ISSN: 0973-7510
E-ISSN: 2581-690X
Heavy metal contamination of soil and water poses serious risks to human health and ecosystems. This study focuses on two bacterial strains, Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2, known for their tolerance and bioaccumulation of lead (Pb), cadmium (Cd), and nickel (Ni). Their potential for bioremediation was examined using Fourier-transform infrared (FTIR) spectroscopy. The bacteria were cultured in nutrient broth containing heavy metals (1000 ppm Pb(NO3)2, 750 ppm CdCl2‚ ·H2O, 200 ppm Ni (NO3 )2 ) to assess changes in biomolecular structures due to metal exposure. FTIR analysis revealed distinct and specific binding interactions between heavy metal ions and bacterial functional groups. These interactions include hydrogen bonding with hydroxyl (O-H) and amine (N-H) groups, ionic interactions with negatively charged phosphate and carboxyl groups, and coordination bonds with carbonyl (C=O) and amino groups in proteins. The evidence for these mechanisms was observed through shifts in key FTIR peaks, such as the O-H and N-H stretching regions (e.g., 3280.1 cm-1), phosphate stretching vibrations (e.g., 1233.7 cm-1), and amide I and II peaks (e.g., 1636.3 cm-1 and 1528.2 cm-1). These interactions provide insights into the mechanisms of metal bioaccumulation and stress adaptation. The findings highlight Enterobacter sp. PGRG2 and Achromobacter insolitus PGRG5 as promising candidates for bioremediation, offering potential solutions for mitigating heavy metal pollution in contaminated environments.
Heavy Metals, FTIR, Bioaccumulation, Soil, Environment
Heavy metal contamination is an expanding environmental hazard, coming from diverse industrial, agricultural, and human activities.1 Due to their persistence and capacity to bioaccumulate, lead (Pb), cadmium (Cd), and nickel (Ni) are some of the most hazardous heavy metals that can accumulate in soil and water, posing serious threats to both human health and ecological systems.2 By attaching to cellular macromolecules and rupturing protein structures, lipid membranes, and nucleic acids, these metals obstruct vital biological functions. Conventional cleanup methods, such ion exchange and chemical precipitation, are frequently pricy, ineffective in large-scale applications, and harmful to the environment.3 As a result, biological techniques-in particular, microbial bioremediation-have become viable and affordable options for cleaning up settings contaminated by metals.4
Bacteria, in particular, have developed a number of defence mechanisms to withstand and eliminate heavy metals.5 Bacteria have the ability to sequester toxic metals through mechanisms such as biosorption, bioaccumulation, and transformation, which can reduce their negative effects.6 Several bacterial strains, notably Pseudomonas aeruginosa, Bacillus spp., various Enterobacter spp., Cupriavidus metallidurans and various modified strains of Escherichia coli. have shown a remarkable ability to live and thrive in heavy metal-rich environments.4,7,8 These strains have special physiological and biochemical characteristics that allow them to interact with metal ions through functional groups such as phosphates, amines, carbonyls, and hydroxyls. This interaction can result in sequestration, cellular absorption, or metal chelation.9
In addition to microbial bioremediation, other emerging techniques such as phytoremediation have shown significant potential for addressing heavy metal contamination. Phytoremediation, which utilizes plants to absorb, sequester, and detoxify heavy metals, offers several advantages including ecosystem restoration, sustainability, and aesthetic value. Similarly, microbial bioremediation is characterized by its versatility and efficiency in diverse environments. While phytoremediation is particularly suitable for surface soils and areas conducive to plant growth, microbial approaches excel in extreme conditions, such as high toxicity or inaccessible subsurface environments. Both methods contribute uniquely to the remediation process and can be combined to create synergistic strategies that maximize effectiveness. By integrating these complementary techniques, more comprehensive and sustainable solutions to mitigate heavy metal pollution can be achieved.
Some primary work has been done which showed that Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2 strains are capable of effectively absorbing and immobilising metals from contaminated environments,10,11 which makes them attractive options for use in bioremediation processes. Aim of this research is to understand better about the molecular interactions between these bacterial strains and heavy metals, especially with regard to the changes in cellular architecture.
Using Fourier-transform infrared (FTIR) spectroscopy, examined the interactions between two bacterial strains-Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2-and Pb, Cd, and Ni in order to determine their metal tolerance and bioaccumulation ability. FTIR has been frequently used to investigate molecular changes in heavy metal-exposed bacterial cells, offering insights into the roles played by functional groups including hydroxyls, phosphates, and amides in metal binding. This work intends to clarify the molecular mechanisms by which these bacterial strains respond to heavy metal stress and contribute to their remediation capability by comparing the FTIR spectra of untreated and metal-treated bacterial cells.
The aim of this research is to: (i) examine the functional group changes that occur in Pb, Cd, and Ni-exposed Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2 strains; and (ii) assess the bacteria’ potential for use as bioremediation agents in environments contaminated with heavy metals.
In this study, Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2, known for their high tolerance and bioaccumulation capacity for Pb, Cd, and Ni, were utilized. The bacterial isolates were stored as glycerol stocks at -80 °C. For preparation of the mother culture, a loopful of each isolate was transferred from the glycerol stock into sterile nutrient broth (pH 7.0). Cultures were incubated at 37 °C with constant shaking at 120 rpm using an orbital shaker. The overnight-grown cultures were subsequently used as inoculum for further experiments.
