ISSN: 0973-7510
E-ISSN: 2581-690X
Aureobasidium sp. strain TD-062, a microcolonial fungus, was isolated from the Thar Desert, India, and it’s aqueous and ethyl acetate extracts were evaluated for antimicrobial activity against clinical isolates of Pseudomonas aeruginosa (sputum), Escherichia coli (pus, blood, and urine), Klebsiella pneumoniae (urine), and Proteus. The ethyl acetate extract was analysed by gas chromatography-mass spectrometry (GC-MS) and examined for toxicity using ProTox-II software. Three compounds, selected based on toxicity and safety profiles, were analysed for molecular docking on BlaR1, a protein that induces antimicrobial resistance in bacteria. The ethyl acetate extract was shown to exhibit antimicrobial activity against clinical pathogens. GC-MS analysis showed that squalene, stigmasterol and delta-tocopherol had lower toxicity profiles. Molecular docking analyses demonstrated that stigmasterol exhibited the highest binding affinity (-8.9 Kcal/mol), compared to positive control clavulanic acid (-6.7 Kcal/mol), suggesting its potential as a potent BlaR1 inhibitor. These findings underscore the bioactive potential of Aureobasidium sp. TD-062 as a promising source of bioactive compounds to combat antimicrobial resistance. The identification of squalene, stigmasterol, and delta-tocopherol as lead compounds for drug development represents a significant advancement in the search for novel antimicrobial agents to address the growing global threat of antimicrobial resistance.
Aureobasidium sp., BlaR1 Protein, Squalene, Delta-tocopherol, Molecular Docking, Bioactive Compounds
Natural products are receiving increasing attention as promising reservoirs for novel drug candidates, given that more than 60% of approved medications trace their origins to natural compounds.1-3 Due to the increasing prevalence of drug resistance, the identification of novel antimicrobial agents is becoming progressively challenging. For a considerable period, microorganisms have been valuable reservoirs of bioactive natural compounds. Through a bioprospecting initiative in the Thar Desert of India, a fungal strain, Aureobasidium pullulans TD-062, was isolated. A. pullulans, a yeast-like fungus, is known for its presence across diverse habitats and environmental conditions.4 Aureobasidum is a yeast-like fungus that is a valuable resource for biotechnological applications that extend beyond geographical constraints.5 Recent research in therapeutics has shifted towards exploring natural products as potential candidates, primarily due to their perceived low toxicity or enhanced bioactivity and tolerance.6
BlaR1 is an integral membrane protein that plays a crucial role in bacterial resistance to β-lactam antibiotics, including widely used drugs such as penicillin, cephalosporins, and carbapenems.7 BlaR1 has been found in gram-positive bacteria, such as Staphylococcus aureus, and gram-negative bacteria, such as Escherichia coli, Klebsiella, pneumonia, and Pseudomonas aeruginosa.8 BlaR1 functions as both a sensor and a signal transducer. It detects the presence of β-lactam antibiotics in the environment and triggers a molecular response that ultimately leads to antibiotic resistance.9
Targeting BlaR1 with bioactive compounds from A. pullulans TD-062 may induce apoptosis and combat drug resistance. In our previous report, antimicrobial and anticancer compounds were detected in ethyl acetate extract using GC-MS.10 Molecular docking studies enable the exploration of protein-ligand interactions, providing insights into ligand-binding mechanisms and the stability of complexes. The objective of the study was: (i) To assess the antimicrobial activity of aqueous and organic solvents strain TD-062 against clinical pathogen and (ii) To evaluate the role of antimicrobial compounds identified through GC-MS analysis using in silico modeling against BlaR1 protein as a target.
Growth dynamics and maintenance
As per our published methodology, Aureobasidium strain TD-062 was inoculated in 100 mL of potato dextrose broth (PDB) and incubated at 37 °C 180-200 rpm for 5 days. After incubation, the culture was centrifuged to separate the supernatant. The supernatant was used as an aqueous extract. In parallel experiments, ethyl acetate (EA), was extracted with equal volume thoroughly agitated for 1 hr. The organic phase dried, and the resulting residue was weighed and mixed with methanol at the concentration of 1 mgmL-1.11
Antimicrobial activity
Agar plugs of the cultures were used to evaluate antimicrobial activity according to Bauer et al. Antimicrobial activity of both aqueous and EA extracts was assessed using the well puncture diffusion method as described previously in our publication,11,12 against clinical pathogens obtained from Jaypee Hospital Noida, 50 µl of clinical pathogens – Pseudomonas aeruginosa –1728 from sputum, Escherichia coli – 1894, 1610, 2646, 2647 from pus, blood, urine samples, Klebsiella pneumoniae – 1862, from urine, and Proteus – 1903, were loaded into wells on Muller Hinton Agar (MHA) media plates, which were then chilled at 5 °C prior to incubation (30 °C, 24 hours). Tetracycline was used as the standard control to compare the antimicrobial activity. The antimicrobial activity was measured by the zones of inhibition. Each experiment was carried out in triplicates and repeated thrice.
