ISSN: 0973-7510
E-ISSN: 2581-690X
Salmonella Paratyphi A, also known as typhoidal Salmonella, is the causative agent of typhoid fever or paratyphoid fever, a threatening, invasive (bacteraemia), and occasionally fatal human disease (also called enteric fever). Even though Salmonella infections can be treated with different antibiotics, developing resistance to many broad-range antibiotics like chloramphenicol, nalidixic acid, ampicillin, and sulfamethoxazole is still a big problem. Therefore, an alternative strategy is needed urgently, which is more effective with minimal systemic side effects for treating diseases caused by Salmonella paratyphi A. In this current study, we describe the isolation, characterization, and in vivo evaluation of Sal11TP, a host-specific bacteriophage with lytic activity against multidrug-resistant Salmonella paratyphi A. Morphological examinations revealed that phage Sal11TP belonged to the order caudovirales of the Siphoviridae family, with an icosahedral head (62.8 nm) and a long tail (104.5 nm in length). Based on the one-step growth curve, Sal11TP has a short latent period (20 min) and burst size (29 PFU/cell). The in vitro stability test showed that it remained stable below 40 °C and pH 6-7 after treatment for 1 h. The ideal multiplicity of infection for phage Sal11TP was 0.001. The therapeutic potential of phage Sal11TP was evaluated using a mouse model. Findings of the study demonstrated a reduction in bacterial loads in the heart and kidney tissues of intraperitoneally infected mice during prophylaxis, as well as a dose-dependent and antibiotic versus phage treatment. The explanation of the characteristics and in vivo research results of phage Sal11TP show that it could be used as an effective bio-control agent to prevent Salmonella paratyphi A infections. Phage Sal11TP’s characteristics and in vivo results suggest it could serve as an effective bio-control agent against Salmonella paratyphi A infections.
Salmonella, Multidrug-Resistance, Phage Therapy, Antibiotics, Therapeutics, Biocontrol
Salmonella is an important food-borne organism that causes life-threatening infections in animals and humans worldwide. Salmonella infection is the most lethal in the United States and the second most common zoonotic pathogen in the European Union.1 Each year, an estimated 26 million cases of typhoid fever and 5 million cases of paratyphoid fever are reported worldwide, resulting in 215,000 deaths.2 According to Global Burden of Disease estimates, India accounted for more than half of the 14.3 million global cases of enteric fever in 2017, with 8.3 million cases and 72,000 deaths.3 Over the past 20 years, the number of cases of enteric fever caused by Salmonella Paratyphi A has gone up. In some parts of southern Asia, it now affects more than 50% of people.4
Salmonella is a genus of Gram-negative bacteria that are rod-shaped (bacillus) and belong to the family Enterobacteriaceae. Salmonella enterica and Salmonella bongori are the two main species of Salmonella. The type of species is S. enterica, which is further subdivided into six sub-species.5 More than 2600 serovars belong to the Salmonella genus. Based on host preference and illness symptoms in humans, Salmonella have been clinically classified as invasive S. Typhimurium (typhoidal Salmonella) or non-invasive S. enteritidis (non-typhoidal Salmonella).6 Salmonella Typhimurium and Salmonella Paratyphi A, commonly known as typhoidal Salmonella, are the causes of typhoid fever or paratyphoid fever, a dangerous, invasive (bacteraemia), and occasionally deadly human disease (also called enteric fever). The primary group includes Salmonella Typhimurium, which causes typhoid fever, and Salmonella paratyphi A, which causes paratyphoid fever (the other two being S. paratyphi C and d-tartrate-negative S. Paratyphi B).7
Humans serve as the only hosts for diseases like Salmonella Typhimurium and paratyphi A. Both of these species typically enter the bloodstream from the gastrointestinal tract, survive and replicate inside macrophages, and lead to chronic infection in 1-4% of patients.8 A persistent fever, headache, abdominal pain, anorexia, a non-productive cough (early in the disease), a relative bradycardia (slow heart rate), and hepatosplenomegaly are all symptoms of paratyphoid fever.9 The main cause of enteric fever is the consumption of food or water contaminated with the feces of patients and carriers in endemic locations. In particular, water plays an important role in urban settings.2
Antibiotics can be used to treat enteric fever. However, resistance to several antibiotics is rising, making treatment more challenging. In the last decade, Salmonella enterica serovar paratyphi A has become a more common cause of enteric fever, and medication resistance rates for this organism appear to be overtaking those for Salmonella enterica serovar Typhimurium, which is worrying.10 Resistance to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole became widespread, leading to the usage of fluoroquinolones in the 1980s as a primary treatment medicine. Resistance to nalidixic acid (a synthetic quinolone) is related to decreased fluoroquinolone susceptibility, which leads to treatment failure and enhanced loss of life.11 Quinolone resistance has increased, mainly in Southeast Asia.12 Further current typhoid fever vaccinations do not protect against S. paratyphi A.13
In the present investigation, phage therapy was explored as an alternative therapeutic to eradicate the disease burden pertaining to the wide spread of Salmonella paratyphi infections. We further reported the methodology for the isolation and characterization of Sal11TP phage in sewage samples routinely collected from SMS Hospital Jaipur, which specifically targets this bacterial strain. Lytic efficiency has been checked using bacterial culture of Salmonella enteric serovar as specific host cells as well as heart and kidney tissues were selected to assess the in vivo efficacy in mouse model.
Bacterial strain and culture conditions
The pathogenic bacteria host Salmonella, was obtained from the local SMS hospital’s microbiology department (the source was clinical isolates of human patients) in Jaipur. Salmonella strain identification and confirmation of antibiotic resistance were confirmed by the VITEK 2 technique (VITEK 2 is a fully automated system that performs bacterial identification and antibiotic susceptibility testing). Overnight bacterial cultures were prepared in tryptic soy broth and incubated in shaking conditions at 35 °C for 18 h. Bacterial strains were streaked on the fresh Tryptic soya agar plate and incubated for 24 hours at 37 °C. After checking growth, plates were wrapped with Parafilm and stored at 4 °C for further use. Before each experiment, bacterial samples were freshly subcultured and 24 h cultures were used. Strains were further processed to determine cfu/ml in an overnight Tryptic soy broth culture. With the help of regular sub-culturing and viability, the purity of the organisms was maintained.