Characterization of bacterial biomass by Fourier Transform Infrared Spectroscopy (FTIR)
Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2 were cultivated in sterile nutrient broth supplemented separately with 1000 ppm Pb (NO3)2, 750 ppm CdCl2·H2O, and 200 ppm Ni(NO3)2. Following growth, the bacterial cells were harvested by centrifugation at 1000 rpm for 10 minutes and washed with phosphate buffer (pH 7.0) to remove residual media. The cell pellets were resuspended in phosphate buffer. For analysis, 0.1 ml of the cell suspension was diluted with 0.9 ml distilled water. The diluted samples were then mixed with KBr to form pellets, which were analyzed using a FTIR spectrophotometer (FTIR630 Agilent) equipped with a standard light source and a TGS detector at the Department of Chemistry, Birla College.
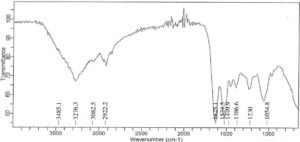
Key functional groups responsible for metal ion adsorption were discovered by the FTIR spectra of Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2, nurtured in the presence and absence of heavy metal salts. The chemical structures on the bacterial biomass have a significant impact on the adsorption behavior.
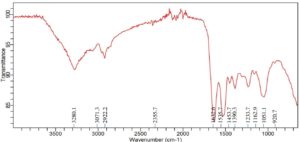
A large absorption band at 3276.3 cm-1 for Achromobacter insolitus PGRG5 (Figure 1) corresponds to O-H or N-H stretching vibrations, which is generally linked to proteins, polysaccharides, or hydroxyl groups found in bacterial cell walls. The C-H stretching vibrations represented by the bands at 2922.2 cm-1 and 2851.4 cm-1 are probably derived from lipids or fatty acids. Asymmetric bands (amide I and amide II) have prominent peaks at 1622.6 cm-1 and 1531.9 cm-1, respectively, indicating their respective protein architectures. Furthermore, peaks located at 1233.7, 1054.8, and 965.4 cm-1 indicate the presence of phosphodiester groups, which may indicate the presence of polysaccharides or nucleic acids from the bacterial cell wall.
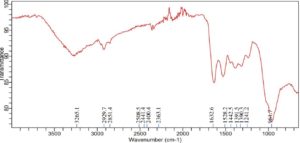
Comparably, the Enterobacter sp. PGRG2 FTIR spectrum (Figure 2) revealed a peak at 3280.1 cm-1, showing O-H stretching, most likely from surface hydroxyls on the bacterial cell surface or hydroxyl groups linked to water. The alkyl chains found in fatty acids and lipids are linked to C-H stretching vibrations, which are represented by peaks at 2922.2 cm-1 and 3071.3 cm-1. The C=O stretching of amide (protein) or carboxyl groups, which is suggestive of the lipid and protein components of the cell wall, is responsible for a significant peak at 1622.6 cm-1. The peaks at 1233.7 and 1051.1 cm-1 indicate stretching vibrations of phosphates, most likely originating from phospholipids or nucleic acids. The band at 920.7 cm-1 can be related to polysaccharide C-C stretching.
FTIR analysis of Achromobacter insolitus PGRG5 with Lead (Pb) treatment
The FTIR analysis of Achromobacter insolitus PGRG5 grown in nutrient broth spiked with 1000 ppm Pb (NO3)2 revealed notable shifts in key functional groups, indicating significant biochemical alterations upon Pb exposure (Figure 3 and Table 1).
Table (1):
FTIR spectrum interpretation and comparison of untreated and Pb treated cells of Achromobacter insolitus PGRG5
Wave number Range (cm-1) |
Treated Cells (with Pb) |
Untreated Cells |
Interpretation |
|---|---|---|---|
3500-3200 |
3280.1 cm⁻¹ (O-H/N-H stretch) |
3485.1 cm⁻¹ (O-H/N-H stretch) |
Shift in O-H/N-H stretch indicates altered hydrogen bonding, possibly due to Pb binding. |
3000-2800 |
2922.2 cm⁻¹ (C-H stretch) |
2922.2 cm⁻¹ (C-H stretch) |
Similar peak for C-H stretch in both samples, indicating minimal change in aliphatic chains of lipids or proteins. |
1650-1500 |
1636.3 cm⁻¹ (C=O stretch, Amide I) |
1625.1 cm⁻¹ (C=O stretch, Amide I) |
Small shift in Amide I peak suggests Pb may alter the secondary structure of proteins. |
1550-1450 |
1524.5 cm⁻¹, 1446.2 cm⁻¹ (Amide II) |
1524.5 cm–¹, 1499 cm⁻¹ (Amide II) |
Similar peaks, but shifts in untreated cells suggest less alteration in protein structure compared to Pb-treated cells. |
1300-1000 |
1386.6 cm⁻¹, 1230 cm⁻¹, 1166.7 cm⁻¹ (C-O stretch, Carbohydrates) |
1386.6 cm⁻¹, 1230 cm⁻¹, 1054.8 cm⁻¹ (C-O stretch, Carbohydrates) |
Significant changes in the carbohydrate region (1300–1000 cm⁻¹), indicating Pb interaction alters polysaccharides in the cell wall. |
A broad peak slightly relocated in comparison to untreated cells near 3280 cm-1, which corresponds to O-H or N-H stretching vibrations, suggests changes in the hydrogen bonding environment. This change suggests that Pb might interact with amine or hydroxyl groups, changing the ways in which they connect. Furthermore, a prominent peak at 2922 cm-1 represents the C-H stretching of methylene groups. This peak is also present in the untreated sample, but with a little shift and decreased intensity, which further suggests a Pb interaction.
Pb contact may cause slight shifts in the protein structure, but a sharp peak at 1636.3 cm-1, which represents C=O stretching from amide I in proteins, stays mostly intact. The treated cells exhibited peaks at 1524.5 cm-1 and 1446.2 cm-1, which suggest alterations in protein structure, possibly as a result of Pb binding. These peaks are generally linked to bending vibrations of N-H and amide II.
