability of the endophytic bacterial isolate to colonize plant tissues has yet to be examined.
Compatibility studies
In terms of technical difficulty and potential effects, programming interactions between multiple bacterial populations represents a new frontier in synthetic biology, it is becoming more and more accepted. Bioinoculants with multiple populations, also known as consortia, may offer some advantages over those with just one population. Such as formation of biofilm by well-established consortium which is comparatively more resistant against invasion by other microbes, distinct compatible populations share essential metabolites for the growth of sub populations under sudden nutrient deprivation conditions and two or more sub populations can efficiently perform metabolically by cooperative division of labor between them. In the present study compatibility among the efficient endophytic bacterial populations was performed to develop a well-established consortium because under a variety of soil and environmental conditions, the consistency, dependability, and efficacy of the microbes may be enhanced through consortium application. Kosakonia radicincitans (NL3E3) was found compatible with the isolates Priestia megaterium, Priestia aryabhattai and Pseudomonas aeruginosa while the growth was inhibited by the isolates Bacillus licheniformis, Klebsiella pneumoniae and Methylorubrum populi. The endophytic bacterial isolate Priestia megaterium (JC3E2) was found compatible with all the tested isolates except Pseudomonas aeruginosa. Bacillus licheniformis (VaR3E1) was found compatible with all the tested isolates except for K. radicintans. The endophytic bacterial isolate Pseudomonas aeruginosa (LS3E1) was found compatible with only B. licheniformis and K. pneumoniae. Klebsiella pneumoniae (LS3E3) was found compatible with P. megaterium, B. licheniformis and M. populi. The pink pigmented methylotrophic bacteria Methylorubrum populi (LL3E1) was found compatible with P. megaterium, B. licheniformis and K. pneumoniae. Based on obtained results different combinations of microbial consortia can be developed and similar observations can be witnessed in many research publications.
The cogitation of fusing various fungal and bacterial species or strains termed as microbial consortia has gained commendation for its effective benefits over single microbe inoculation. The consortia showed efficacy in mitigating biotic and abiotic stress also in managing many crop diseases and pests,24 on the other hand single strain application were observed to be more prone to environmental stress and sensitive to encountered indigenous microbes25 and limit their activity to only one or two growth promoting traits.26 In contrast, group of compatible microbes known as consortia having multifaceted growth promoting and stress mitigating attributes will have wider adaptability to various environmental conditions and so functional flexibility.27,28
Five bacterial isolates, chosen from in vitro screening for consortium development, underwent mutual compatibility testing using the cross streak method. The combinations of TRB-1 with VSB-1, EkRB-1 with TRB-1, and EkRB-1 with VSB-1 showed no clearance zone at their intersection points, indicating compatibility among these isolates.29
It was also reported that endophytes Methylobacterium oryzae CBMB20 and Bacillus megaterium LNL6 did not produce any zone of clearance on the plates preceded with Bradyrhizobium japonicum MN110 which were in accordance with the present investigation.30 positive interaction between Mesorhizobium sp. and Piriformospora indica with different Pseudomonas sp. in plate culture method was also been proved.23
Endophytic colonization studies
In the present study, endophytic competence (infection and persistence characteristics) of the inoculated bacterial isolates were examined as survival in the intracellular plant niche after inoculation likely involve a specific adaptation to new environment with competition against indigenous endophytic microbes. The bacterial isolates were found to colonize plants at the levels ranging from 4.30 to 5.26 Log10 CFU g-1. Similar bacterial titers were observed in corn and sorghum having the average colonization levels of 6.1 ± 0.1 log10 CFU g-1 (fresh weight)17 and in prairie plant out of 86 endophytes 28 were found to inhabit its host tissues after 42 days of inoculation in population range of 3.5 to 7.7 log10 CFU g-1 (fresh weight).31 In tomato and pomato endophytic bacterial population was reported in range of 3.0 and 5.0 log10 CFU g-1 of plant tissue.32
Maximum colonization of inoculated isolates was observed in roots followed by stem and least in leaves. Similar results were found in Alyssum bertolonii when plant colonization studies were conducted for 23 different taxonomic groups by inoculating in stelire grown plant seedlings and reportedly more number of recovered endophytes were found in roots than in stem and leaves.23 In the present investigation among the recovered isolates the population density of Bacillus licheniformis was found to be highest recording maximum of 5.26 Log10 CFU g-1 in roots of maize while the lowest population density was shown by Kosakonia radicincitus both in individual inoculation as well as in consortium.