Phage enrichment and isolation
The bacteriophages were isolated from sewage water samples of the local wastewater treatment plant (location: 26.8943°N, 75.8126° E), recipients of wastewater from a local hospital in Jaipur, and kept at 4 °C before processing. In total, 15 samples were collected. A modified version of Twest and Kropinski was adopted for bacteriophage enrichment from collected water samples.14 The phages were enriched by mixing 50 ml of wastewater with the same amount of double-strength Tryptic soy broth (TSB) and 5 ml of a single bacterial strain cultured overnight. After incubation overnight at 30 °C, 10 ml of the mixture was shaken with 1% v/v chloroform and left at room temperature for 30 min. to kill the bacteria, centrifuged at 6000 rpm at 4 °C for 15 minutes, and filtered through a disposable sterile syringe filter of pore size 0.45 µm. The filtrate thus obtained was termed phage lysate, which was further used for phage detection.
Phage detection
The presence of bacteriophage in phage lysate was detected by an enrichment spot test. This test was performed by spreading a loop of host bacteria on the surface of the TSA agar plate. After 30 min, the fluid in the plates was getting dry, and then 10 µl of the clarified enrichment was placed on the bacterial lawn. Plates were observed for the zone of killing after overnight incubation at 30 °C.
Purification of phage
The method used for isolation, purification, and enumeration was the double agar overlay assay (DLA) by Adams.15 The bottom layer was formed with TSA containing 1.5% agar, and then separate freshly prepared soft agar (0.5% agar in TSB) was put in a water bath maintained at 48 °C. Then sterile, filtered phage lysates were diluted in SM buffer to five different dilutions (10-5-10-9). 100 µl of each diluted phage and 150 µl of target bacteria were allowed to rest for 10 min for phage absorption. Later, this mixture of phage and host was added to a tube containing 2.5 ml of soft agar and spread on pre-warmed TSA plates, which were allowed to harden for 30 minutes. The plates were incubated in a single layer overnight at 37 °C. The plates were observed for plaque morphologies (large, small, clear, halo, and double zone) after overnight incubation. A sterile inoculation loop was used for the purification of phage. Materials from the center of the plaque were scraped using a loop and transferred aseptically to the TSB broth containing the respective host. They were then incubated at 30 °C for about 18 hours. Later, the incubation mixture was centrifuged at 5000 rpm for 30 min, and the supernatant was filtered through a 0.45 µm membrane filter. The filtrate was stored in 50 mL sterile amber bottles. The plaque assay was again carried out. Thus, the cycle was repeated three times to ensure the purity of the phages. All the phage lysate was stored in SM buffer at 4 °C. All phage stocks were stored in SM buffer at 4 °C or in a 50% glycerol solution at -70 °C.
Phage enumeration by double agar overlay assay
The concentration of infective phage (titre) in a phage stock can be established by pouring serial dilutions of phage lysate. 100 µl of diluted phage and 150 µl of target bacteria (OD 0.6 or 108 cfu/ml) along with 2.5 ml of soft agar were poured on a fresh TSA plate. The plates were left for 30 minutes to allow the top agar layer to solidify. Then they were incubated at 37 °C for 18-24 hours. After overnight incubation, the plates were checked for plaque formation. Plaques appeared in the form of localized clear, halo, or translucent zones. Plates with 30-300 plaques were selected for titre determination. The plaque number was counted on plates for phage titre using the following formula:
Phage titre = Number of plaques x 10 x reciprocal of counted dilution
The concentration is expressed as a plaque forming unit (pfu), analogous to the colony forming unit.
Determination of host range
Host ranges of Sal11TP were performed by using different bacterial cultures (Table 1). It was done using the method of Elizabeth Kutter with some modifications.16 Each bacterial strain was tested for lysis and cultured in TSB up to the mid-log phase, and then serial dilutions of all the strains were prepared in a 0.9% saline solution. All bacteria were diluted to the same concentration (108 cfu/ml) by using the formula:
Colony forming unit calculated as cfu/ml = No. of colonies x dilution / Volume of plating dilution (ml)
100 µl of each bacterial culture was mixed with 2.5 ml of 0.5% soft agar in different sterile test tubes and poured onto the surface of the TSA plates. The plates were left to dry, and then 10 µl of phage lysate dilutions (108 pfu/ml) was applied at the center of the plate and incubated overnight at 37 °C to observe whether there was a lysis zone on the plate.
Transmission electron microscopy
The morphological characterization of bacteriophage was carried out through transmission electron microscope (TEM) with some modifications.17 High titer phage suspension in SM buffer was absorbed for 5 minutes on copper-coated grids (3 mm; 300 mesh). Negative staining was performed to visualize the bacteriophage particles under TEM. For staining, 2% (w/v) phosphotungstic acid (PTA) was added to bacteriophage particles for 10 s. After 20 minutes, the grid was investigated under a 200 kV TEM (FEI Tecnai S Twin) (SAIF, AIIMS, Delhi, India).
Determination of optimal Multiplicity of Infection (MOI) of phage
The multiplicity of infection (MOI) is the ratio of virus particles (pfu/ml) to host cells (cfu/ml) at which the highest phage titer was observed in each infection medium. The MOI experiment was carried out in accordance with Delbruck,18 with minor modifications. In separate sterile test tubes, 100 µl of phage lysate 105 pfu/ml and 100 µl of each overnight fresh bacterial culture 104-108 cfu/ml were mixed with 5 ml of broth. The tubes were left in a shaker incubator (110 rpm) for 6 h at 37 °C before centrifuging for 15 min at 6000 rpm. The collected supernatant was filtered through a 0.45 µl sterile syringe filter. The phage titre of the filtrate phage lysate was determined using a double agar overlay assay. The phage-to-bacterium ratio at which the highest phage titer was obtained was considered the optimal multiplicity of infection for that phage.