The polysaccharide and carbohydrate areas show additional notable alterations. Peaks at 1386.6, 1230, and 1166.7 cm-1 imply that Pb contact has changed the bacterial cell wall’s polysaccharide composition, possibly compromising its structural integrity.
The comparison of treated and untreated bacterial cells highlights several key changes upon Pb exposure:
O-H and N-H stretching region (3500-3200 cm-1)
Changes in the O-H and N-H stretching region (3500-3200 cm-1) indicate that Pb contact is changing hydrogen bonding, which is probably affecting the structures of proteins or polysaccharides.
C-H stretching region (2922.2 cm-1)
This region does not alter much, suggesting that it has little influence on proteins or lipids that are aliphatic.
Amide I region (C=O stretching, 1636.3 cm-1)
Minor shifts suggest that Pb contact has caused minute changes in the secondary structure of proteins.
Regions rich in polysaccharides and carbohydrates (1300-1000 cm-1)
Significant shifts in peak positions and intensity point to significant effects on the components of the bacterial cell wall, most likely as a result of Pb binding.
These spectral changes strongly suggest that Pb binds to proteins and polysaccharides in the bacterial cell wall, altering molecular structure and organization. Such binding could interfere with essential cellular functions, contributing to the toxic effects of Pb on bacterial growth and metabolism.
FTIR analysis of Achromobacter insolitus PGRG5 with Cadmium (Cd) treatment
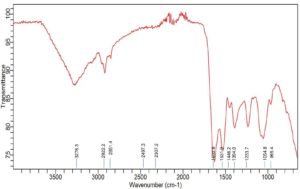
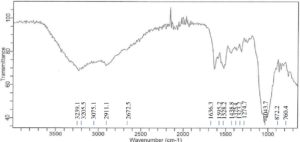
Accompanying the bacterial growth in nutrient broth supplemented with 750 ppm CdCl2·H2O, the FTIR analysis of Achromobacter insolitus PGRG5 revealed unique spectrum changes that indicate important interactions between Cd and functional groups in the bacterial cell wall (Figure 4 and Table 2). The positions and intensities of the peaks at 3276.3 cm-1 and 3485.1 cm-1, which represent O-H and N-H stretching vibrations, respectively, changed slightly. These alterations show how Cd interacts with hydroxyl and amine groups, probably changing hydrogen bonds or changing the bacterial cell wall’s structure.
Table (2):
FTIR spectrum interpretation and comparison of untreated and Cd treated cells of Achromobacter insolitus PGRG5/span>
Wave number Range (cm-1) |
Functional Group/Vibration |
Untreated cells |
Treated cells (with Cd) |
Observations |
|---|---|---|---|---|
3276.3 |
O-H/N-H stretching (Proteins/Carbohydrates) |
Strong broad peak |
Shifted to 3485.1 cm⁻¹ and 3276.3 cm⁻¹ |
Shift in O-H/N-H stretch indicates altered hydrogen bonding, possibly due to Pb binding. |
2922.2 |
C-H stretching (Lipids) |
Strong peak |
Similar peak at 2922.2 cm⁻¹ |
Similar peak for C-H stretch in both samples, indicating minimal change in aliphatic chains of lipids or proteins. |
2851.4 |
C-H stretching (Lipids) |
Strong peak |
Diminished or reduced intensity |
Small shift in Amide I peak suggests Pb may alter the secondary structure of proteins. |
1622.6, 1531.9 |
Amide I and II (Proteins) |
Defined peaks, indicative of protein structure |
Slight shift to 1625.1 cm⁻¹ and 1524.5 cm⁻¹ |
Similar peaks, but shifts in untreated cells suggest less alteration in protein structure compared to Pb-treated cells. |
1233.7, 1054.8, 965.4 |
Fingerprint region (Polysaccharides/Nucleic Acids) |
Strong, defined peaks |
Slight shifts to 1230 cm⁻¹, 1054.8 cm⁻¹ |
Significant changes in the carbohydrate region (1300–1000 cm⁻¹), indicating Pb interaction alters polysaccharides in the cell wall. |
There were also noticeable changes in the amide bands. Protein secondary structures are linked to peaks at 1625.1 cm-1 and 1524.5 cm-1. These peak positions imply that conformational changes in proteins have resulted from Cd binding to peptide bonds or amino acid residues. While C=O stretching from carboxyl groups was largely absent, this region of the spectrum might have been affected indirectly by the interaction of Cd with adjacent bonds or functional groups.
Subtle changes in wavenumbers and intensities were observed for similar peaks located in the areas of 1230 cm-1 and 1054.8 cm-1. These modifications can be the result of Cd interfering with the structures of polysaccharides or nucleic acids. Cd affects the bacterial cell membrane and may denature proteins, as evidenced by the broadening and drop in intensity, particularly in the protein and lipid areas.
In conclusion, the FTIR analysis shows that the addition of Cd causes notable changes in the components of the bacterial cell wall, namely impacting proteins (amide bands), lipids (C-H stretching), and polysaccharides. These changes strongly imply that Cd interacts with important functional groups inside the bacterial cell, resulting in biochemical and structural alterations that compromise protein stability and membrane integrity.
FTIR analysis of Achromobacter insolitus PGRG5 with Nickel (Ni) treatment
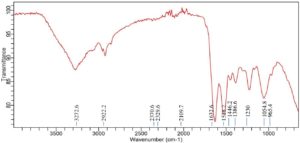
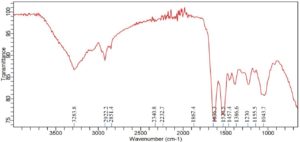
Significant spectrum variations were found in the FTIR study of Achromobacter insolitus PGRG5 cultured in nutritional broth injected with 200 ppm Ni (NO3)2. These changes suggest that Ni and the components of the bacterial cell interact biochemically (Figure 5 and Table 3). The stretching vibrations of O-H or N-H groups, which are generally connected to hydroxyl groups in proteins, lipids, or carbohydrates, or amine stretching in proteins, are represented by a unique peak that was detected at 3272.6 cm-1. This peak’s shift and decrease in intensity point to disruptions in hydrogen bonding in biomolecules, like proteins and polysaccharides, in reaction to Ni exposure. This suggests that stress has altered the structure of the bacterial cell wall or membrane.
