ISSN: 0973-7510
E-ISSN: 2581-690X
The “limón sutil” (Citrus aurantifolia) has been widely cultivated and well established for many years in Piura, northwestern Peru, because of its exceptional climate and soil conditions. However, decline and death of C. aurantifolia trees caused by different phytopathogens remain a common problem which has been observed in the last decades. It is known that the microbiota of soil plays an important role with their host and could be the starting point to understand the causes of citrus decline. In this study, we identified through culture-independent methods the bacterial and fungal microbiota associated to C. aurantifolia, C. jambhiri and C. volkameriana rhizospheres in the main areas of Piura. By using a 16S rRNA and ITS-metabarcoding analysis, we evaluated the taxonomic diversity between healthy trees and with decline symptoms and how this diversity could influence the health status of citrus trees. More than 600 and 200 bacterial and fungal ASVs were identified, respectively. Our metabarcoding analysis was able to identify Proteobacteria, Cyanobacteria, Firmicutes, Bacteroidota and Acidobacteriota prokaryotic phyla, while fungal phyla included Ascomycota, Basidiomycota and Glomeromycota. In addition, there were differences between microbial diversity indices in rhizospheres evaluated. Finally, bacterial and fungal genera were shared among the different citrus rhizospheres. These results have allowed us to obtain a preliminary identification of microbiota in the citrus rhizospheres of healthy trees and with decline symptoms.
Citrus aurantifolia, limón sutil, Rhizosphere, Decline, Microbiota, Bacterial and Fungal Diversity, Non-culturable, Metabarcoding, Next Generation Sequencing
Citrus aurantifolia (Christm.) Swingle, known in Peru as “limón sutil” (the Key lime is called “limón” in much of South America and “sutil” from phonetic derivative “ceuti” gentilic term from Ceuta),1 is an important citrus plant widely cultivated in coast and Amazonian regions of Peru. The “limón sutil” has been well established for many years in Piura, northwestern Peru, because of its exceptional agrometeorological and soil conditions, which allow for growing, flowering and fruiting throughout yearly. It is a highly valued plant for its culinary, industrial, nutritional, antioxidant and antimicrobial properties.2,3
In 2022, Piura was main producer region with 204,000 t, allowing to achieve US$ 33 million in exports.4
The “limón sutil” grafted onto “limón rugoso” rootstocks (C. jambhiri or C. volkameriana), thrives in Piura in full sun in a warm and humid climate with moderate annual rainfall. Although it is very well adapted to unfavorable conditions such as drought, it succumbs to adverse climatic events such as “El Niño” phenomenon.5 The diseases can be a limiting factor in these scenarios, mainly due to the proliferation of phytopathogens at the ground level and the opportunism of other microorganisms in the upper parts of the plant.5-8
This has led to identifying a typical problem of progressive decline and death of C. aurantifolia trees in Piura over the last two decades. It is believed to be caused by pathogens mainly from soil. Phytophthora spp., Fusarium spp., Rhizoctonia solani and Macrophomina phaseolina have been associated with root rot,6 and infestations of nematodes have also been described.9 On the other hand, Citrus Tristeza Virus has been associated with citrus decay, whilst Huanglongbing (HLB), no presence in Peru, have made it possible to reinforcement the national surveillance of phytoplasmas-transmitting vectors.10,11 Overall, different aspects have to be considered in the management and control of citrus decline.
It is known that certain soil microorganisms, interact with plants in an extraordinary manner generating physiological benefits for the host.12 However, it is still poorly understood how microorganisms couple to plant tissues.13 Microbial colonization of plant tissues generally begins with the establishment of microorganisms in the rhizosphere.14-16 Some studies have shown that the microorganisms in the rhizosphere can influence in citrus plant physiology and health through different mechanisms as nutrient supply, phytohormone production and antimicrobial compounds synthesis.14,16-18 All these features are mainly due to the formation of a complex community known as microbiome.
The microbiome of rhizosphere has a high taxonomic diversity of bacteria and fungi that inhabit it. Culture-independent approaches have shown that microbiota diversity of rhizosphere is highly underestimated. Next Generation Sequencing (NGS) have demonstrated that only a minority of bacteria (< 5%) are culturable and a significant proportion of bacterial taxa detected by these technologies are unidentified.19-22 A few studies have characterized the microbiome of citrus rhizosphere23 and how plant health can be impaired when certain pathogens indirectly alter the host-microbiome interaction at the rhizosphere level.24 In this study, by culture-independent methods, we characterized and compared the bacterial and fungal microbiota associated with rhizospheres of C. aurantifolia, C. jambhiri and C. volkameriana as a starting point to shed light on the possible causes about decline of citrus trees in Piura.
Sampling and plant material
The estates evaluated were located producing-areas of citrus in Chira valley, Sullana, Piura (Figure S1; Table S1). In this valley, the “limón sutil” grafted onto a “limón rugoso” rootstock (C. aurantifolia/C. jambhiri) is the predominant plantation. Two kinds of plant status were evaluated:i) healthy trees with foliage, trunk and production remarkable and ii) decline or dieback trees (Figure 1). Also, C. jambhiri and C. volkameriana healthy trees, were evaluated.