From our study it was also interpreted that the inoculated bacterial isolate could have movement in plant tissues during growth stages as particular tissue specific isolate was observed to re-isolated also from other tissues of plant. As in case of Priestia megaterium and Priestia aryabhattai which are leaf colonizers at first but they were re-isolated from all the three tissues. Bacillus licheniformis which was initially root colonizer could only re-isolated from roots and stem but not the leaf. Pseudomonas aeruginosa was also re-isolated from only roots and stem indicating that non colonizer of leaf. Klebsiella pneumoniae which was initially stem colonizer was re-isolated from all the three tissues while Methylorubrum populi, a leaf colonizer was isolated from only roots and leaves but not stem (Table 2).
It is likely that endophytes alter the functional characteristics that enable them to interact with the host plant and swiftly respond to adverse growth conditions. The fact that distinct endophyte communities are found in various stages of the host life cycle and environmental conditions suggests that particular functional groups of bacteria are likely to be active in response to a particular stress.33
Qualitative examination of recovered isolates with inoculated isolates
Antibiotic sensitivity and carbon fermenting ability of microorganisms were taken as qualitative traits for assaying re-isolated bacteria to confirm its identity as prior inoculated bacteria that proves their invasiveness and persistence in stable endophytic colonization. Prior to the inoculation the antibiotic sensitivity and carbon fermentation tests were done to the bacterial isolates and same tests were repeated for that isolates resembling morphologically identical with the inoculated bacteria even after re-isolation. And the recovered isolates showing similar patterns as of inoculated bacteria in all the tests were confirmed for their endophytic colonization. The re-isolated strains RI7T2S and RI3T11R were identified as Priestia megaterium as their antibiotic sensitivity assay has shown that all the tested antibiotics were in similar pattern with inoculated P. megaterium. The isolates RI5T12R, RI12T4R and RI15T12R were phenotypically identified as Bacillus licheniformis and the trend of antibiotic sensitivity of all the three isolates were found similar with the inoculated isolate B. licheniformis. The isolates RI6T10S, RI2T9L were identified as Klebsiella pneumoniae and the re-isolated strains RI8T7L and RI10T12L were phenotypically identical with Methylorubrum populi and the results of antibiotic sensitivity assay has also obtained in similar with the inoculated isolate. Except for four recovered isolates (RI11T9S, RI12T4R, RI13T10L and RI14T9S) rest of all re-isolated bacterial strains have shown similar pattern of carbon source utilization as of their inoculated bacterial isolates. Similar tests were used earlier to prove the endophytic colonization ability and results has demonstrated that carbon source utilization tests of re-isolated prairie plant endophytes identified LB030 as M. testaceum, PD039 as Clavibacter michiganensis subsp. insidiosus, and SG041 as Curtobacterium citreum.31
Quantitative examination of recovered isolates with inoculated isolates
Molecular assay was employed for quantitative examination of re-isolates by comparing the 16S rRNA gene sequences of recovered isolates with 16S rRNA gene sequences of inoculated bacterial isolates by PCR amplification. The recovered isolate RI5T12R was identified as Bacillus licheniformis the isolate RI6T10S was found to have high similarity with the gene sequence of Klebsiella pneumoniae. The results indicated that the recovered strain RI10T12L was found to have 100% similarity with the inoculated isolate Methylorubrum populi. The recovered isolate RI3T11R was found to have 97.06% sequence similarity with Priestia megaterium and 97.07% sequence similarity with P. aryabhattai. Hence the isolates B. licheniformis, K. pneumoniae, M. populi, P. megaterium and P. aryahattai were confirmed as endophytic colonizers as they have fulfilled Koch’s postulates.