Following our earlier published protocol,10 EA extract fractions showing antimicrobial activity against the selected target pathogens were further subjected to GC-MS analysis.
ProTox-II Toxicity prediction and molecular docking modelling
The toxicity of compounds obtained from GC-MS was predicted using ProTox-II,13 a machine learning-based online tool. The 2D structure of the compounds were searched via PubChem, and uploaded in mol format and a toxicity profile was obtained.14
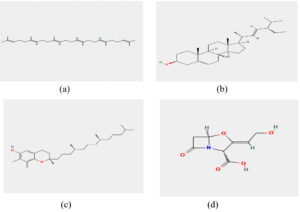
Based on toxicity analysis, three compounds were used: squalene, stigmasterol, and delta-tocopherol as ligands in molecular docking studies. Clavulanic acid was selected as the control which inhibits beta-lactamase and is commonly used in antibiotic formulations.15 Their chemical structures were obtained from the PubChem database in SDF format
(Figure 1 a-d).16-19 Open Babel version 2.3.2 was used to convert the structure compatible with PDB format.20 Molecular docking was performed using Auto dock Vina 1.5.6 software with selected protein BlaR1.21 The Research Collaboratory for Structural Bioinformatics (RCSB),22 provided the protein structures of BlaR1 (PDB ID: 8 EXP) Figure 2, which were then saved in the PDBQT file format, water, hetatm, side chains, and ligand molecules were then removed from the protein structure using the BIOVIA discovery Studio 2021 tool.23 PyMOL 2.5.0 was then used to predict the protein’s tertiary structure.24 Active site prediction for the targeted blar1 proteins was carried out using CASTp 3.0.25
Figure 1. Chemical structures obtained from PubChem: (a) Squalene, (b) Stigmasterol, (c) Delta-tocopherol, and (d) Clavulanic acid16-19
Unexplored environments have been suggested as a strategy in bioprospecting for unique chemical diversity. Aureobasidium sp. TD-062 was isolated from Thar Desert’s red rocky soil, (GenBank accession number JAKSGJ000000000). The aqueous and EA extracts demonstrated antimicrobial activity. As shown in Table 1, the aqueous extract exhibited activity against clinical pathogens – P. aeruginosa – 1728, and E. coli – 1894, 2647 whereas the EA extract showed activity against P. aeruginosa – 1728, and E. coli – 1894, 1610, 2646, 2647, but no activity against Proteus. Zones of inhibition were comparable to or more than that of positive control tetracycline.
In our previous work, GC-MS analysis of compounds from second purification cycle of ethyl acetate extract showed the presence of three bioactive compounds categorized as tocopherols and triterpenes.10
Table (1):
Antimicrobial activity of aqueous and ethyl acetate extracts from Aureobasidium sp. TD-062 against clinical isolates
| Extract | Inhibition zone (mm) Mean ± SD | ||||||
|---|---|---|---|---|---|---|---|
| Pseudomonas aeruginosa (1728) | Klebsiella pneumoniae (1862) | Proteus (1903) | E. coli (1894) | E. coli (1610) | E. coli (2646) | E. coli (2647) | |
| Ethyl acetate TD-062 | 23 ± 0.5 | 0 | 0 | 22 ± 0.5 | 20 ± 0.5 | 21 ± 0.5 | 18 ± 0.5 |
| Aqueous TDs-062 | 18 ± 0.5 | 0 | 0 | 19 ± 0.5 | 0 | 0 | 17 ± 0.5 |
| Control | 26 ± 0.4 | 24 ± 0.4 | 25 ± 0.8 | 21 ± 0.4 | 21 ± 0.4 | 24 ± 0.4 | 22 ± 0.12 |
Toxicity prediction of bioactive compounds
The Protox-II toxicity prediction analysis was used to evaluate the safety profiles of compounds obtained from the second purification cycle of the ethyl acetate extract. As shown in Table 2, the bioactive compounds analyzed were squalene, stigmasterol, and delta-tocopherol. Squalene (molecular weight 410) demonstrated a favorable safety profile, with predicted inactivity against hepatotoxicity, neurotoxicity, nephrotoxicity, respiratory toxicity, cardiotoxicity, carcinogenicity, mutagenicity, cytotoxicity, and clinical toxicity. However, it exhibited activity against CYP2C9, indicating a potential for drug interactions. Stigmasterol and delta-tocopherol exhibited promising safety profiles, with minimal risks for neurotoxicity, nephrotoxicity, cardiotoxicity, and carcinogenicity. However, both stigmasterol and delta-tocopherol were found to interact with CYP2C9, a liver enzyme essential for the metabolism of various drugs. This interaction indicates a potential for drug-drug interactions, as CYP2C9 modulation. Both compounds showed inactivity against other key cytochrome P450 enzymes, reducing the likelihood of widespread metabolic disruptions.