Table (3):
FTIR spectrum interpretation and comparison of untreated and Ni treated cells of Achromobacter insolitus PGRG5
Wavenumber (cm⁻¹) |
Functional Group/Assignment |
Treated Cells (Pb) |
Untreated Cells |
Interpretation |
|---|---|---|---|---|
3272.6 |
O-H/N-H Stretching (Hydrogen Bonding) |
Peak at 3272.6 cm⁻¹ |
Peak at 3276.3 cm⁻¹, additional peak at 3485.1 cm⁻¹ |
Indicates disruption in hydrogen bonding in treated cells, possibly affecting proteins and polysaccharides. |
2922.2 |
C-H Stretching (Lipids) |
Peak at 2922.2 cm⁻¹ |
Peak at 2922.2 cm⁻¹ |
No significant change in lipid content or structure between treated and untreated cells. |
1632.6 |
Amide I (C=O Stretching) (Proteins) |
Peak at 1632.6 cm⁻¹ |
Peak at 1625.1 cm⁻¹ |
Shift indicates protein denaturation or conformational changes in treated cells. |
1528.2 |
Amide II (N-H Bending) (Proteins) |
Peak at 1528.2 cm⁻¹ |
Peak at 1544.5 cm⁻¹ |
Shift suggests changes in protein secondary structures due to Pb stress. |
1386.6 |
C-O/C-C Stretching (Polysaccharides) |
Peak at 1386.6 cm⁻¹ |
Peak at 1386.6 cm⁻¹ |
Minor changes in intensity indicate slight modifications in polysaccharide structure. |
1230 |
C-O Stretching (Polysaccharides) |
Peak at 1230 cm⁻¹ |
Peak at 1230 cm⁻¹ |
Stable polysaccharide content with minor variations between treated and untreated cells. |
1054.8 |
C-O Stretching (Polysaccharides) |
Peak at 1054.8 cm⁻¹ |
Peak at 1054.8 cm⁻¹ |
Similar intensity suggests stable polysaccharide content in both treated and untreated cells. |
2370.6 / 2329.6 |
Nitrile or Carbonate Stretching |
Peaks at 2370.6 and 2329.6 cm⁻¹ |
No significant peaks observed |
Unique peaks indicate the presence of stress-related metabolites or intermediates in treated cells. |
A peak reflecting C-H stretching vibrations from methylene (-CH) groups in lipids, located at 2922.2 cm-1, exhibited very little change. This suggests that either the lipid composition is not significantly altered by Ni exposure, or the changes are too small to be noticed at this concentration. The amide I band (C=O stretching vibrations) from proteins is linked to a strong peak at 1632.6 cm-1, which implies that Ni exposure leads to denaturation or conformational changes in proteins, potentially damaging protein structures. A peak at 1528.2 cm-1, which corresponds to amide II (N-H bending and C-N stretching) vibrations, provides more support for this. The amide II band shift indicates that stress has altered the secondary structure of proteins, most likely as a result of Ni-induced protein aggregation or unfolding.
The presence of further peaks at 1386.6, 1230, and 1054.8 cm-1, which indicate C-O and C-C stretching from polysaccharides and carboxylate groups, suggests that the bacterial cell wall’s polysaccharide content is still largely stable. On the other hand, modest variations in peak intensity point to small structural alterations in the polysaccharide upon exposure to Ni. Peaks at 2370.6 cm-1 and 2329.6 cm-1 may indicate the development of stress-related metabolic by-products or external pollutants under Ni stress. These peaks may potentially correspond to asymmetric stretching of nitrile or carbonate groups. These peaks signify the production of particular metabolites or intermediates associated with stress-response systems or detoxification activities.
Finally, the FTIR study shows that Achromobacter insolitus PGRG5 undergoes notable metabolic changes as a result of Ni exposure. The amide I (1632.6 cm-1) and amide II (1528.2 cm-1) bands exhibit notable shifts that draw attention to conformational changes due to stress or denaturation of proteins. Modest changes in the O-H/N-H stretching region (3272.6 cm-1) indicate that there may have been hydrogen bonding disruptions in the bacterial cell components, which most likely affected the lipids and polysaccharides. Furthermore, distinct peaks in the 2100-2400 cm-1 range suggest the emergence of stress-related chemicals, which could be important for survival strategies and detoxification in the presence of heavy metal stress. These results advance knowledge of bacterial cells’ potential for bioremediation of Ni-contaminated environments by shedding light on the biochemical responses of these cells to Ni exposure.
FTIR analysis of Enterobacter sp. PGRG2 with Lead (Pb) treatment
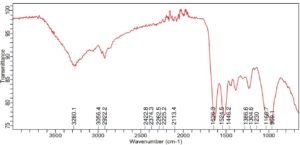
Significant alterations in the spectrum profile are revealed by the FTIR analysis of Enterobacter sp. PGRG2 subjected to 1000 ppm of Pb (NO3)2, as shown in Figure 6 and Table 4 suggesting important molecular interactions between Pb and bacterial cell components. The O-H stretching region was found to change to a lower wavenumber (3265.1 cm-1), indicating possible interaction between Pb ions and hydroxyl groups within the bacterial cells. This interaction likely affects the hydrogen bonding environment, impacting the overall structural integrity of the cell wall or membrane.