Thermal stability test
The stability of the isolated phages was tested at various temperatures. Sterile Eppendorf tubes containing 1 ml of pure phage lysate (1×108) were maintained in a water bath for 1 hour at 4 °C, 25 °C, 37 °C, 42 °C, 50 °C, 60 °C, 70 °C and 90 °C. The titre of the bacteriophage was then determined using the double layer test method following treatment. The experiment was done in triplicate.
pH stability test
The pH stability tests were carried out using a modified approach developed.19 With the addition of 1 M HCl and 1 M NaOH, fresh TSB was adjusted to a pH range of 2 to 12. 1 ml of phage lysate was mixed with 9 ml of pH-adjusted media and incubated at 37 °C for 3 h. The control was TSB medium (pH 7) containing phage lysate. Following incubation, phage titre was assessed for each aliquot using the DLA method.
One step growth curve of the bacteriophage
One-step growth experiments were performed using a modified method.20 Before starting the experiment, a fresh overnight culture of bacteria in TSB was adjusted to an optical density (OD 0.5 at 600 nm) to reach the mid-exponential phase to give 108 cfu/ml and then 1 ml of these bacterial cells were infected with phage pfu/ml at which the optimal MOI was obtained. The phage-bacterium mixture was then allowed to adsorb for 5 minutes at 37 °C and then centrifuged at 12000 x g for 1 min to remove the free phage particles. After the supernatant was removed, the pellet containing the phage-infected bacterial cells was resuspended in 1.2 ml of fresh TSB broth and incubated with shaking at 120 rpm and 37 °C. 200 µl of sample was taken at 10 minute intervals for 40 min and was tittered by the DLA method. This assay was performed in triplicate. The latent period and burst size were determined accordingly.21 The latent period was identified as the time between phage absorption and the initial rise in plaque number. The burst size was identified by dividing the average of pfu/infected-cell in the post-rise period of the growth curve by the average of pfu/infected cells in the pre-rise period of the growth curve.
Animal model and experimental design
Swiss albino mice (male) of age 6-8 weeks (weight 20-25 g) were used in the study. Randomly bred healthy animals were distributed in six groups, each containing 9 animals, were as follows: I: healthy mice treated intraperitoneally with saline (control); II: infected mice induced intraperitoneally by S. paratyphi A (Bacteria), III: Bacteria treated mice intraperitoneally with Gentamycin (simultaneous), IV: Bacteria treated mice intraperitoneally with Bacteriophage (simultaneous), V: Bacteria treated mice with intraperitoneally with Bacteriophage (later), VI: Bacteria treated mice intraperitoneally with Bacteriophage along with Gentamycin (simultaneous). Experiments were carried out with the approval of Institutional Animal Ethics Committee (IAEC), University of Rajasthan (UDZ/IAEC/I/05/07/2017). The control group was given 200 µl of phosphate buffered saline (pH 7.4). Treatment groups, as specified above, were infected with 200 µl of 106 cfu/ml of Salmonella paratyphi A. The mode of induction was intraperitoneal for Salmonella paratyphi A. Groups assigned to be treated by phages were administered by 109 pfu/ml (Concurrent or delayed). For dose efficacy tests, phages were administered to respective groups at 106 pfu/ml, 107 pfu/ml, and 108 pfu/ml challenged by 106 CFU/ml of infectious agent.
Measurement of bacterial load in tissues
Following intraperitoneal infections, phage therapy was allowed after 24 hours before scheduled euthanasia mediated by cervical dislocation. Tissue samples (0.5 cm) were obtained after dissecting the treated mice and suspended in 1 ml of sterile cold phosphate buffer saline (PBS). The obtained tissue samples were homogenized using a bead beater for 6 minutes at 30 Hz. Homogenized samples (20 µl) were added to a 96-well plate containing 180 µl of PBS. Following proper mixing, serial dilution was performed by adding 20 µl from each previous dilution, forming a concentration of 10-1 to 10-6. Each dilution was plated in triplicate on a TSB grid plate and incubated for 12 h at 37 °C. Colony forming units (cfu) were recorded with respect to the dilution factor and represented in cfu/g tissue.
Bacterial strain
The Salmonella strain collected from the SMS hospital was identified to be Salmonella enterica Serovar enterica (Salmonella paratyphi A). Throughout the research, this natural S. paratyphi A strain was used as a typical Salmonella enterica strain for phage isolation, propagation and evaluation. This strain showed resistance to (>16 mg/l cefuroxime), (>16 mg/l cefuroxime Axetil), (<=2 mg/l Amikacin), (<=1 mg/l Gentamicin), (>=32 mg/l Nalidixic acid), (>1 mg/l Ciprofloxacin) and (>1 mg/l Tigecycline) as analyzed by the clinical laboratory service at Jaipur.
Phage isolation and detection
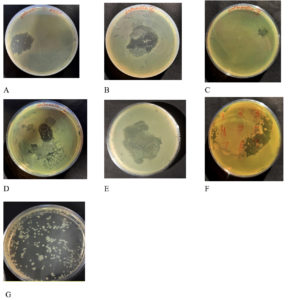
Only seven out of the 15 water samples produced a clear lytic zone by enrichment and spot assay analysis (Figure 1). Sample 1, sample 2, sample 4, sample 5, sample 9 and sample 11 were positive for lytic zone produced against host specific S. paratyphi A. On the basis of the larger area of lytic zone produced after prolonged incubation, sample 11 phage lysate was further processed for purification and enumeration and the phage obtained was named Sal11TP.
Figure 1. Enrichment and spot assay analysis produced the distinct lytic zone in seven plates (A-G) out of the 15 water samples. Samples 1, 2, 4, 5, 6, 9, and 11 (Sal11TP) all tested positive for lytic zone generated against a particular host S. Paratyphi A
Plaque enumeration
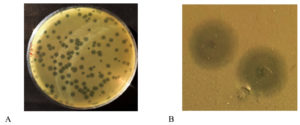
The DLA method was used to assess the phage titre after the first purification, and the result of this determination was 138 plaques at 10-5 dilution (100 µl purified phage). Thus, by using the formula above, 1.38 x 108 pfu/mL phage titre was obtained (Figure 2A). The propagation and subsequent enumeration were repeated thrice, and on each occasion, the phage titre of Sal11TP was 108 pfu/ml.