Figure 1. Agricultural property “Eloy”, Sullana, Piura (4.5 ha). A) The picture shows a plantation of C. aurantifolia/C. jambhiri healthy trees of nine years old. B) A C. aurantifolia/C. jambhiri tree with decline symptoms, notice the difference in the foliage compared to the healthy trees on its sides. The samples were uptake from roots
For each tree, four sub-samples were collected around the trunk at 25 cm depth using a shovel previously disinfected with 1.5% NaClO. The sub-samples were homogenized together until reaching one sample of 200 g (soil and roots) stored in zip lock bags. All samples were stored at 4°C and processed within 24 h.
Five samples for each citrus were collected: 1) C. aurantifolia/C. jambhiri healthy trees (code 01ML); 2) C. aurantifolia / C. jambhiri decline symptoms trees (code 05ML); 3) C. jambhiri healthy trees (code 02ML) and 4) C. volkameriana healthy trees (code 06ML). A total of twenty samples for four kinds of rhizospheres were analyzed in this study.
DNA extraction, metabarcoding and sequencing
The samples of roots were recovered and pooled (from the five samples of each citrus tree) and washed using 100 ml PBS (1%) buffer in 250 ml bottles with strong shaking. The soil adhered that was washed off from the roots (rhizosphere) was stored into 50 ml tubes. The metagenomics DNA was extracted from 250 mg rhizosphere using a DNeasy® PowerSoil Kit (QIAGEN, Germany) following the manufacturer’s guidelines. The DNA concentrations were quantified using a BioPhotometer UV/Vis Spectrophotometer (Eppendorf, Germany) at a wavelength of 260 nm and adjusted to 10 ng/µl for subsequent analyzes. Aliquots of 25 µl of DNA were used for PCR amplification of V4 and ITS1 regions belonging to the 16S rRNA and ITS genes using primers from Walters et al.25 The samples were purified using calibrated AMpure XP beads (Beckman Coulter, USA). Then, purified PCR products were used to prepare Illumina DNA library. The sequencing was performed at MR DNA (www.mrdnalab.com, USA) using Illumina MiSeq Systems, following the manufacturer’s guidelines.
Bioinformatics and statistical analysis
Raw reads generated using 2 × 250 Illumina Miseq, were evaluated with FastQC v. 0.11.2 and demultiplexed on the Galaxy web platform (http://usegalaxy.org).26 Removals of barcodes and primers were established according to the configuration of HEADCROP parameters (30 bp) of Trimmomatic v. 0.36.27 The quality filtration was processed using the DADA2 workflow for bacteria and or fungi,28 implemented in R v. 3.4.3 software. The filtered sequences were assigned to six taxonomy levels (from phylum to genus) through assignTaxonomy, using the SILVA database v. 138 for bacteria and UNITE ITS database v.8.2 for fungi. The final result was an amplicon sequence variant (ASV) table. The taxonomic analysis of the microbial diversity was developed through the Phyloseq v. 1.24.2 package.29 The statistical significance of Chao1, Shannon and Simpson alpha diversity indices were evaluated in R v. 3.4.3. The rarefaction curves were obtained by Vegan v. 2.5.3. Venn diagrams were elaborated by Mothur software.30
Bacterial diversity analysis
The diversity indices were used to calculate the alpha diversity of bacterial communities of each rhizosphere (Table 1). The 02ML sample showed the highest indices (Chao1 = 540.0, Shannon = 3.75 and Simpson = 0.97).
Table (1):
Alpha diversity indices of bacterial richness associated to citrus tree rhizospheres
Rhizospheres |
Status |
Code |
Chao1 |
Shannon |
Simpson |
|---|---|---|---|---|---|
C. aurantifolia/C. jambhiri |
healthy trees |
01ML |
340.0 |
3.23 |
0.95 |
C. aurantifolia/C. jambhiri |
decline trees |
05ML |
100.0 |
1.81 |
0.79 |
C. jambhiri |
healthy trees |
02ML |
540.0 |
3.75 |
0.97 |
C. volkameriana |
healthy trees |
06ML |
190.0 |
2.50 |
0.88 |
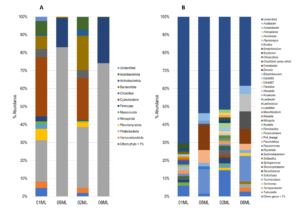
The bacterial communities in the four groups of rhizosphere were mainly represented by following phyla: Proteobacteria (01ML = 23.09%, 05ML = 83.07%, 02ML = 33.86% and 06ML = 74.07%), Bacteroidota (01ML = 7.33%, 05ML = 0.66%, 02ML = 19.10% and 06ML = 0.25%), Firmicutes (01ML = 2.70%, 05ML = 16.27%, 02ML = 0.46% and 06ML = 25.69%), Cyanobacteria (01ML = 33.02%, 05ML = 0.0%, 02ML = 23.34% and 06ML = 0.0%) and Acidobacteriota (01ML = 7.38%, 05ML = 0.0%, 02ML = 7.13% and 06ML = 0.0%). Other phyla detected were Actinobacteriota, Chloroflexi, Myxococcota, Nitrospirota, Planctomycetota and Verrucomicrobiota in lower abundances (Figure 2a).