Table (4):
FTIR spectrum interpretation and comparison of untreated and Pb treated cells of Enterobacter sp. PGRG2
Peak assignment |
Wave number (cm-1) untreated cell |
Wave number (cm-1) Pb treated cells |
Interpretation |
|---|---|---|---|
O-H Stretching (Hydroxyl) |
3280.1 |
3265.1 |
Shift to a lower wavenumber indicates interaction of Pb with hydroxyl groups (altered hydrogen bonding). |
C-H Stretching (Alkyl) |
3071.3, 2922.2 |
2929.7, 2851.4 |
Shift and broadening suggest Pb affects lipid structures, possibly binding with lipophilic components. |
C=O Stretching (Carbonyl) |
1622.6 |
1632.6 |
Slight shift, indicating Pb interaction with carbonyl groups, potentially altering protein conformation. |
Phosphate Stretching |
1233.7, 1051.1 |
1241.2, 961.7 |
Shift in phosphate peaks suggests Pb binding to phosphate groups in nucleic acids or membrane phospholipids. |
Other Functional Groups |
1535.7, 1390.3, 1162.9, 920.7 |
1528.2, 1391.5, 1241.2, 961.7 |
Minor shifts indicate Pb influences polysaccharides and protein secondary structures in cell wall. |
The C-H stretching peaks show both a shift and a small broadening, with positions of 2929.7 cm-1 and 2851.4 cm-1, respectively. These alterations suggest that Pb ions may interact with lipophilic components of the bacterial membrane, affecting the lipid structure or content. These changes are suggestive of lipid bilayer disturbances, which may compromise the integrity and functionality of the membrane.
There is also a minor change in the C=O stretching vibration, which is located at 1632.6 cm-1. This suggests that carbonyl groups, which are found in proteins or other components of cell walls, may coordinate with Pb ions. This coordination may cause structural changes in the bacterial cell wall or change the conformation of proteins, which would impair the functionality of critical activities.
In addition, there are slight changes in the peaks at 1241.2 cm-1 and 961.7 cm-1 that are linked to phosphate groups. These modifications imply that Pb binds to phosphate groups, which are probably present in membrane phospholipids or nucleic acids. The structural and functional roles of phospholipid bilayers or nucleic acids may be impacted by such interactions, which could further affect the metabolic processes and viability of the bacterial cell.
Further slight alterations in the 1500 cm-1 to 1000 cm-1 area suggest that Pb has an impact on the bacterial cell wall’s polysaccharide and protein secondary structures. These changes could be the result of Pb-induced changes to structural polysaccharides and proteins’ conformations, which would increase the observed harmful effects.
Enterobacter sp. PGRG2 cells treated and untreated with Pb exhibit different FTIR patterns, which indicate that Pb treatment causes notable changes in important functional group vibrations. These alterations show that Pb binds to phosphate, carbonyl, and hydroxyl groups, changing the structure of proteins, lipids, and nucleic acids. These interactions risk the integrity of the cell wall and membrane, which may impair essential cellular processes and exacerbate the harmful effects of Pb on bacterial growth and metabolism.
FTIR analysis of Enterobacter sp. PGRG2 with Cadmium (Cd) treatment
As shown in Figure 7 and Table 5, the FTIR analysis of Enterobacter sp. PGRG2 cultured in nutritional broth supplemented with 750 ppm of CdCl2·H2O reveals notable metabolic changes within the bacterial cells. When compared to untreated cells, there is a discernible shift in the O-H/N-H stretching area at 3239.1 cm-1, which implies that Cd ions (Cd2+) are interacting with amine or hydroxyl groups in the bacterial proteins or polysaccharides. This shift may result in modifications to cell walls or membrane structures caused by stress because it represents shifts in the hydrogen bonding or direct coordination of Cd2+ with these functional groups.
Table (5):
FTIR spectrum interpretation and comparison of untreated and Cd treated cells of Enterobacter sp. PGRG2
Wave number (cm-1) |
Functional Group |
Untreated cells |
Cd treated cells |
Changes observed |
|---|---|---|---|---|
3280.1-3239.1 |
O-H/N-H stretching |
3280.1 (broad) |
3239.1 (Shifted) |
Shift in O-H/N-H band suggests interaction of Cd with hydroxyl or amine groups, possible hydrogen bonding. |
2922.2-2911.1 |
C-H stretching (aliphatic chains, lipids) |
2922.2 |
2911.1 |
Shift indicates alteration in lipid components, likely due to Cd binding to membrane structures. |
1635.7-1639.3 |
Amide I (C=O stretching in proteins) |
1635.7 |
1636.3 |
Slight shift and reduction in intensity, indicating possible protein structural modification by Cd. |
1535.7-1528.2 |
Amide II (N-H bending, C-N stretching) |
1535.7 |
1528.2 |
Shift and weakening suggest protein structure changes, potentially from denaturation or folding disruption. |
1233.7-1274.7 |
P=O stretching (phosphates, nucleic acids) |
1233.7 |
1274.7 |
Shift indicates interaction of Cd with phosphates or nucleic acids, potentially altering their structure. |
1051.1-1043.7 |
C-O stretching (carbohydrates, polysaccharides) |
1051.1 |
1043.7 |
Shift suggests that Cd affects carbohydrate/polysaccharide structures. |
920.7 |
C-C stretching (polysaccharides) |
920.7 |
Absent |
Disappearance suggests structural changes in polysaccharides due to Cd. |
1595.2 (new) |
Interaction with carboxyl/amine groups |
Absent |
1595.2 |
New peak indicating Cd binding to carboxyl or amine groups, likely altering protein or membrane structures. |
872.2-760.4 (new) |
Metal-binding groups (phosphates/carboxylates) |
Absent |
872.2,760.4 |
New peaks indicating Cd interaction with specific groups such as phosphate or carboxylates. |
A shift in the C-H stretching vibration at 2911.1 cm-1 was seen in the lipid area, suggesting that Cd2+ interacts with the lipophilic elements of the bacterial membrane. This shift reflects abnormalities in the membrane’s lipid content or structure, perhaps as a result of Cd-induced disturbance of membrane integrity.