It has been proved that the original inoculated strain could be recovered from host by fulfilling Koch’s postulates in rice inoculated with various strains of Azorhizobium caulinodans using rep-PCR genomic fingerprinting,34 Clavibacter, Cellulomonas, Curtobacterium, and Microbacterium were proved to have promising ability to colonize in prairie and agronomic plants by 16S rRNA gene sequencing.31 Three new rhizobial endophytic bacterial strains of which two are phylogenitically close to Burkholderia cepacia complex while other strain is in affinity with Rhizobium leguminosarum bv. phaseoli were reported to colonize the rice root tissues using gusA reporter gene tagged construct.35
The results provided empirical evidence for the colonization of maize endophytic bacteria. It has demonstrated that nonpathogenic endophytes can be recovered from maize plants following inoculation by different delivery methods. It was also interpreted that the inoculated bacterial isolate could have movement in plant tissues during growth stages as particular tissue specific isolate was observed to re-isolated also from other tissues of plant. Compatibility among the efficient endophytic bacterial populations was performed to develop a well-established consortium because under a variety of soil and environmental conditions, the consistency, dependability, and efficacy of the microbes may be enhanced through consortium application. By employing qualitative methods like antibiotic sensitivity and carbon fermentation and quantitative method like 16S rRNA sequencing the inoculated bacterial isolates including the isolates B. licheniformis, K. pneumoniae, M. populi, P. megaterium and P. aryahattai were confirmed for their endophytic colonization. Culturing of endophytes by classic microbiological methods to study their intracellular colonization may find difficulty in practice and also to get accurate results stronger reliance on advanced techniques like fluorescent tagging, fish and metagenomics could be used for further study of endophytic ecology of microorganisms.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
Not applicable.
ETHICS STATEMENT
Not applicable.
- Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43(10):895-914.
Crossref - Kavamura VN, Santos SN, da Silva JL, et al. Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiol Res. 2013;168(4):183-91.
Crossref - Compant S, Duffy B, Nowak J, Clement C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71(9):4951-4959.
Crossref - Naveed M, Mitter B, Yousaf S, Pastar M, Afzal M, Sessitsch A. The endophyte Enterobacter sp. FD17: a maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol Fertil Soils. 2014;50:249-262.
Crossref - Chandran V, Shaji H, Mathew L. 7-Endophytic microbial influence on plant stress responses. Microbial Endophytes. 2020:161-193.
Crossref - Ullah A, Nisar M, Ali H, et al. Drought tolerance improvement in plants: an endophytic bacterial approach. Appl Microbiol Biotechnol. 2019;103(18):7385-7397.
Crossref - Sandhya V, Shrivastava M, Ali SZ, Prasad VSSK. Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russian Agricultural Sciences. 2017;43:22-34.
Crossref - Paul D, Lade H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: a review. Agron Sustain Dev. 2014;34:737-752.
Crossref - Pandey C, Bajpai VK, Negi YK, Rather IA, Maheshwari DK. Effect of plant growth promoting Bacillus spp. on nutritional properties of Amaranthus hypochondriacus grains. Saudi J Bio Sci. 2018;25(6):1066-1071.
Crossref - Luo J, Tao Q, Jupa R, et al. Role of vertical transmission of shoot endophytes in root-associated microbiome assembly and heavy metal hyperaccumulation in Sedum alfredii. Environ Sci Technol. 2019;53(12):6954-6963.
Crossref - Miliute I, Buzaite O, Baniulis D, Stanys V. Bacterial endophytes in agricultural crops and their role in stress tolerance: a review. Zemdirbyste-Agriculture. 2015;102(4):465-478.
Crossref - Marag PS, Suman A. Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol Res. 2018;214:101-113.
Crossref - Deyett E, Rolshausen PE. Temporal dynamics of the sap microbiome of grapevine under high Pierce’s disease pressure. Front Plant Sci. 2019;10:1246.