Plaque morphology
The isolated phage Sal11TP produced circular individual plaques against host bacteria on the double layer agar plate (Figure 2B). This bacteriophage formed a bull’s eye shaped clear plaque with a 2 mm wide clear center (with some spots of bacterial growth) that was surrounded by a 4 mm wide opaque halo zone.
Figure 2. A) Plaques obtained by double layer agar assay B) Plaque morphology: On a double layer agar plate with S. Paratyphi A lawn, Sal11TP bacteriophage produced bulls’ eye shaped clear plaques having an inner circle of a diameter of 2 millimetres with spots or rings of growth in the middle of clear regions of complete lysis surrounded by a large halo of a diameter of 4 mm
Host range of Sal11TP phage
The bacterial strains used for host range analysis are listed in Table 1. The ability of the Sal11TP bacteriophage to infect and form lytic zones against various bacteria was examined using a spot test, and Sal11TP was able to infect 3 out of the 8 Salmonella clinical isolates of human patients from SMS hospital while failing to establish any lytic zone on the remaining isolates. Sal11TP phage did not cross the genus border and did not lyse any of the other bacteria examined, including Methicillin-resistant Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, and E. coli.
Table (1):
The host range test for the isolated bacteriophage Sal11TP was conducted on numerous bacterial strains
Bacterial culture Salmonella enteric Serovar |
Spot test |
Source |
|---|---|---|
Paratyphi A 01 |
+ |
Clinical isolate |
Paratyphi A 02 |
+ |
Clinical isolate |
Paratyphi A 03 |
– |
Clinical isolate |
Paratyphi A 04 |
+ |
Clinical isolate |
Typhimurium 01 |
– |
Clinical isolate |
Typhimurium 02 |
– |
Clinical isolate |
Typhimurium 03 |
– |
Clinical isolate |
Typhimurium 04 |
- |
Clinical isolate |
Methicillin-resistant Staphylococcus aureus (MRSA) |
- |
Clinical isolate |
Klebsiella pneumonia |
– |
Sewage isolate |
Pseudomonas aeruginosa |
– |
Sewage isolate |
Escherichia coli |
– |
Sewage isolate |
+ produced clear lytic zone; – Was unable to produce lytic zone
Sewage isolates: Strains isolated from raw sample of local sewage treatment plant at Jaipur
Transmission electron microscopy
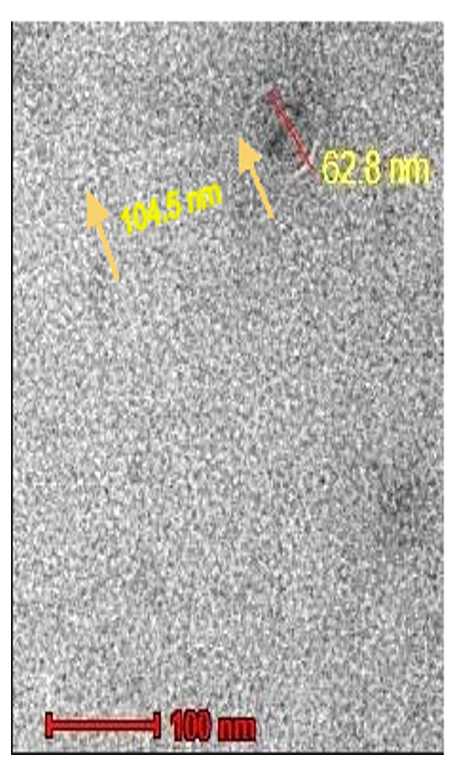
An electron microscope was used to examine negatively stained pure phage Sal11TP. Sal11TP virions have an icosahedral head 62.8 nm in diameter and a noncontractile tail 104.5 nm in length, indicating that they are members of the Siphoviridae family of the order caudovirales, according to the current ICTV classification system (Figure 3).
Figure 3. Transmission electron micrograph of the Sal11TP phage. Sal11TP belongs to the Siphoviridae family. The diameter of its head is 62.8 nm, and the length of its non-contractile tails is 104.5 nm
Optimal multiplicity of infection
The bacteriophage titer was determined to be 1.05 x 108 and 1.52 x 108 pfu/ml after 6 h of incubation when the MOI was 0.1 and 0.01, respectively. The maximum bacteriophage titer, 2.84 x 108 pfu/ml, was observed when MOI was 0.001 (Table 2). Therefore, the optimal MOI of S. paratyphi A infected bacteriophage Sal11TP was determined to be 0.001.
Table (2):
Optimal multiplicity of infection of phage Sal11TP
PFU of phage Sal11TP (pfu/ml) |
CFU of host S. paratyphi A strain |
Multiplicity of infection (MOI) |
Sal11TP phage titers |
|---|---|---|---|
108 |
106 |
100 |
4.1 x 107 |
108 |
107 |
10 |
5.0 x 106 |
108 |
108 |
1 |
9.7 x 107 |
107 |
108 |
0.1 |
1.03 x 108 |
106 |
108 |
0.01 |
1.52 x 108 |
105 |
108 |
0.001 |
2.84 x 108 |
Thermal and pH stability test
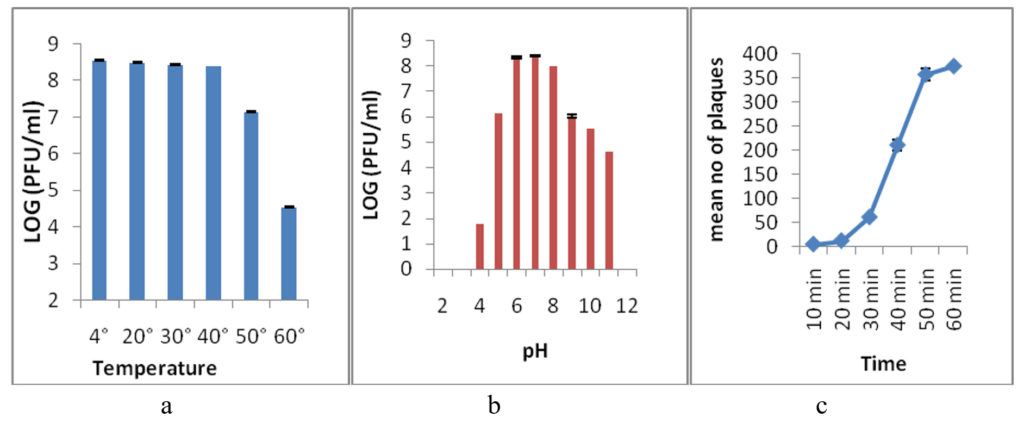
The thermal stability test was performed to analyze the heat resistance of the Sal11TP phage. After 60 minutes, the phage was stable at 4°C, 20°C, 30°C, and 40°C, but lowered its infectivity by one log at 50°C and three logs at 60°C, and lost its infectivity completely at 70°C and higher temperatures (Figure 4a). A pH stability test was done for 3 hours at different pH levels to find the best pH for the Sal11TP phage. Sal11TP was stable at pH values between 6.0 and 7.0. At pH 5.0, the phage’s titer drops by two orders of magnitude. At pH 4.0, it was down to 10 pfu/ml, and at pH 3.0 and 2.0, it was no longer infectious. After pH 8.0, the phage titer dropped a lot and reached 104 pfu/ml at pH 11.0, but it disappeared completely at pH 12.0 (Figure 4b).