A peak at 1636.3 cm-1, which is the amide I band (C=O stretching), is still in close proximity to the spectrum of the untreated bacterial cells. The modest decrease in intensity, however, suggests that there may have been modifications to the structure of the protein after exposure to Cd2+. This could indicate partial denaturation of the protein or structural rearrangement. Furthermore, a novel signal was detected at 1595.2 cm-1, which can be related to Cd interactions with amine or carboxyl groups. This novel peak indicates that Cd2+ binds to functional groups that are involved in the sequestration of metals, potentially forming complexes with proteins or carboxyls.
The small shift in the amide II band (N-H bending and C-N stretching) at 1528.2 cm-1 suggests that there may be changes in the secondary structure of proteins. These changes could involve partial unfolding or folding of proteins as a result of the interaction with Cd2+.
The carbohydrate/phosphate area also showed a change, with a peak at 1043.7 cm-1 moving from 1051.1 cm-1. This implies that the polysaccharides or nucleic acid constituents of the bacterial cell wall may be reacting with Cd ions. In the treated cells, two additional peaks were seen at 872.2 cm-1 and 760.4 cm-1. These likely result from Cd2+ interacting with phosphate or other metal-binding functional groups, underscoring Cd’s interaction with particular biomolecules.
General Trends and Observations in Enterobacter sp. PGRG2 Under Cd Stress:
Broadening of O-H/N-H Region
The improved hydrogen bonding or direct contact between Cd2+ and hydroxyl or amine groups is suggested by the broadening and shift of the O-H and N-H stretching bands in the Cd-treated spectra. The bacterial cell wall, proteins, and water molecules are probably all impacted by this interaction, which modifies the biochemical environment.
Disruption of Lipid and Protein Structures
The alterations in the C-H stretching area (2911.1 cm2+) and amide I/II bands show that Cd2+ impacts both lipid and protein structures. Lipid disruption most likely takes place at the membrane level, and variations in amide bands indicate that the secondary structure of the protein may have changed, possibly resulting in denaturation or unfolding of the protein.
Emergence of New Peaks
New peaks that show up at 1595.2 cm2+, 872.2 cm2+, and 760.4 cm2+ suggest that Cd2+ interacts with specific biomolecules such amine, phosphate, or carboxyl groups. These interactions are probably caused by Cd binding to proteins, polysaccharides, or other biomolecules that sequester metals; this could be a function of the detoxification or stress-reaction processes of the bacterial cell.
Phosphate and Carbohydrate Interaction
Changes in carbohydrate-related peaks and a shift in the phosphate area (from 1233.7 cm2+ to 1274.7 cm2+) imply that Cd2+ interacts with polysaccharides, phospholipids, or nucleic acids inside the bacterial cell. These interactions may compromise these essential biomolecules’ structural and functional integrity, which could have an effect on important cellular functions.
Significant molecular changes are revealed by the FTIR analysis of Enterobacter sp. PGRG2 treated with 750 ppm of CdCl2·H2O. These changes mainly involve interactions between Cd2+ and hydroxyl, amine, carboxyl, and phosphate groups. The membrane integrity, protein conformation, and general biochemical stability of the bacterial cell are all impacted by these interactions, which result in structural alterations in proteins, lipids, and polysaccharides. The emergence of new peaks in the Cd-treated cells suggests that Cd2+ may bind to specific biomolecules, possibly as part of the cell’s stress response or detoxification processes, which could play a crucial role in understanding the bacterial adaptation and survival mechanisms under heavy metal stress.
FTIR analysis of Enterobacter sp. PGRG2 with Nickel (Ni) treatment
Upon exposure to Ni, Enterobacter sp. PGRG2 cultured in nutritional broth supplemented with 200 ppm of Ni(NO3)2 underwent significant metabolic changes, as revealed by the FTIR measurement (Figure 8 and Table 6). A tiny change to 3283.8 cm-1 in the O-H or N-H stretching region indicates that Ni interacts with amine or hydroxyl groups, which are probably present in proteins. This interaction suggests that changes in Ni binding or hydrogen bonding to these functional groups may affect the structure or function of the protein.