Crossref - Moturu US, Nunna T, Avula VG, Jagarlamudi VR, Gutha RR, Tamminana S. Investigating the diversity of bacterial endophytes in maize and their plant growth-promoting attributes. Folia Microbiol. 2023;68(3):369-379
Crossref - Suman A, Shasany AK, Singh M, Shahi HN, Gaur A, Khanuja SPS. Molecular assessment of diversity among endophytic diazotrophs isolated from subtropical Indian sugarcane. World J Microbiol Biotechnol. 2001;17:39-45.
Crossref - Barzanti R, Ozino F, Bazzicalupo M, Gabbrielli R, Galardi F, Gonnelli C, Mengoni A. Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microbial Ecol. 2007;53(2):306-316.
Crossref - Cappuccino JC, Sherman N. In: Microbiology. A Laboratory Manual,3rd eds, Benjamin/Cumming Pub. Co. New York. 1992;125-179.
- Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology:nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci USA. 1999;96(12):6814-6819.
Crossref - Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463-5467.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Moturu US, Nunna T, Avula VG, Jagarlamudi VR, Gutha RR, Thamminana S. Evaluation of water stress resilient endophytic bacteria for seed vigor index and antagonistic activity in maize (Zea mays L.). Biol forum. 2021;13:611-619.
- Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. Plos one. 2011;6(6):e20396.
Crossref - Mansotra P, Sharma P, Sharma S. Bioaugmentation of Mesorhizobium cicer, Pseudomonas spp. and Piriformospora indica for sustainable chickpea production. Physiol Mol Biol of Plants. 2015;21(3):385-393.
Crossref - Nunes PS, Junior GV, Mascarin GM, et al. Microbial consortium of biological products: do they have a future? Biol Control. 2024;188:105439.
Crossref - Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18(11):607-621.
Crossref - Bhatt P, Bhatt K, Sharma A, Zhang W, Mishra S, Chen S. Biotechnological basis of microbial consortia for the removal of pesticides from the environment. Crit Rev Biotechnol. 2021;41(3):317-338.
Crossref - Bhatia SK, Yoon JJ, Kim HJ, et al. Engineering of artificial microbial consortia of Ralstonia eutropha and Bacillus subtilis for poly (3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production from sugarcane sugar without precursor feeding. Bioresour Technol. 2018;257:92-101.
Crossref - Xu C, Yu H. Insights into constructing a stable and efficient microbial consortium. Chinese J Chem Eng. 2021;30:112-120.
Crossref - Deepa J, Mathew SK. Compatibility studies on different endophytic microbes of tomato antagonistic to bacterial wilt pathogen. Inter J Advanced Biol Res. 2017;7(1):190-194.
- Subramanian P, Kim K, Krishnamoorthy R, Sundaram S, Sa T. Endophytic bacteria improve nodule function and plant nitrogen in soybean on co-inoculation with Bradyrhizobium japonicum MN110. Plant Growth Regul. 2015;76:327-32.
Crossref - Zinniel DK, Lambrecht P, Harris NB, et al. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol. 2002;68(5):2198-2208.
Crossref - Kobayashi DY, Palumbo JD. Bacterial endophytes and their effects on plants and uses in agriculture. Bacon CW and White JF (eds). Microbial endophytes. Marcel Dekker, New York, USA, 2000;199-233.
- Kandel SL, Joubert PM, Doty SL. Bacterial endophyte colonization and distribution within plants. Microorganisms. 2017;5(4):77.
Crossref - Stoltzfus JR, So R, Malarvithi PP, Ladha JK, De Bruijn FJ. Isolation of endophytic bacteria from rice and assessment of their potential for supplying rice with biologically fixed nitrogen. Plant Soil. 1997;194:25-36.
Crossref - Singh RK, Mishra RPN, Jaiswal HK, et al. Isolation and identification of natural endophytic rhizobia from rice (Oryza sativa L.) through rDNA PCR-RFLP and sequence analysis. Curr microbiol. 2006;52:345-349.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.