At genera level, 01ML rhizosphere was mainly represented by Bradyrhizobium (2.50%), Nitrospira (2.40%), Pir4_lineage (2.28%), Bryobacter (2.07%), Sulfurifustis (1.91%), Pseudolabrys (1.76%), Steroidobacter (1.52%), Chryseolinea (1.50%) and Pseudomonas (1.40%). The 02ML rhizosphere had the highest bacterial diversity and was mainly represented by Steroidobacter (3.22%), Sphingomonas (2.71%), Edaphobaculum (2.50%), Ellin6055 (2.16%), Acidibacter (1.82%), Lacibacter (1.76%), Terrimicrobium (1.45%), Stenotrophobacter (1.43%) and Pseudolabrys (1.43%). In the 05ML rhizosphere, Cronobacter (13.78%) and Pseudomonas (13.58%) were the most representative genera, followed by Klebsiella (7.12%), Acinetobacter (4.07%), Kosakonia (1.94%), Bacillus (1.78%) and Lactococcus (1.63%) as markers genera. Finally, 06ML rhizosphere comprised Pseudomonas (14.24%), Clostridium sensu stricto (9.81%), Cronobacter (8.19%) as the most representative genera, while Aeromonas (9.73%), Terrisporobacter (2.15%), Trabulsiella (2.13%), Paenibacillus (1.69%), Paraclostridium (1.64%) and Solibacillus (1.29%) were markers genera. Others such as Acinetobacter (4.57%) and Klebsiella (3.45%) were also present (Figure 2b).
Figure 2. Relative abundance of bacterial microbiota. A) Barplots showing relative abundance of the bacterial phylum. B) Barplots showing relative abundance of the bacterial genera (metabarcoding analysis based 16S rRNA) present in the different kinds of citrus rhizospheres analyzed in this study
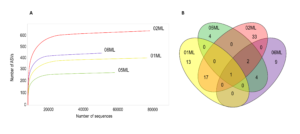
Over 600 bacterial ASVs were detected. The 02ML sample had the highest abundance (Figure 3a). The highest number of shared genera was observed between 01ML and 02ML, in addition, Pseudomonas was the only common genus the four rhizospheres evaluated in this study (Figure 3b).
Figure 3. Rarefaction curves and Venn diagram of shared bacterial genera of citrus rhizospheres. A) Shown the number of bacterial ASVs observed for different citrus rhizosphere. The rhizosphere of C. jambhiri trees (02ML) showed the highest number of ASVs. B) Pseudomonas was the only genus shared for all rhizospheres
Fungal diversity analysis
The diversity indices were used to calculate the alpha diversity of fungal communities of each rhizosphere (Table 2). The 02ML sample showed the highest indices (Chao1 = 181.9 and Shannon = 1.97), while the 05ML sample had the highest Simpson index (Simpson = 0.69).
Table (2):
Alpha diversity indices of fungal richness associated to citrus tree rhizospheres
Rhizospheres |
Status |
Code |
Chao1 |
Shannon |
Simpson |
|---|---|---|---|---|---|
C. aurantifolia/C. jambhiri |
healthy trees |
01ML |
41.0 |
1.40 |
0.58 |
C. aurantifolia/C. jambhiri |
decline trees |
05ML |
37.0 |
1.80 |
0.69 |
C. jambhiri |
healthy trees |
02ML |
181.9 |
1.97 |
0.60 |
C. volkameriana |
healthy trees |
06ML |
37.0 |
1.57 |
0.65 |
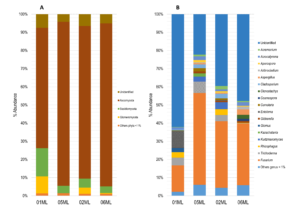
The fungal communities were mainly represented by following phyla: Ascomycota (01ML = 66.32%, 05ML = 90.32%, 02ML = 84.01% and 06ML = 89.76%), Basidiomycota (01ML = 15.64%, 05ML = 4.11%, 02ML = 5.0% and 06ML = 3.64%) and Glomeromycota (01ML = 9.29%, 05ML = 0.0%, 02ML = 3.33% and 06ML = 0.47%) (Figure 4a).