One step growth curve
The change in the number of phages during one cycle of replication was used to figure out the Sal11TP phage’s latent period and burst size. After the first steady period of 20 min, the phage titer started to go up (Figure 4c). So, the latent period for Sal11TP was found to be 20 min, and the size of the burst was approximated to be around 29 phages per infected cell.
Figure 4. Thermal and pH stability of isolated phage Sal11TP at various temperatures and pH. a-b) Phage Sal11TP had stable activity from 4°C to 40°C temperature after treatment for 1 hour and produced the most plaque at pH 6-7. c) One-step growth curve had a latency period of 20 min and a burst size of 29 phages per infected cell. The values shown correspond to the means and SD of three independent biological repeats (n = 3)
Prophylaxis study
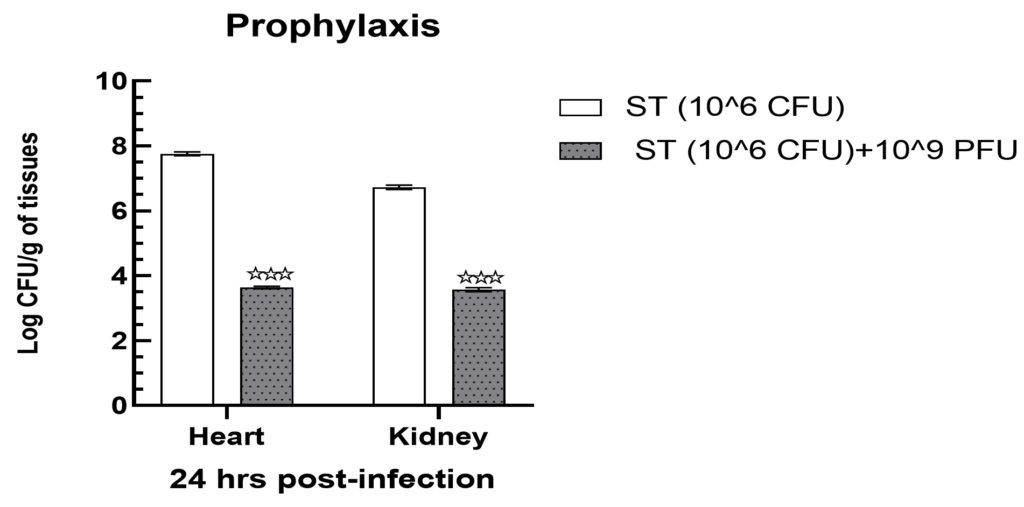
Mice infected with 106 cfu in group II showed a substantial increase in bacterial load in heart tissues, which was recorded as 7.76 ± 0.03 log cfu/g. Likewise, renal tissues of the same group showed a slightly lower but still statistically significant reduction in bacterial load, which was found at 6.73 ± 0.03 log cfu/g (Figure 5). Following treatment with 109 pfu of phage, the bacterial load in the heart and kidney tissues of group IV demonstrated a significant reduction (p < 0.001) up to 3.64 ± 0.02 and 3.57 ± 0.03 log cfu/g of tissues samples (Figure 5).
Figure 5. Prophylaxis assay indicated a significantly declined bacterial load in heart and kidney tissue samples when treated with 109 PFU. Levels of significance were measured between Group II and Group IV, where ** is expressed as P < 0.01 and
*** is expressed P < 0.001
Phage dose response study
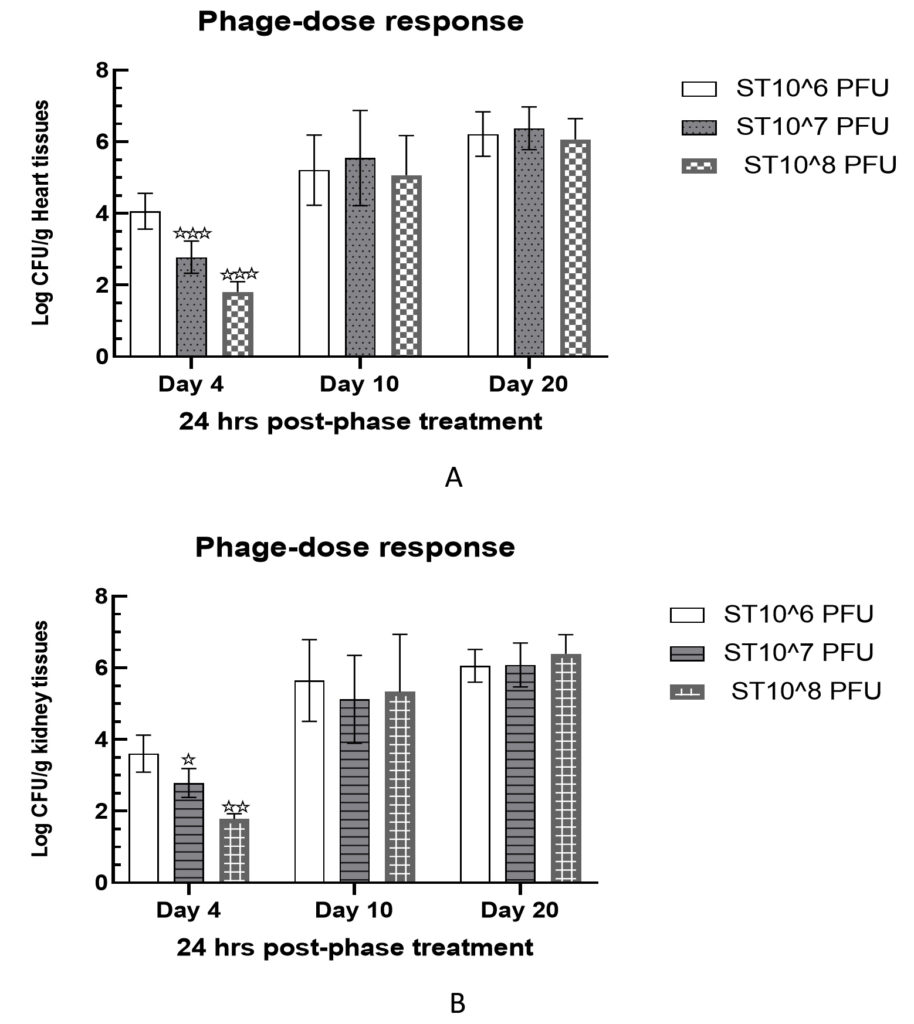
Intraperitoneally treated mice with 106 cfu of Salmonella paratyphi A in group V were examined for varying phage doses for a period of 20 days. All treated mice demonstrated a significant reduction in bacterial load in heart and kidney tissue samples on day 4 post-infection. In heart tissues, bacterial load was measured as 4.06 ± 0.23, 2.78 ± 0.21, and 1.81 ± 0.14 log cfu/g against phage doses of 106 pfu, 107 pfu, and 108 pfu, on the 4th day, respectively. This clearly indicated a dose dependent loss of bacteria cells in heart tissue samples. Similar observations were also recorded for kidney tissue samples on the day 4th post-infection which were recorded as 3.61 ± 0.24, 2.79 ± 0.19, and 1.78 ± 0.07 log cfu/g against 106 pfu, 107 pfu, and 108 pfu, respectively (Figure 6).