Table (6):
FTIR spectrum interpretation and comparison of untreated and Ni treated cells of Enterobacter sp. PGRG2
Wavenumber (cm⁻¹) |
Functional Group |
Untreated Cells |
Ni-Treated Cells |
Changes Observed |
|---|---|---|---|---|
3280.1-3283.8 |
O-H/N-H stretching |
3280.1 (broad) |
3283.8 (shifted) |
Slight shift in O-H/N-H stretching band, indicating interaction of Ni with hydroxyl or amine groups. |
2922.2-2922.2 |
C-H stretching (aliphatic chains, lipids) |
2922.2 |
2922.2 |
No shift, suggesting minimal interaction of Ni with lipid aliphatic chains. |
1635.7-1636.3 |
Amide I (C=O stretching in proteins) |
1635.7 |
1636.3 |
Minimal shift, indicating slight interaction of Ni with protein structures. |
1535.7-1527.5 |
Amide II (N-H bending, C-N stretching) |
1535.7 |
1527.5 |
Shift and reduction in intensity suggest Ni-induced changes in protein secondary structure. |
1233.7-1230 |
P=O stretching (phosphates, nucleic acids) |
1233.7 |
1230 |
Minor shift, indicating Ni’s interaction with phosphate or nucleic acid groups. |
1051.1-1043.7 |
C-O stretching (carbohydrates, polysaccharides) |
1051.1 |
1043.7 |
Significant shift, indicating Ni’s impact on carbohydrate or polysaccharide structure. |
920.7-1155.5 (new) |
C-C stretching (polysaccharides) |
920.7 |
1155.5 |
New peak at 1155.5 cm⁻¹, suggesting Ni-induced changes in polysaccharide structures. |
2851.4 (new) |
C-H symmetric stretching |
Absent |
2851.4 |
New peak indicating Ni’s interaction with membrane lipids or proteins, causing structural changes. |
1867.4 (new) |
Metal-carbonyl or metal interaction |
Absent |
1867.4 |
New peak possibly indicating strong interaction between Ni and carbonyl or metal-binding groups. |
1386.6 (new) |
C-H bending (lipids/proteins) |
Absent |
1386.6 |
New peak suggesting Ni’s interaction with lipids or proteins, altering the cellular structure. |
The peak at 2922.2 cm-1, which represents aliphatic lipid chains, does not exhibit any discernible shift or alteration in intensity, suggesting that Ni has no effect on the lipid composition of bacterial membranes. On the other hand, the amide I band shift to 1636.3 cm-1 implies that Ni and the protein backbone may interact somewhat, which could result in minute changes in protein structure. At 1527.5 cm-1, a more pronounced shift is seen, suggesting greater interactions with protein amide groups. This shift suggests that exposure to Ni may alter the secondary structure of proteins, including α-helices and β-sheets.
A minor shift in the phosphate-related peak to 1230 cm-1 indicates that Ni may interact with the phosphate groups in membrane phospholipids or nucleic acids, potentially changing their structural integrity. A more noticeable shift to 1043.7 cm-1 emphasises the effect of Ni on polysaccharide structures and suggests modifications to the makeup of the bacterial cell wall.
The observation of a new peak at 2851.4 cm-1, which is indicative of C-H symmetric stretching, is intriguing as it implies that Ni undergoes interactions with membrane lipids, potentially leading to structural alterations or an increase in membrane fluidity. A second new peak, located at 1867.4 cm-1, suggests the creation of Ni-carbonyl bonds or complexes and is probably associated with metal-carbonyl or other metal-binding interactions.
There are two more additional peaks that can be seen at 1155.5 cm-1 and 1386.6 cm-1, respectively. These correspond to structural alterations in polysaccharides and C-H bending, respectively. These peaks indicate that exposure to Ni modifies intracellular lipid or protein structures as well as the integrity of the cell wall (via disrupting polysaccharides).
In conclusion, the FTIR study shows that Enterobacter sp. PGRG2 is exposed to Ni, which causes a number of biochemical alterations. These alterations mainly impact protein structures, polysaccharides, and, to a lesser degree, membrane lipids. These changes shed light on how bacterial cells react to heavy metal stress and may be essential to comprehend the mechanisms underlying bioremediation in Ni-contaminated environments.
The FTIR analysis provided critical insights into the molecular interactions between heavy metal ions and the functional groups present in the bacterial cell components of Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2. These interactions and the resultant structural changes correlate directly with specific cellular responses, including the upregulation of stress proteins such as chaperones (e.g., GroEL and DnaK) to refold or degrade denatured proteins, as indicated by shifts in amide I and II peaks. Disruptions in lipid membranes, reflected in changes in C-H stretching bands, trigger lipid remodeling to stabilize the membrane and minimize metal ion influx. Similar observations have been made by Jaafar et al. in Shewanella oneidensis exposed to lead nitrate, where lipid remodeling was a key adaptation to mitigate membrane damage under heavy metal stress.12 Alterations in polysaccharide-associated peaks suggest enhanced exopolysaccharide production to bind and sequester extracellular metals, reducing their intracellular toxicity. This aligns with findings of Gupta et al. in Rhizobium sp. and Azotobacter sp., where increased exopolysaccharide secretion was observed as a defensive mechanism to immobilize heavy metals like lead and nickel.13 Shifts in phosphate vibrations imply interactions with nucleotides or membrane phospholipids, potentially activating DNA-binding proteins or repair pathways to counteract genotoxic effects. This is consistent with the work of D’Souza et al,14 which highlighted phosphate-related changes in Padina tetrastromatica (Hauck). Additionally, the emergence of new FTIR peaks indicates the production of stress-related metabolites involved in detoxification or metal chelation. Similar, work is also reported by Manasi et al. for Halomonas species, which produce polyamines to chelate metals and mitigate intracellular toxicity.15 Collectively, these molecular adaptations highlight the robustness of these bacteria under heavy metal stress and their suitability for bioremediation applications. Similar applications have been demonstrated in Rhodococcus erythropolis, by Canizo et al. which has been utilized in biosorption filters,16 and Pseudomonas putida, effectively integrated into bioreactors for wastewater treatment.17 In real-world scenarios, these strains can be implemented for soil detoxification by direct introduction or in combination with organic amendments, support phytoremediation by enhancing metal uptake and growth of hyperaccumulating plants, and treat industrial wastewater through integration into bioreactors. Additionally, immobilizing these bacteria on matrices like alginate beads can create effective biosorption filters for water treatment, while their bioaccumulation potential can stabilize and recover metals at e-waste sites. These findings demonstrate their utility in mitigating heavy metal pollution across various contaminated environments.