Table (2):
Toxicity prediction of compounds obtained from Aureobasidium sp. TD-062 based on Protox-II analysis
Target |
Squalene |
Stigmasterol |
Delta-tocopherol |
Clavulanic acid |
|---|---|---|---|---|
Molecular weight |
410 |
412 |
416 |
199 |
Number of hydrogen bond acceptors |
0 |
1 |
2 |
6 |
Number of hydrogen bond donors |
0 |
1 |
1 |
2 |
Hepatotoxicity |
Not-active |
Active |
Active |
Not-active |
Neurotoxicity |
Not-active |
Not-active |
Not-active |
Not-active |
Nephrotoxicity |
Not-active |
Not-active |
Not-active |
Active |
Respiratory toxicity |
Not-active |
Active |
Active |
Not-active |
Cardiotoxicity |
Not-active |
Not-active |
Not-active |
Not-active |
Carcinogenicity |
Not-active |
Not-active |
Not-active |
Not-active |
Immunotoxicity |
Not-active |
Not-active |
Not-active |
Not-active |
Mutagenicity |
Not-active |
Not-active |
Not-active |
Not-active |
Cytotoxicity |
Not-active |
Not-active |
Not-active |
Not-active |
Bbb-barrier |
Active |
Active |
Active |
Not-active |
Nutritional toxicity |
Not-active |
Not-active |
Not-active |
Active |
Clinical toxicity |
Not-active |
Not-active |
Not-active |
Active |
Cytochrome CYP1A2 |
Not-active |
Not-active |
Not-active |
Not-active |
Cytochrome CYP2C19 |
Not-active |
Not-active |
Not-active |
Not-active |
Cytochrome CYP2C9 |
Active |
Active |
Active |
Not-active |
Cytochrome CYP2D6 |
Not-active |
Not-active |
Not-active |
Not-active |
Cytochrome CYP3A4 |
Not-active |
Not-active |
Not-active |
Not-active |
Cytochrome CYP2E1 |
Not-active |
Not-active |
Not-active |
Not-active |
Clavulanic acid exhibits low risks for hepatotoxicity, neurotoxicity, cardiotoxicity, and respiratory toxicity but shows activity for nephrotoxicity and clinical toxicity. Inactive for most cytochrome P450 enzymes and unable to cross the blood-brain barrier, it presents a favorable profile for combating antimicrobial resistance with minimal metabolic interactions.
Active site prediction for the targeted blar1 proteins was carried out using CASTp 3.0,21 which identified 22 amino acid residues at the predicted site, with this, the grid box identified active site for subsequent docking studies, as illustrated in Table 3.
Table (3):
Prediction of active site for targeted blar1 protein with CASTp server
Protein |
Volume (SA) |
Area (SA) |
Aa residues at predicted active site |
Total aa residue in chain a |
|---|---|---|---|---|
Blar1 |
14251.82 |
7625.88 |
22 |
226 |
Docking studies on BlaR1 protein
Molecular docking was performed using autodock 1.5.6 to assess the binding affinities of the bioactive compounds from Aureobasidium sp. TD-062 with the BlaR1 protein, a potential target for antimicrobial resistance. Listed in Table 4. stigmasterol exhibited the strongest binding affinity (-8.9 kcal/mol), suggesting a stable and effective interaction within the blar1 binding pocket. delta-tocopherol showed a moderate binding affinity (-7.9 kcal/mol), making it a promising candidate for optimization. Squalene demonstrated the weakest binding affinity (-7.2 kcal/mol) among the compounds, suggesting it may require further optimization for efficacy.