Figure 6. Delayed response of phage therapy against S. paratyphi A challenged mice. Bacterial load was measured in heart (A) and kidney (B) tissues samples.
Levels of significance were measured between Group II and Group IV, where * is expressed as P < 0.05 and ** P < 0.01 is expressed *** P < 0.001
Assessment of antibiotic efficiency compared to novel phage therapy
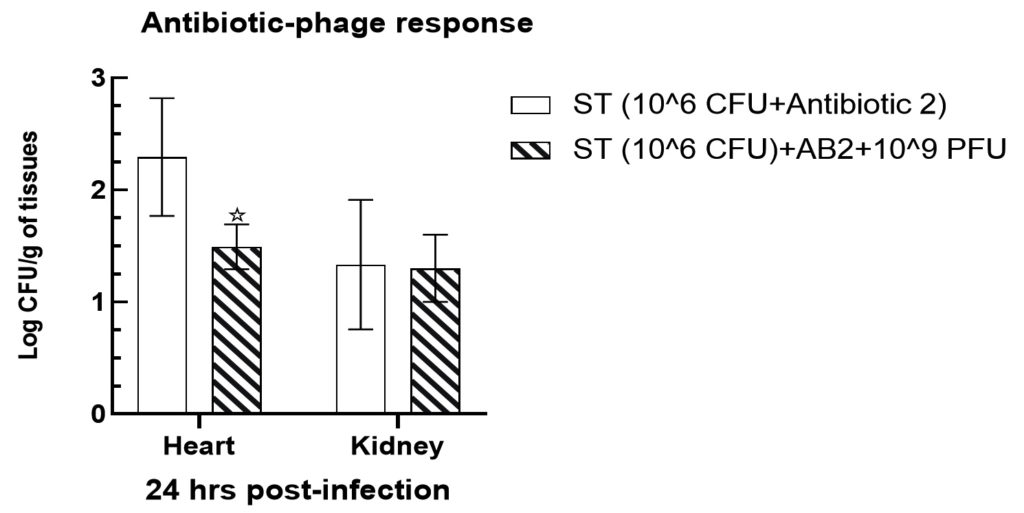
Mice infected intraperitoneally with 106 cfu of Salmonella paratyphi A were exhibited to have a significant decline (p < 0.05) bacterial load in the heart tissues in group V up to 1.49 ± 0.09 and 1.3 ± 0.14 log cfu/g, respectively, when compared with the gentamycin treated mice tissues such as 2.29 ± 0.25 and 1.33 ± 0.27 log cfu/g in group III (Figure 7). However, no significant alterations were observed in kidney tissues when compared with heart tissue samples. The effectiveness of phage therapy was significantly enhanced when investigated along with antibiotic treatment in heart tissues, while no significant alteration was observed in the bacterial load in the kidney tissues in group VI animals (p > 0.05).
A significant and widely dispersed foodborne pathogen, Salmonella is a leading cause of illness worldwide, with symptoms ranging from gastroenteritis to enteric fever. Salmonella paratyphi A is one of the main causative organisms of enteric fever. Antibiotics are frequently used to treat Salmonella infections; however, because of the unrestricted use of antibiotics, the problem of drug resistance is becoming worse.22 Bacteriophages have gradually been considered by scientists as a new biological therapy for the prevention and treatment of bacterial diseases due to their rapid propagation, host specificity, low mutation rates,23 abundance in the environment and absence of any noticeable side effects while being applied for treatment in humans and animals.24-26 To expand the phage arsenal, it is urgently required to isolate new, sensitive phages and analyze their physiological and genetic properties.