Furthermore, FTIR analysis of Pb (NO3)2, CdCl2·H2O, and Ni (NO3)2 exposed to various heavy metal concentrations, along with Enterobacter sp. PGRG2 and Achromobacter insolitus PGRG5, revealed significant biochemical alterations within bacterial cells. Modifications in the amide I and II peaks and the O-H, N-H, and C-H stretching bands indicated that these alterations were mostly associated with interactions between metal ions and significant biomolecules such proteins, lipids, and polysaccharides. When exposed to heavy metals, the structural components of proteins, lipid membranes, and polysaccharides were considerably altered in both Achromobacter insolitus and Enterobacter sp. This implies that metal ions interact with phosphate and carbonyl groups, denaturize proteins, and disrupt hydrogen bonds, all of which are critical for the integrity of cell membranes and metabolic activities.
Enterobacter sp. PGRG2 showed obvious interactions with Pb and Cd, especially impacting proteins and cell wall components, while Achromobacter insolitus PGRG5 clearly changed membrane structures and protein conformations in response to Ni and Cd. These results provide important insights into the application of these bacterial isolates in the bioremediation of heavy metal-contaminated settings by demonstrating their ability to bind metals and their possible processes of response to metal stress. The findings emphasize how crucial it is to research these microbial interactions in order to create efficient bioremediation plans that would lessen the amount of heavy metal contamination in the environment.
ACKNOWLEDGMENTS
The authors would like to thank and acknowledge the Ministry of Environment, Forest and Climate Change (MoEF) for providing the funding to conduct the research, as well as Jaypee Institute of Information Technology, Noida, for providing the infrastructure and support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RG, SD and PG conceptualized and designed the study. RG and PG performed acquisition and data interpretation. RG and PG wrote and revised the manuscript. SD and PG reviewed the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by The Ministry of Environment, Forest & Climate Change (MoEF&CC).
DATA AVAILABILITY
The genome sequence of Achromobacter insolitus PGRG5 and Enterobacter sp. PGRG2 strains has been deposited in GenBank under the accession number CP129898 and CP127837 (11), respectively. Both the assembly and raw reads are available at DDBJ/ENA/GenBank under the BioProject numbers PRJNA979826 and PRJNA981674 (11), respectively.
ETHICS STATEMENT
Not applicable.
- Briffa J, Sinagra E, Blundell R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon. 2020;6(9):e04691.
Crossref - Alengebawy A, Abdelkhalek ST, Qureshi SR, Wang MQ. Heavy Metals and Pesticides Toxicity in Agricultural Soil and Plants: Ecological Risks and Human Health Implications. Toxics. 2021;9(3):42.
Crossref - Ali N, Rampazzo RCP, Costa ADT, Krieger MA. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int. 2017;2017(1):9306564.
Crossref - Pande V, Pandey SC, Sati D, Bhatt P, Samant M. Microbial Interventions in Bioremediation of Heavy Metal Contaminants in Agroecosystem. Front Microbiol. 2022;13:824084.
Crossref - Li X, Sun M, Zhang L, Finlay RD, Liu R, Lian B. Widespread bacterial responses and their mechanism of bacterial metallogenic detoxification under high concentrations of heavy metals. Ecotoxicol Environ Safety. 2022;246:114193.
Crossref - Pham VHT, Kim J, Chang S, Chung W. Bacterial biosorbents, an efficient heavy metals green clean-up strategy: prospects, challenges, and opportunities. Microorganisms. 2022;10(3):610.
Crossref - Alotaibi BS, Khan M, Shamim S. Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species. Microorganisms. 2021;9(8):1628.
Crossref - Mandal S, Saha KK, Mandal NC. Molecular Insight into Key Eco-Physiological Process in Bioremediating and Plant-Growth-Promoting Bacteria. Front Agron. 2021;3:664126.
Crossref - Pagnucco G, Overfield D, Chamlee Y, et al. Metal tolerance and biosorption capacities of bacterial strains isolated from an urban watershed. Front Microbiol. 2023;14:1278886.
Crossref - Garg R, Dang S, Gauba P. Genomic analysis of heavy metal-resistant Enterobacter sp. PGRG2 isolated from electronic waste of NCR region, India. Afr J Bio Sc. 2024;6(13):5468-5496.
Crossref - Garg R, Dang S, Gauba P. Genomic characterization of Enterobacter sp. PGRG2 and Achromobacter insolitus PGRG5: bacterial strains isolated from soil present near electronics manufacture industry for heavy metal remediation. Microbiol Resour Announc. 2024;13(9):e00617-24.
Crossref - Jaafar R, Al-Sulami A, Al-Taee A. Bioaccumulation of cadmium and lead by Shewanella oneidensis isolated from soil in Basra governorate, Iraq. Afr J Microbiol Res. 2016;10(12):370-375.
Crossref - Gupta P, Diwan B. Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnol Rep. 2017;13:58-71.
Crossref - D’Souza L, Devi P, Divya Shridhar MP, Naik CG. Use of Fourier Transform Infrared (FTIR) spectroscopy to study cadmium-induced changes in Padina tetrastromatica (Hauck). Anal Chem Insights. 2008;3:117739010800300001.
Crossref - Manasi, Mohapatra S, Rajesh N, Rajesh V. Impact of heavy metal lead stress on polyamine levels in Halomonas BVR 1 isolated from an industry effluent. Sci Rep. 2017;7(1):13447.
Crossref - Canizo BV, Agostini E, Oller ALW, Dotto GL, Vega IA, Escudero LB. Removal of crystal violet from natural water and effluents through biosorption on bacterial biomass isolated from rhizospheric soil. Water Air Soil Pollut. 2019;230(8):1-14.
Crossref - Lin YH, Cheng YS. Phenol degradation kinetics by free and immobilized Pseudomonas putida BCRC 14365 in batch and continuous-flow bioreactors. Processes. 2020;8(6):721.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.