Table (4):
BlaR1 protein interactions with selected compounds obtained from Aureobasidum sp. TD-062
Protein and ligand |
Molecular docking Binding affinity (kcal/mol) |
|---|---|
Blar1 with squalene |
-7.2 |
Blar1with stigmasterol |
-8.9 |
Blar1 with delta-tocopherol |
-7.9 |
Blar1 with clavulanic acid |
-6.7 |
Antimicrobial activity
The extracts from Aureobasidium sp. TD-062 exhibited significant antimicrobial activity against clinical pathogens. The results suggest that the extracts contain bioactive compounds capable of inhibiting microbial growth, with activity levels comparable or more than that of standard antibiotic tetracycline, considering that the extracts used were not completely purified, unlike tetracycline. Authors suggest that this may be due to the synergistic activities of the compounds in the extracts.
Toxicity prediction and safety profiles
The toxicity prediction analysis revealed that the compounds squalene, stigmasterol, and delta-tocopherol from Aureobasidium sp. TD-062 has promising safety profiles. Squalene, a component of shark liver oil, serves as an intermediate metabolite in the synthesis of cholesterol.26 It is recognized as a skin protectant, is resistant to lipid peroxidation and possesses antimicrobial activity on E. coli B-8208.27 Squalene is predicted to cross the blood-brain barrier (BBB) without neurotoxic effects, highlighting its potential CNS compatibility.28 However, squalene shows activity against CYP2C9, which is a liver enzyme critical for metabolizing drugs, such as warfarin, and plays a key role in drug detoxification.29 Its variations or inhibition can affect drug efficacy, safety, and interactions, indicating the possibility of drug-drug interactions while being inactive for other cytochrome P450 enzymes that are crucial for metabolizing drugs, hormones, and xenobiotics.30 They catalyze oxidation reactions, aid detoxification, drug clearance, and biosynthesis. Variations in CYPs can affect drug metabolism, efficacy, and safety.31 These findings suggest minimal risk across multiple endpoints.
Stigmasterol, also referred to as stigmasterin, is a kind of steroid that belongs to the tetracyclic triterpene class.32 Stigmasterol has the potential to possess antiparasitic qualities against specific parasitic strains, including Leishmania and Trypanosoma congolense.33 Vitamin E, also known as tocopherol, is a collective term for a set of fat-soluble substances comprising four tocopherols and four tocotrienols.34 Delta-tocopherol has demonstrated antimicrobial activity on Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA).35
Stigmasterol and delta-tocopherol exhibited encouraging safety profiles, with minimal predicted risks for neurotoxicity, nephrotoxicity, cardiotoxicity, and carcinogenicity, supporting their potential for therapeutic applications. These findings are significant as they suggest that both compounds may have a wide safety margin when used in clinical or pharmaceutical formulations.
The interaction of both compounds with CYP2C9, a crucial liver enzyme involved in drug metabolism, is another key finding. CYP2C9 plays a vital role in the metabolism of various drugs, including anticoagulants such as warfarin and nonsteroidal anti-inflammatory drugs (NSAIDs).36 The modulation or inhibition of CYP2C9 by stigmasterol and delta-tocopherol could lead to altered drug metabolism, resulting in either enhanced toxicity or reduced efficacy of co-administered medications. This indicates a potential for drug-drug interactions that could have clinical implications.
Clavulanic acid, a β-lactam compound, inhibits β-lactamase enzymes, enhancing the efficacy of antibiotics like penicillin and cephalosporins against resistant pathogens such as Staphylococcus aureus and Escherichia coli.37 Toxicity predictions indicate a favorable safety profile with low risks for hepatotoxicity, neurotoxicity, cardiotoxicity, and respiratory toxicity, though nephrotoxicity and clinical toxicity remain concerns, particularly in patients with pre-existing kidney conditions. Its inactivity against most cytochrome P450 enzymes reduces the likelihood of drug-drug interactions, and its inability to cross the blood-brain barrier minimizes central nervous system-related risks.