Regarding the genera identified, the 01ML rhizosphere was mainly represented by Fusarium (14.48%), Entoloma (9.22%), Trichoderma (4.32%), Rhizophagus (2.95%) and Glomus (2.29%). The 02ML rhizosphere was represented by Fusarium (36.69%), Kurtzmanomyces (3.90%), Trichoderma (3.39%), Rhizophagus (3.20%), Cladosporium (2.42%) and Acremonium (1.82%). The 05ML rhizosphere, the genus Fusarium was the most representative (50.60%), followed by Trichoderma (6.32%), Kurtzmanomyces (2.69%), Cladosporium (2.14%), Acrocalymma (1.97%) and Kazachstania (1.83%) as genus marker and Curvularia (1.81%). Finally, 06ML rhizosphere was represented by Fusarium (33.94%), Aspergillus (3.04%), Clonostachys (2.11%), Acrocalymma (2.08%) and Arthrocladium (1.50%). Figure 4b summarizes the abundance of the fungal genera identified.
Figure 4. Relative abundance of fungal microbiota. a) Barplots showing relative abundance of the fungal phylum and b) Barplots showing relative abundance of the fungal genera (metabarcoding analysis based ITS) present in the different kinds of citrus rhizospheres analyzed in this study
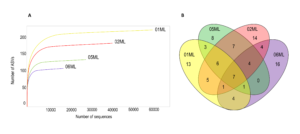
Over 200 fungal ASVs were detected. The 01ML sample had a greater richness (Figure 5a). Finally, Acremonium, Acrocalymma, Aspergillus, Coprinellus, Fusarium, Gibberella and Trichoderma were the seven common genera in the four kinds of rhizospheres (Figure 5b).
Figure 5. Rarefaction curves and Venn diagram of shared fungal genera of citrus rhizospheres. A) Rarefaction curve showing the number of fungal ASVs observed for different citrus rhizosphere. The rhizosphere C. aurantifolia/C. jambhiri healthy trees (01ML) showed the highest number of ASVs. B) Seven fungal genera were shared in the different kinds of citrus rhizospheres
Some microorganisms identified by our metabarcoding analysis were similar to those reported in different citrus species while others had not been described before. Such variability in the rhizospheric microbiota may be associated to geographic location and plant genotype.31,32 Furthermore, the microbial structure of the rhizosphere could be modulated due to the plant health status.24
Proteobacteria are the most abundant and important phylum within the microbial communities found in the citrus rhizosphere compared to those microbes reported in the bulk soil.33 Proteobacterial species have been described as dominant in the rhizosphere of citrus.23,24,34,35 and of other plant species of economic importance such as grapevine, cocoa, rice, etc. However, we could appreciate some differences in microbiological abundance according to plant health status. Proteobacteria phylum was decreased in the rhizosphere of C. aurantifolia/C. jambhiri healthy tree compared to those that showed decline symptoms. This relative difference could explain the modulation of the rhizospheric microbiota due to the presence of diseases in citrus.24 A high diversity of bacterial phyla in the rhizosphere of C. aurantifolia/C. jambhiri healthy tree could suggest that the microbiota may be regulated by the balance of interactions that occur at the root and upper part of plant, which reflects its healthy status (Figure 2a). Proteobacteria, Cyanobacteria, Bacteroidota, Actinobacteriota, Firmicutes, Planctomycetota, Verrucomicrobiota and other phyla were detected in healthy plants, and many species belonging to those taxa are known to be beneficial microorganisms. Otherwise, the abundances of the phyla for C. jambhiri and C. volkameriana showed differences. Although both species did not show decline symptoms, the low diversity of C. volkameriana may be associated with genotype or location. Furthermore, the cultivation conditions for this species (rootstocks) could influence its microbiological diversity, since the type of irrigation, fertilization and pest management differ from those of C. aurantifolia/C. jambhiri healthy trees.
The main rhizobacteria genera identified in C. aurantifolia/C. jambhiri, C. jambhiri and C. volkameriana healthy trees have been reported as endophytic microorganisms, growth promoters, inducers of systemic resistance bioremediators. Some endophytes from the genus Sphingomonas have been described in citrus plants in China.36 The genus Paenibacillus harbors some species described as growth suppressants of citrus pathogenic fungi.37
Interestingly, three genera of the Enterobacteriaceae family were prominent in the rhizosphere of C. aurantifolia/C. jambhiri (05ML) decline symptoms trees evaluated by metabarcoding analysis. Cronobacter, Klebsiella, and Kosakonia can behave as pathogens in humans and be food contaminants. However, some Cronobacter strains are capable of endophytically colonizing tomato and corn roots. In addition, this genus comprises species with phosphate solubilizing properties and producers of 3-IAA.38 While some Klebsiella strains have been isolated from the rhizospheres of different crops and show endophytic properties, tolerance to salt stress and promote growth in sugarcane and wheat.39,40 Kosakonia species have shown endophytic capacity in rice roots.41
Cronobacter and Klebsiella have been found in the rhizosphere of citrus,42,43 while Kosakonia has been isolated from citrus leaves and branches.44 There is no evidence that Cronobacter and Klebsiella are associated with the development of decline symptoms in citrus trees, however, these bacteria appears to have an environmental role and have been found in water, soil and plant material.45 Although some species of Kosakonia are benefical, K. cowanii has been described as pathogen in soybean leaves.46 Also, a K. radicincitans strain has been associated with bacterial wilt in banana plants,47 but no evidence showing the same action in citrus. Studies that include the isolation, characterization and analysis are required to evaluate the pathogenic effect that these Enterobacteriaceae species could develop when interacting with C. aurantifolia trees.