This study describes the isolation and identification of a lytic Sal11TP phage from hospital sewage that was highly specific and had a constrained host range against multidrug-resistant Salmonella paratyphi A. In our study, we have isolated 7 phages against Salmonella from a hospital sewage wastewater treatment plant in Jaipur. Due to its high lytic effectiveness, phage Sal11TP was chosen for additional biological and morphological characterization as well as in vivo testing. For phage therapy, the majority of phages were separated from various environments, such as sewage, freshwater, soil, and more typically, where the bacterial hosts are present.27,28 Sewage was observed to contain the highest number of phages, at 1010 ml-1, indicating that it is an important source for phage isolation.29 Previously, vB_SenS_SE1 Salmonella phage was isolated from a wastewater treatment plant in Beijing.30 A virulent bacteriophage (PSDA-2) against Salmonella enteric Serovar Typhimurium was isolated from the sewer sewage.31
To date, multiple Salmonella bacteriophage families have been identified, including Siphoviridae, Podoviridae, and Myoviridae.32 Morphological examination by TEM showed that Sal11TP had an icosahedral, non-enveloped head of 62.8 nm and a non-contractile tail of 104.5 nm. According to the International Committee on Taxonomy of Viruses (ICTV), Sal11TP can be classified into the order Caudovirales of the family Siphoviridae. On the other hand, phage SE-W109 was discovered to be a Siphovirus with a smaller, icosahedral head (62.2 nm) and a non-contractile tail attached to a less extensive baseplate structure (116 nm).33 Sal11TP’s ideal pH was 7, but it could also thrive in the ranges between pH 4 to 11. Sal11TP may be able to survive in more hostile environments than SaFB14 and φst1 because the lytic phage of SaFB14 grows well only between pH 3-10, unlike φst1 (pH 4-9).34 The phage Sal11TP demonstrated good temperature tolerance because it could withstand temperatures up to 40°C without losing infectivity. However, after an hour of incubation, the number of phages started to decline at greater than 50°C while above 80°C, there was a complete loss of infectiousness.
For the following experiments, the MOI produced the highest phage titer, which was considered ideal.35 In the current study, Sal11TP’s optimal MOI was found to be 0.001, yielding a lysate concentration of 2.84 x 108/mL (Table 2). According to the one step growth curve, the latency time of Sal11TP was 20 min, similar to vB_SPuM_SP116,36,37 SS3e38 less than Salmonella phage φst1 (50 min), vB_SenS_SE1 (40 min) and burst size was 29 phages per infected cell more than SaFB14 (23 phages/cell), φst1 (22 phages/cell) and vB_SenS_SE1 (19 phages/cell) while less than PSDA-2 120 pfu/cell.39 Phage Sal11TP has a short latent period and a large burst size, allowing it to effectively remove host bacteria in a short period of time, making it a good candidate for biocontrol.
In this investigation, we successfully demonstrated phage therapy at the dose ranges between 106-109 pfu against intraperitoneally infected mice with S. paratyphi A, which specifically reduced bacterial infections in the selected vital organs. In this study, phage therapy with varying dose regimens demonstrated a significant reduction in bacterial load in tissue samples of the heart and kidneys during different time intervals, which revealed clinical efficacy on the 4th day of exposure. Accordingly, time dependent efficacy was also observed by Wang et al. for the complete eradication of imipenem-resistant P. aeruginosa infections using bacteriophage phages in mice. All mice were infected intraperitoneally with S. paratyphi A died when phage therapy was delayed between 10 and 20 days.40 Similar findings were reported for E. coli infections when intraperitoneal phage therapy was delayed in mice.41 The bar graph profile exhibited that antibiotic efficiency was enhanced significantly in the phage treatment group VI, which may pave new avenues for developing novel biocontrol agents against this bacterial strain. Our results well corroborate that antibiotic efficiency was improved when combined phage therapy was used in mice.42 This isolated and characterized phage played an important role in the prevention and control of bacterial infections induced by S. paratyphi A. Phage therapy is an effective biocontrol agent as well as synergistically efficacious with the gentamycin antibiotic in the early days of infection because phages specifically bind to the surface receptor of a specific host bacterial strain through receptor binding proteins (RBP).43 This is preliminary experiment that may further enhance the safety and efficacy of phage therapy by editing its novel genome and utilizing genetic engineering tools to improve susceptibility to a broad range of challenging bacterial strains at the onset of infection.
This study establishes phage Sal11TP as a promising bio-control agent against multidrug-resistant Salmonella paratyphi A major cause of invasive and potentially fatal enteric fever. The phage’s stability under physiological conditions, short replication cycle, and potent lytic activity highlight its suitability for therapeutic applications. In vivo experiments demonstrated significant bacterial load reduction in infected tissues, confirming its efficacy in both prophylactic and therapeutic settings. These findings provide a strong basis for advancing phage Sal11TP as an alternative strategy to combat antibiotic-resistant S. Paratyphi A infections. Further research should explore optimizing delivery methods and evaluating its potential in combination with antibiotics for enhanced therapeutic outcomes.
ACKNOWLEDGMENTS
The authors are grateful to the Centre for Advance Studies (CAS), Department of Zoology, University of Rajasthan, Jaipur, for providing the Animal House facility for in vivo study. The authors are also thankful to the funding agency, the Council of Scientific and Industrial Research (CSIR), for supporting this study through a fellowship.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NM and AS conceptualized the study. AS, NM and IM design the experiments. IM carried out the in vivo evaluation experiments. PG performed isolation and characterization of the bacteriophage and data analysis. KS assisted in TEM analysis. PG and KS performed result interpretation. NM supervised the study. PG wrote the manuscript. IM revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by the Council of Scientific and Industrial Research (CSIR), India, in the form of Junior/Senior Research Fellowship.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Ethical Committee CPCSEA/IAEC, University of Rajasthan, under approval number UDZ/IAEC/I/05/07/2017.
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food borne outbreaks in 2017. EFSA Journal. 2018;16(12):e05500.
Crossref - McAteer J, Derado G, Hughes M, et al. Typhoid fever in the US Pediatric Population, 1999-2015: opportunities for improvement. Clin Infect Dis. 2021;73(11):e4581-9.
Crossref - Stanaway JD, Reiner RC, Blacker BF, et al. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(4):369-381.
- Ochiai RL, Wang X, von Seidlein L, et al. Salmonella paratyphi A rates, Asia. Emerg Infect Dis. 2005;11(11):1764.
Crossref - Kusumaningrum HD, Riboldi G, Hazeleger WC, Beumer RR. Survival of foodborne pathogens on stainless steel surfaces and cross-contamination to foods. Int J Food Microbiol. 2003;85(3):227-236.