Binding affinity with BlaR1 protein
The results from the molecular docking studies indicated that stigmasterol exhibited the strongest binding affinity to BlaR1, suggesting its potential as a highly effective inhibitor of this protein, which plays a central role in β-lactam antibiotic resistance.38 This strong interaction supports the idea that stigmasterol could be a promising candidate for further development as a therapeutic agent targeting BlaR1. In contrast, delta-tocopherol displayed a moderate binding affinity, indicating its potential as an inhibitor as well, but suggesting that structural optimization may be necessary to enhance its binding strength and efficacy.39 On the other hand, squalene showed the weakest binding affinity among the compounds tested. Despite this, it still holds promise as a candidate for further studies, as modifications or optimizations may improve its binding affinity and inhibitory activity against BlaR1. These findings underscore the need for these compounds and explore their potential as novel therapeutic agents in combating antibiotic resistance.
Antimicrobial compounds from Aureobasidium TD-062 have exhibited activity against clinical pathogens, with good safety profiles. Amongst them, stigmasterol has demonstrated potential in inhibiting BlaR1, with high binding affinity of -8.9 kcal/mol, thereby indicating its effectiveness in control of both Gram-positive and Gram-negative pathogens via inhibition of beta-lactamase activity. Hence, we conclude that natural compounds continue to offer promise in targeting antimicrobial resistance.
ACKNOWLEDGMENTS
The authors are thankful to Jaypee Institute of Information Technology, Noida, India, for providing the necessary infrastructure facilities to carry out the work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Ibeyaima A, Dwivedi AK, Saini N, Gupta S, Sarethy IP. Saccharothrix sp. TD-093 from the Thar Desert, India: Metabolite Fingerprinting of Antimicrobial Compounds and In silico Analysis. Curr Microbiol. 2017;74(3):334-343.
Crossref - Ahmad S, Ruby T, Shahzad MI, Rivera G, Carriola DVN, Khan AA. Antimicrobial, antioxidant, antiviral activity, and gas chromatographic analysis of Varanus griseus oil extracts. Arch Microbiol. 2022;204(8):531.
Crossref - Ahmad S, Ali K, Ahmad K, et al. Chemical composition and antimicrobial study of Crossobamon orientalis body oil. Heliyon. 2024;10(6):e28225.
Crossref - Bozoudi D, Tsaltas D. The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation. 2018;4(4):85.
Crossref - Campana R, Fanelli F, Sisti M. Role of melanin in the black yeast fungi Aureobasidium pullulans and Zalaria obscura in promoting tolerance to environmental stresses and antimicrobial compounds. Fungal Biol. 2022;126(11-12):817-825.
Crossref - Huang M, Lu JJ, Ding J. Natural products in cancer therapy: past, present, and future. Nat Prod Bioprospect. 2021;11(1):5-13.
Crossref - Frederick TE, Peng JW. A gratuitous β-lactamase inducer uncovers hidden active site dynamics of the Staphylococcus aureus BlaR1 sensor domain. PLoS One. 2018;13(5):e0196945.
Crossref - Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088-17.
Crossref - Llarrull LI, Toth M, Champion MM, Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. J Biol Chem. 2011;286(44):38148-38158.
Crossref - Chaturvedi S, Sarethy IP. GC-MS-based metabolite fingerprinting reveals the presence of novel anticancer compounds in the microcolonial fungus Aureobasidium sp. TD-062 from the under-explored Thar Desert. Nat Prod Res. 2024.
Crossref - Chaturvedi S, Sarethy IP. Virtual screening of compounds from microcolonial fungal strain TD-062 obtained from the Thar Desert of India. Curr Trends Biotechnol Pharm. 2021;15(6):62-66.
Crossref - Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-496.
Crossref - Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46(W1):W257-W263.
Crossref - Kim S, Chen J, Cheng T, et al. PubChem 2023 update. Nucleic Acids Res. 2023;51(D1):D1373-D1380.
Crossref - Lopez-Agudelo VA, Gomez-Rםos D, Ramirez-Malule H. Clavulanic acid production by Streptomyces clavuligerus: insights from systems biology, strain engineering, and downstream processing. Antibiotics. 2021;10(1):84.
Crossref - National Center for Biotechnology Information. PubChem Compound Summary for CID 638072, Squalene. https://pubchem.ncbi.nlm.nih.gov/compound/Squalene. Accessed November 19, 2024.