Pseudomonas, Bacillus, and Acinetobacter are cosmopolitan microbes acting as endophytes in roots, stems, leaves, flowers, and fruits. Furthermore, those microorganisms have been proven to be promoters of plant growth, phytohormones producers and inducers of systemic resistance.48,49 These genera were identified in healthy and sick rhizospheres in our study. Pseudomonas and Acinetobacter were predominant in the rhizosphere of C. volkameriana healthy trees, whereas a lower abundance was founded in trees with decline symptoms (Figure 2b). Moreover, the genus Bacillus was present in decline symptoms trees according to our study. However, Bacillus strains were isolated from roots of healthy trees (data not shown). In the other hand, different species of Pseudomonas have pathogenic effects on citrus leaves and fruits.50,51 Interestingly, we found that the relative abundance of the genus Pseudomonas was abundant in diseased plants (Figure 2b) and this could be a key factor associated with the decline symptoms.
Our metabarcoding analysis showed that Ascomycota was the predominant fungal phylum in citrus rhizospheres. The Rhizophagus and Glomus genera, known as arbuscular mycorrhizal fungi, were unique to healthy trees, and the genus Entoloma, a beneficial ectomycorrhizal basidiomycete, was also found in the rhizosphere of healthy trees (Figure 4b). Some species of Rhizophagus and Glomus would have beneficial effects on citrus. A study evaluated the effect R. intraradices co-inoculated with a rhizobacteria on seeds and seedlings of citrus Poncirus trifoliata, found that both microorganisms improved the performance in the uptake of phosphorus and nitrogen, in addition, the physiological characteristics were improved, compared to control.52 Watanarojanaporn et al.53 isolated thirteen mycorrhizae from the rhizosphere of citrus orchards in Thailand, seven of them belonged to the genus Glomus. G. etunicatum was found to have a better colonization capacity in citrus roots and antagonism to Phytophthora nicotianae. This shows that arbuscular mycorrhizal fungi detected in rhizospheres, could be associated to healthy physiological status.
Some Trichoderma strains have shown positive effects to control Penicilium digitatum, Alternaria alternata and Colletotrichum gloeosporioides by producing antifungal enzymes in sweet orange fruits.54,55 Clonostachys, Cladosporium, Aporospora, Acrocalymma, and Acremonium are fungal genera found in citrus plant with beneficial features. C. rosea is an entomopathogen of citrus-pests.56 Bioactive metabolites from C. cladosporoides, isolated from citrus orchards in Florida, inhibited the growth in vitro of Liberibacter crescens 44. Acrocalymma has been found to inhabit citrus tree roots with symptoms of HLB.57 Although there is no evidence that Aporospora and Acremonium inhabit citrus roots, these fungi behave as endophytes in other plants. Whereas, Kazachstania, found both in the rhizospheres of healthy and sick trees, comprises yeasts that control pathogens in lemon fruits.58
Certain species of the genus Aspergillus have been associated with postharvest diseases affecting Citrus limón and Citrus sinensis fruits.59,60 However, it is unknown whether microorganisms of this genera cause roots rot. A study found that Curvularia spicifera, causes fruit rot of Citrus reticulata61 and Gibberella species have not been reported to affect citrus, but they could be pathogens of some grasses.62-64
Many species of the genus Fusarium cause root rot, blight and dieback in citrus.7 F. solani causes dry rot in C. aurantifolia in India,65 while F. sarcochroum, F. oxysporum, F. citricola, F. salinense, F. ensiforme and F. siculi affect lemon trees in Europe.7 Fusarium is a relevant pathogen in top harmful fungi of citrus. Our results showed a high abundance of the genus Fusarium in healthy trees and those with decline symptoms. A high number of Fusarium (50.6%) in trees with decline symptoms (Figure 4b) is an interesting characteristic to consider when evaluating the role of those microbes in the disease. Javier-Alva5 carried out pathogenicity studies where Phytophthora parasitica was found to cause a primary infection triggering decline symptoms in C. aurantifolia seedlings. Javier-Alva5 suggests that Fusarium species and another fungi subsequently would promote the development of the decline. Besides, Handique et al.66 suggests that Phytophthora alters microbial community structure in rhizosphere of citrus. In our study, the genus Fusarium was present in different citrus species with and without decline symptoms. Furthermore, we isolated species of F. solani and P. parasitica in healthy trees (data not shown), which would indicate that the presence of both pathogens in the citrus production fields in Piura is still an agricultural challenge that needs attention.