Crossref - Su LH, Chiu CH. Salmonella: clinical importance and evolution of nomenclature. Chang Gung Med J. 2007;30(3):210.
- Okoro CK, Kingsley RA, Connor TR, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44(11):1215-1221.
Crossref - Sanderson KE, Liu SL, Tang L, Johnston RN. Salmonella Typhi and Salmonella Paratyphi A. In Molecular medical microbiology 2015;2:1275-1306.
Crossref - Gokmen V, Morales F. Encyclopedia of Food Safety. 2014.
- Centers for Disease Control and Prevention. CDC health information for international travel 2014: The yellow book. Oxford University Press. 2013. https://wwwnc.cdc.gov/travel/page/yellowbook-home
- Day J. Jaundice in a returned traveler from Nepal. Infectious Diseases. 2010;2:1085-1087.
Crossref - Chau TT, Campbell JI, Galindo CM, et al. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007;51(12):4315-4323.
Crossref - Ferri FF. Ferri’s Clinical Advisor 2021 E-Book: 5 Books in 1. Elsevier Health Sciences. 2020.
- Clokie MRJ, Kropinski AM. Bacteriophages. In: Bacteriophages: Methods and Protocols. Humana Press; 2009:1-6.
Crossref - Adams MH. Bacteriophages. Bacteriophages. 1959.
Crossref - Kutter E. Phage host range and efficiency of plating. Methods Mol Biol. 2009;501:141-149.
Crossref - Goodridge L, Gallaccio A, Griffiths MW. Morphological, host range, and genetic characterization of two coliphages. Appl Environ Microbiol. 2003;69(9):5364-5371.
Crossref - Delbruck M. The growth of bacteriophage and lysis of the host. J Gen Physiol. 1940;23(5):643-660.
Crossref - Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol. 2009;59(3):274-281.
Crossref - Bao H, Zhang H, Wang R. Isolation and characterization of bacteriophages of Salmonella enterica serovar Pullorum. Poult Sci. 2011;90(10):2370-2377.
Crossref - Hyman P, Abedon ST. Practical methods for determining phage growth parameters. Methods Mol Biol. 2009;502:175-202.
Crossref - Cogan TA, Humphrey TJ. The rise and fall of Salmonella enteritidis in the UK. J Appl Microbiol. 2003;94(s1):114-119.
Crossref - Kim KP, Klumpp J, Loessner MJ. Enterobacter sakazakii bacteriophages can prevent bacterial growth in reconstituted infant formula. Int J Food Microbiol. 2007;115(2):195-203.
Crossref - Hua Y, Luo T, Yang Y, et al. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol. 2018;8:2659.
Crossref - Harada LK, Silva EC, Campos WF, et al. Biotechnological applications of bacteriophages: State of the art. Microbiol Res. 2018;212-213:38-58.
Crossref - Keen EC, Dantas G. Close encounters of three kinds: bacteriophages, commensal bacteria, and host immunity. Trends Microbiol. 2018;26(11):943-954.
Crossref - Hyman P. Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals. 2019;12(1):35.
Crossref - Ross A, Ward S, Hyman P. More is better: selecting for broad host range bacteriophages. Front Microbiol. 2016;7:1352.
Crossref - Tamaki H, Zhang R, Angly FE, et al. Metagenomic analysis of DNA viruses in a wastewater treatment plant in tropical climate. Environ Microbiol. 2012;14(2):441-452.
Crossref - Lu M, Liu H, Lu H, Liu R, Liu X. Characterization and genome analysis of a novel Salmonella phage vB_SenS_SE1. Curr Microbiol. 2020;77(7):1308-1315.
Crossref - Karthik L, Kumar G, Keswani T, Bhattacharyya A, Chandar SS, Bhaskara RKV. Protease inhibitors from marine Actinobacteria as a potential source for antimalarial compound. PloS one. 2014;9(3):e90972.
Crossref - Li M, Li M, Lin H, Wang J, Jin Y, Han F. Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-a and its endolysin. Arch Virol. 2016;161(2):377-84.
Crossref - Phothaworn P, Supokaivanich R, Lim J, et al. Development of a broad-spectrum Salmonella phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand. Food Microbiol. 2020;92:103586.
Crossref - Tang F, Zhang P, Zhang Q, et al. Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb Pathog. 2019;126:193-198.
Crossref - Sun WJ, Liu CF, Yu L, et al. A novel bacteriophage KSL-1 of 2-Keto-gluconic acid producer Pseudomonas fluorescens K1005: isolation, characterization and its remedial action. BMC Microbiol. 2012;12(1):1-8.
Crossref - Wong CL, Sieo CC, Tan WS, et al. Evaluation of a lytic bacteriophage, Φst1, for biocontrol of Salmonella enterica serovar Typhimurium in chickens. Int J Food Microbiol. 2014;172:92-101.
Crossref - Bao H, Shahin K, Zhang Q, et al. Morphologic and genomic characterization of a broad host range Salmonella enterica serovar Pullorum lytic phage vB_SPuM_SP116. Microb Pathogen. 2019;136:103659.
Crossref - Kim SH, Park JH, Lee BK, et al. Complete genome sequence of Salmonella bacteriophage SS3e. 2012.
Crossref - Sun Z, Mandlaa, Wen H, Ma L, Chen Z. Isolation, characterization, and application of bacteriophage PSDA-2 against Salmonella Typhimurium in chilled mutton. PLoS One. 2022;17(1):e0262946.
Crossref - Wang J, Hu BE, Xu M, et al. Therapeutic effectiveness of bacteriophages in the rescue of mice with extended spectrum b-lactamase-producing Escherichia coli bacteremia. Int J Mol Med. 2006;17(2):347-355.
Crossref - Wang J, Hu B, Xu M, et al. Use of bacteriophage in the treatment of experimental animal bacteremia from imipenem-resistant Pseudomonas aeruginosa. Int J Mol Med. 2006;17(2):309-317.
Crossref - Engeman E, Freyberger HR, Corey BW, et al. Synergistic killing and re-sensitization of Pseudomonas aeruginosa to antibiotics by phage-antibiotic combination treatment. Pharmaceuticals. 2021;14(3):184.
Crossref - Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010:8(5):317-327.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.