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5280794, Stigmasterol. https://pubchem.ncbi.nlm.nih.gov/compound/Stigmasterol. Accessed November 19, 2024.
- National Center for Biotechnology Information. PubChem Compound Summary for CID 92094, Delta-Tocopherol. https://pubchem.ncbi.nlm.nih.gov/compound/Delta-Tocopherol. Accessed November 19, 2024.
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5280980, Clavulanic Acid. https://pubchem.ncbi.nlm.nih.gov/compound/Clavulanic-Acid. Accessed November 19, 2024.
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminf. 2011;3:33.
Crossref - Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-461.
Crossref - Alexander JAN, Worrall LJ, Hu J, et al. Structural basis of broad-spectrum β-lactam resistance in Staphylococcus aureus. Nature. 2023;613:375-382.
Crossref - Biovia DS. Discovery Studio Visualizer. San Diego: Biovia; 2021. https://www.3ds.com/products/biovia/discovery-studio. Accessed November 19, 2024.
- Lilkova E, Petrov D, Doumanov J, Ilieva N, Veselinov R. The PyMOL Molecular Graphics System, Version 2.0. Schrodinger, LLC. 2015. https://pymol.org/. Accessed November 19, 2024.
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(W1):W363-W367.
Crossref - Widyawati T, Syahputra RA, Syarifah S, Sumantri IB. Analysis of antidiabetic activity of squalene via in silico and in vivo assay. Molecules. 2023;28(9):3783.
Crossref - Dmitrieva A, Vesnina A, Dyshlyuk L. Antioxidant and antimicrobial properties of squalene from Symphytum officinale and chlorogenic acid from Trifolium pratense. AIP Conf Proc. 2022; 2636:020005.
Crossref - Bhat MP, Rudrappa M, Hugar A, et al. In-vitro investigation on the biological activities of squalene derived from the soil fungus Talaromyces pinophilus. Heliyon. 2023;9(11):e14871.
Crossref - Tegenge MA, Von Tungeln LS, Mitkus RJ, et al. Pharmacokinetics and biodistribution of squalene-containing emulsion adjuvant following intramuscular injection of H5N1 influenza vaccine in mice. Regul Toxicol Pharmacol. 2016;81:113-119.
Crossref - Kusama M, Maeda K, Chiba K, Aoyama A, Sugiyama Y. Prediction of the effects of genetic polymorphism on the pharmacokinetics of CYP2C9 substrates from in vitro data. Pharm Res. 2009;26(4):822-835.
Crossref - Tassies D, Freire C, Pijoan J, et al. Pharmacogenetics of acenocoumarol: cytochrome P450 CYP2C9 polymorphisms influence dose requirements and stability of anticoagulation. Haematologica. 2002;87(11):1185-1191
- Zhang X, Wang J, Zhu L, et al. Advances in stigmasterol on its anti-tumor effect and mechanism of action. Front Oncol. 2022;12:1101289.
Crossref - Bansal R, Sen SS, Muthuswami R, Madhubala R. Stigmasterol as a potential biomarker for amphotericin B resistance in Leishmania donovani. J Antimicrob Chemother. 2020;75(4):942-950.
Crossref - Parker RS, Swanson JE. A novel 5′ -carboxychroman metabolite of g-tocopherol secreted by HepG2 cells and excreted in human urine. Biochem Biophys Res Commun. 2000;269(2):580-583.
Crossref - Hensley K, Benaksas EJ, Bolli R, et al. New perspectives on vitamin E: g-tocopherol and carboxyethylhydroxychroman metabolites in biology and medicine. Free Radic Biol Med. 2004;36(1):1-5.
Crossref - Kingsley LJ, Wilson GL, Essex ME, Lill MA. Combining structure- and ligand-based approaches to improve site of metabolism prediction in CYP2C9 substrates. Pharm Res. 2015;32(3):986-1001.
Crossref - Bush K, Bradford PA. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247.
Crossref - Alghamdi BA, Al-Johani I, Al-Shamrani JM, et al. antimicrobial resistance in methicillin-resistant Staphylococcus aureus. Saudi J Biol Sci. 2023;30(4):103604.
Crossref - Vlasiou MC, Nikolaou G, Spanoudes K, Mavrides DE. β-Tocotrienol and δ-Tocotrienol as additional inhibitors of the main protease of feline infectious peritonitis virus: An In silico analysis. Vet Sci. 2024;11(9):424.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.