This is the first study that describes the diversity of the microbiota associated with the rhizospheres of C. aurantifolia, C. jambhiri and C. volkameriana in Piura by using culture-independent technique. Over 600 ASVs bacterial and 200 ASVs fungal were identified by metabarcoding analysis. In addition, we were able to establish the differences between the microbiota associated with healthy trees and those with decline symptoms, a problem that has affected “limón sutil” crops in Piura for more twenty years. The diversity of bacterial and fungal identified allowed us to have insight into the structure and behavior of the microbiota that interact with citrus rhizospheres and how they differ according to the health status of host. The use of native bacterial and fungal strains isolated from healthy trees could be evaluated in future to find out their capabilities in improving the health status of the Citrus aurantifolia trees in crops of Piura.
Additional file: Additional Table S1 and Figure S1.
ACKNOWLEDGMENTS
The authors are thankful to the Innóvate Perú and Somatito EIRL for the financial support. The authors are also thankful to incabiotec sac Research Company for the development of the lab activities, and Engineer James Leigh, general manager of Somatito EIRL for providing the facilities and permits field sampling.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ENM and EM conceived and designed the experiments. ENM and MSB carried out the sampling. ENM and MSB carried out the lab experiments. CCM led the processing of sequencing data and bioinformatic analysis. MSB and ENM wrote the manuscript with contributions from CCM and EM. All authors read and approved the final manuscript for publication.
FUNDING
The research was co-supported by Innóvate Perú and the SOMATITO EIRL company with agreement N°010-FIDECOM-INNOVATEPERU-PIMEN-2017.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Arrizabalaga C. limón sutil. Revista de Historia de la Lengua Espanola. 2014;(9):151-157.
Crossref - Lim TK. Citrus x aurantiifolia. Edible Medicinal And Non-Medicinal Plants: Volume 2, Fruits. Springer Netherlands. 2012:742-754.
Crossref - Tavallali H, Bahmanzadegan A, Rowshan V, Tavallali V. Essential Oil Composition, Antioxidant Activity, Phenolic Compounds, Total Phenolic and Flavonoid Contents from Pomace of Citrus aurantifolia. J Med Plants byprod. 2021;10(spl):103-116.
Crossref - Ministerio de Comercio Exterior y Turismo (MINCETUR). Reporte de Comercio Regional Anual 2022. Piura. 2022. https://cdn.www.gob.pe/uploads/document/file/4334490/RCR%20Piura%20-%20Anual%202022.pdf?v=1680020694
- Javier-Alva J. Fungicidas y enmiendas orgánicas en el control de Phytophthora spp. en limonero Citrus aurantifolia (L.) Swingle injertado sobre Citru sjambhiri Lush, bajo riego tecnificado. Dissertation. Universidad Nacional Agraria La Molina;1998.
- Javier-Alva J, Mattos L. Nuevo metodo para aislar “Phytophthora parasitica” Dastur de raicillas de limonero patron Rugoso “Citrus jambhiri” Lush bajo riego por aspersion. Universalia. 2006;11(1):19-31.
- Sandoval-Denis M, Guarnaccia V, Polizzi G, Crous PW. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia. 2018;40:1-25.
Crossref - Verniere C, Cohen S, Raffanel B, Dubois A, Venard P, Panabieres F. Variability in Pathogenicity among Phytophthora spp. Isolated from Citrus in Corsica. J Phytopathol. 2004;152(8-9):476-483.
Crossref - Gómez J, Martín A. Problemas nematológicos en cultivos de costa, sierra y selva del país. Rev Peru Entomol. 1967;10(1):32-39. https://sisbib.unmsm.edu.pe/BVRevistas/entomologia/v10/pdf/a06v10.pdf
- Bederski K, Roistacher CN, Silvestre OP, Müller GW. Long-term Cross Protection of Severe Stem Pitting Citrus tristeza virus in Peru. Int Organ Citrus Virol Conf Proc (1957 2010). 2010;17(17).
Crossref - Mercado W, Guadalupe K, Vega Alegre K. Possible economic losses on the oranges production chain of Peru due to introduction of Huanglongbing (HLB): Simulation of prospective scenarios to 2045. Sci Agropecu. 2023;14(4):419-434.
Crossref - Passera A, Compant S, Casati P, et al. Not Just a Pathogen? Description of a Plant-Beneficial Pseudomonas syringae Strain. Front Microbiol. 2019;10:1409.
Crossref - Rosier A, Medeiros FHV, Bais HP. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil. 2018;428(1):35-55.
Crossref - Faddetta T, Abbate L, Alibrandi P, et al. The endophytic microbiota of Citrus limon is transmitted from seed to shoot highlighting differences of bacterial and fungal community structures. Sci Rep. 2021;11(1):7078.
Crossref - Zarraonaindia I, Owens SM, Weisenhorn P, et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio. 2015;6(2):e02527.
Crossref - Zhang Y, Trivedi P, Xu J, Roper MC, Wang N. The Citrus Microbiome: From Structure and Function to Microbiome Engineering and Beyond. Phytobiomes. 2021;5(3):249-262.
Crossref - Agrahari RK, Singh P, Koyama H, Panda SK. Plant-microbe Interactions for Sustainable Agriculture in the Postgenomic Era. Curr Genomics. 2020;21(3):168-178.
Crossref - Vives-Peris V, Gomez-Cadenas A, Perez-Clemente RM. Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingobium sp. Plant Cell Rep. 2018;37(11):1557-1569.
Crossref - Bhargava P, Khan M, Verma A, et al. Metagenomics as a Tool to Explore New Insights from Plant-Microbe Interface. In: Varma A, Tripathi S, Prasad R, eds. Plant Microbe Interface. 2019:271-289.
Crossref - Finkel OM, Castrillo G, Herrera Paredes S, Salas Gonzalez I, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155-163.
Crossref - Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37(5):634-663.
Crossref - Muller T, Ruppel S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol Ecol. 2014;87(1):2-17.
Crossref - Xu J, Zhang Y, Zhang P, et al. The structure and function of the global citrus rhizosphere microbiome. Nat Commun. 2018;9(1):4894.
Crossref - Zhang Y, Xu J, Riera N, Jin T, Li J, Wang N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome. 2017;5(1):97.
Crossref - Walters W, Hyde ER, Berg-Lyons D, et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2015;1(1):e00009-15.
Crossref - Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3-W10.
Crossref - Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-2120.
Crossref - Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13(7):581-583.
Crossref - McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. Plos ONE. 2013;8(4):e61217.
Crossref - Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75(23):7537-7541.
Crossref - Edwards J, Johnson C, Santos-Medellin C, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112(8):E911-920.
Crossref - Peiffer JA, Spor A, Koren O, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110(16):6548-6553.
Crossref - Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012;6(2):363-383.
Crossref - Cherni M, Ferjani R, Mapelli F, Boudabous A, Borin S, Ouzari HI. Soil parameters drive the diversity of Citrus sinensis rhizosphere microbiota which exhibits a potential in plant drought stress alleviation. Appl Soil Ecol. 2019;135:182-193.
Crossref - Wu Y, Qu M, Pu X, Lin J, Shu B. Distinct microbial communities among different tissues of citrus tree Citrus reticulata cv. Chachiensis. Sci Rep. 2020;10(1):6068.
Crossref - Munir S, Li Y, He P, et al. Core endophyte communities of different citrus varieties from citrus growing regions in China. Sci Rep. 2020;10(1):3648.
Crossref - Lai K, Chen S, Hu M, et al. Control of postharvest green mold of citrus fruit by application of endophytic Paenibacillus polymyxa strain SG-6. Postharvest Biol Technol. 2012;69:40-48.
Crossref - Schmid M, Iversen C, Gontia I, et al. Evidence for a plant-associated natural habitat for Cronobacter spp. Res Microbiol. 2009;160(8):608-614.
Crossref - Singh RP, Jha P, Jha PN. The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J Plant Physiol. 2015;184:57-67.
Crossref - Wei CY, Lin L, Luo LJ, et al. Endophytic nitrogen-fixing Klebsiella variicola strain DX120E promotes sugarcane growth. Biol Fertil Soils. 2014;50(4):657-666.
Crossref - Mosquito S, Bertani I, Licastro D, et al. In Planta Colonization and Role of T6SS in Two Rice Kosakonia Endophytes. Mol Plant Microbe Interact. 2019;33(2):349-363.
Crossref - Gholami D, Goodarzi T, Aminzadeh S, Alavi SM, Kazemipour N, Farrokhi N. Bacterial Secretome Analysis in Hunt for Novel Bacteriocins with Ability to Control Xanthomonas citri subsp. Citri. Iran J Biotechnol. 2015;13(3):10-19.
Crossref - Handique U. Characterization of the Structure and Function of the Microbiome in Citrus Rhizosphere. Thesis for Degree of Doctor of Philosophy. University of Florida; 2017.
- Blacutt A, Ginnan N, Dang T, et al. An In Vitro Pipeline for Screening and Selection of Citrus-Associated Microbiota with Potential Anti-”Candidatus Liberibacter asiaticus” Properties. Appl Environ Microbiol. 2020;86(8):e02883.
Crossref - Healy B, Cooney S, O’Brien S, et al. Cronobacter (Enterobacter sakazakii): An Opportunistic Foodborne Pathogen. Foodborne Pathog Dis. 2009;7(4):339-350.
Crossref - Krawczyk K, Borodynko-Filas N. Kosakoniacowanii as the New Bacterial Pathogen Affecting Soybean (Glycine max Willd.). Eur J Plant Pathol. 2020;157(3):173-183.
Crossref - Mohd Suhaimi NS, Yap KP, Ajam N, Thong KL. Genome sequence of Kosakonia radicincitans UMEnt01/12, a bacterium associated with bacterial wilt diseased banana plant. FEMS Microbiol Lett. 2014;358(1):11-13.
Crossref - Compant S, Clement C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42(5):669-678.
Crossref - Gaiero JR, McCall CA, Thompson KA, Day NJ, Best AS, Dunfield KE. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am J Bot. 2013;100(9):1738-1750.
Crossref - Beiki F, Busquets A, Gomila M, Rahimian H, Lalucat J, Garcia-Valdes E. New Pseudomonas spp. Are Pathogenic to Citrus. PLoS ONE. 2016;11(2):e0148796.
Crossref - Oueslati M, Mulet M, Gomila M, et al. New species of pathogenic Pseudomonas isolated from citrus in Tunisia: Proposal of Pseudomonas kairouanensis sp. nov. and Pseudomonas nabeulensis sp. nov. Syst Appl Microbiol. 2019;42(3):348-359.
Crossref - Wang P, Wu SH, Wen MX, Wang Y, Wu QS. Effects of combined inoculation with Rhizophagus intraradices and Paenibacillus mucilaginosus on plant growth, root morphology, and physiological status of trifoliate orange (Poncirus trifoliata L. Raf.) seedlings under different levels of phosphorus. Scientia Horticulturae. 2016;205:97-105.
Crossref - Watanarojanaporn N, Boonkerd N, Wongkaew S, Prommanop P, Teaumroong N. Selection of arbuscular mycorrhizal fungi for citrus growth promotion and Phytophthora suppression. Sci Hortic. 2011;128(4):423-433.
Crossref - Abo-Elnaga HIG. Control of green mould of citrus by using Trichoderma harzianum, humic acid or garlic. Arch Phytopathol Plant Protec. 2013;46(9):1118-1126.
Crossref - Ferreira FV, Herrmann Andrade AM, Calabrese CD, Bello F, Vazquez D, Musumeci MA. Effectiveness of Trichoderma strains isolated from the rhizosphere of citrus tree to control Alternaria alternata, Colletotrichum gloeosporioides and Penicillium digitatum A21 resistant to pyrimethanil in post-harvest oranges (Citrus sinensis L. (Osbeck)). J Appl Microbiol. 2020;129(3):712-727.
Crossref - Dao HT, Beattie GAC, Rossman AY, Burgess LW, Holford P. Four putative entomopathogenic fungi of armoured scale insects on Citrus in Australia. Mycol Progress. 2016;15(5):47.
Crossref - Ginnan NA, Dang T, Bodaghi S, et al. Bacterial and Fungal Next Generation Sequencing Datasets and Metadata from Citrus Infected with ‘Candidatus Liberibacter asiaticus’. Phytobiomes Journal. 2018;2(2):64-70.
Crossref - Perez MF, Contreras L, Garnica NM, et al. Native Killer Yeasts as Biocontrol Agents of Postharvest Fungal Diseases in Lemons. PLoS One. 2016;11(10):e0165590.
Crossref - Kotan R, Dikbas N, Bostan H. Biological control of post harvest disease caused by Aspergillus flavus on stored lemon fruits. Afr J Biotechnol. 2009;8(2):209-214.
- Scuderi G, Bonaccorsi A, Panebianco S, Vitale A, Polizzi G, Cirvilleri G. Some strains of Burkholderia gladioli are potential candidates for postharvest biocontrol of fungal rots in citrus and apple fruits. J Plant Pathol. 2009;91(1):207-213.
- Garganese F, Sanzani SM, Mincuzzi A, Ippolito A. First report of Curvularia spicifer a causing brown rot of citrus in Southern Italy. J Plant Pathol. 2015;97(3):541-551. https://pubag.nal.usda.gov/catalog/5381306. Accessed August 29, 2020.
- Hsuan HM, Salleh B, Zakaria L. Molecular Identification of Fusarium Species in Gibberella fujikuroi Species Complex from Rice, Sugarcane and Maize from Peninsular Malaysia. Int J Mol Sci. 2011;12(10):6722-6732.
Crossref - Schmale DG, Shah DA, Bergstrom GC. Spatial Patterns of Viable Spore Deposition of Gibberella zeae in Wheat Fields. Phytopathology®. 2005;95(5):472-479.
Crossref - Zhang D, Liu Y, Guo Y, et al. Fine-mapping of qRfg2, a QTL for resistance to Gibberella stalk rot in maize. Theor Appl Genet. 2012;124(3):585-596.
Crossref - Ravi Chandran M, Reddi Kumar M. Studies on cultural, morphological variability in isolates of Fusarium solani (Mart.) Sacc., incitant of dry root-rot of Citrus. Curr Biot. 2012;6(2):152-162. https://www.cabidigitallibrary.org/doi/pdf/10.5555/20123350108.
- Handique M, Bora P, Ziogas V, Srivastava AK, Jagannadham PTK, Das AK. Phytophthora Infection Reorients the Composition of Rhizospheric Microbial Assembly in Khasi Mandarin (Citrus reticulata Blanco). Agronomy. 2024;14(4):661.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.