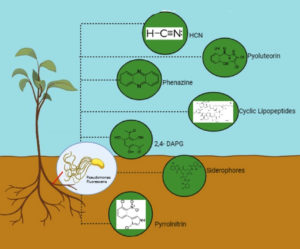
Concerning sustainable agriculture, plant growth promoting rhizobacteria (PGPR), which are a subgroup of “fluorescent pseudomonads,” are crucial. They are widely known for supporting plant health through a variety of methods. The use of fluorescent pseudomonads in agri-biotechnology has gained traction due to their potential for safeguarding plants from a variety of phytopathogens. Fluorescent pseudomonads being commercialized as bioinoculants for the treatment of various plant diseases is currently regarded as highly successful on a global scale. Fluorescent pseudomonads are being employed as efficient bio-control agents (BCAs) against an array of phytopathogens. Due to their capacity to generate a wide range of secondary metabolites, they offer enormous promise as BCA. Thus, this review’s goal is to outline and evaluate the functions of fluorescent pseudomonads’ secondary metabolites in reducing phytopathogens and improving plant health. Prominent secondary metabolites linked to biocontrol through fluorescent pseudomonads include phenazines (PHZ), 2, 4-diacetylphloroglucinol (DAPG), pyoluteorin (PLT), pyrrolnitrin (PRN), cyclic lipopeptides (CLPs), and volatile organic compounds (VOCs), including hydrogen cyanide (HCN). The antifungal, antibacterial, antiviral, antitumor, and antinematicidal effects of these metabolites are well-established.
Bio-control Agent (BCA), Eco-friendly Management, Fluorescent Pseudomonads, Phytopathogens, Secondary Metabolites
Phytopathogens have taken advantage of crop plants at different phases of their development ever since agriculture began. There are many different agrochemicals on the market for controlling diseases and pests, and some of them are harmful for the environment. Demands for safer-to-consume organic products have increased recently on the global market.1 A closer examination of plant diseases reveals a long history of agricultural losses brought on by phytopathogens. Phytopathogens are the primary cause of a number of serious epidemics that have resulted in a significant loss of human life. For instance, Phytophthora infestans caused potato blight epidemics known as the “Great Irish Famines” (1739 and later 1845-1849) claimed 750,000 lives and drove two million others to immigrate to United States.2
An estimated 10-20% of the world’s agricultural output is impacted by phytopathogens, depriving 800 million people of a sufficient diet.3 Diseases brought on by bacteria, nematodes that parasitize plants, fungus, and viruses inflict billions of dollars’ worth of losses in terms of economic damage annually. Pests and diseases cause more than 25% of crop loss globally, according to estimates from the Food and Agriculture Organization (FAO) more recently.4 Due to agriculture’s primary role in these countries’ economic growth, A decrease in production is more likely to occur in developing nations.5 With differing degrees of effectiveness, many strategies for managing phytopathogens have been developed for more than a century. Nonetheless, the cornerstone of crop protection among them has remained chemical control based on synthetic agents. Synthetic insecticides gained traction as efficient ways to manage a wide range of pests due to their rapid efficacy and simplicity of usage. But its unrestrained usage also caused harm to human health and ecological collapse.6 Nevertheless, the impact of bacteria on plant phenotype—which is linked to modifications in the secondary metabolite profiles of plants—has not been well examined in many theoretical or experimental research.7 Therefore, in modern agriculture, protecting crops from harmful phytopathogens and eliminating them using more environmentally friendly methods is of utmost importance. Numerous different types of microbes live in and communicate with one another in the soil, which is a living system. The “rhizosphere,” which is the region immediately surrounding a plant’s roots, is thought to have the greatest influence on microbial interactions.8 Numerous PGPRs, or beneficial rhizobacteria, have their home in the rhizosphere.9 These bacteria actively colonize plant roots and play a major part in plant growth promotion (PGP).10 Many genera of bacteria, such as Serratia, Arthrobacter, Acinetobacter, Burkholderia, Enterobacteria, Pseudomonas, Rhizobium, and Azospirillum11,12 as well as representatives from a wide range of bacterial taxa are included in PGPR.13 For the past few decades, Many of these PGPR have been used as biocontrol agents (BCA), particularly in formulations that are cell-based. Eco-friendly methods of controlling phytopathogens to preserve the sustainability of agro-ecosystems are largely supported. Poor shelf life and inconsistent performance are the main causes. There is a dearth of information regarding the functioning of the real biocontrol factors, such as the metabolites generated by PGPR in field conditions. The capacity of “Fluorescent pseudomonas” to suppress a broad range of phytopathogens sets them apart from other members of the PGPR group.14 Due to their advanced biocontrol capabilities, these rhizosphere bacteria are utilized to create bioinoculants that protect crops against various phytopathogens.15 Signals from the environment and the plant host help microorganisms and plants to create complex connections. Both harmful and helpful bacteria can have an indirect or direct impact on these interactions, and the intricate chemical signaling also has an impact on the growth and development of plants.16
Because they have evolved the ability to biosynthesize a variety of secondary chemicals, they have an advantage over other rhizospheric bacteria in the selection process. The fluorescent pseudomonas must be taken advantage of in order to manage the phytopathogens with their variety of secondary metabolites. Understanding the mechanisms of action of these metabolites while accounting for environmental, ecological, and gene regulatory factors is crucial for this Thus, the focus of this work is on fluorescent pseudomonas’ secondary metabolites, investigating their production, structures, and physiological roles in connection to the biological control of phytopathogens.
About the Fluorescent pseudomonas
Pseudomonas is a member of the gammaproteobacteria class and the Pseudomonads family (Order: Pseudomonadales), which includes 236 species that have been scientifically described. Plants that are resistant to disease react to biotic stress caused by microorganisms by boosting the activity of antioxidant enzymes like peroxidase (POD) and superoxide dismutase (SOD) to lessen oxidative stress linked to disease. Under a range of stress situations, malondialdehyde (MDA) is a potent indication of cell damage resulting from membrane lipid peroxidation, and its concentration might indirectly represent the extent of peroxide damage.17 The name’s origin was first recorded as Greek pseudo meaning “false” and monas meaning “a single unit” in the 7th edition of Bergey’s Manual. Consequently, single-celled organisms were referred to as “monas” in the early history of microbiology.18 Nevertheless,19 named the typed species “Pseudomonas aeruginosa” after seeing that this strain may be identified by its ability to manufacture colors (aerugo is the Latin word for verdigris, the blue-greenish copper rust). Therefore, it is thought that one distinguishing characteristic shared by all fluorescent Pseudomonas is the production of luminous pigment. Typically found in agricultural soils, saprophytic fluorescent pseudomonas interacts with plants in a variety of ways.9 Medium chain length polyhydroxy alkenoates (mcl-PHA) are accumulated by numerous species in the genus as a carbon store material. Fluorescent pseudomonas’ antagonistic action is mostly associated with the synthesis of lipopeptides, lytic enzymes, antibiotic chemicals, and siderophores. Fluorescent pseudomonas also synthesizes a variety of volatile organic chemicals, including several types of molecules involved in antagonistic interactions with other species and in inducing systemic reactions in plants.20 The genus includes rod-shaped, Gram-negative, nonspore-forming, catalase-positive organisms that can breathe aerobically (some strains can also breathe anaerobically using nitrate as the terminal electron acceptor and/or fermenting arginine)21; They also have a high genomic G+C content (59-68%) and metabolic flexibility. Pseudomonas aeruginosa, P. aureofaciens (now P. chlororaphis), P. cichorii, P. fluorescens, P. putida, P. syringae, and P. viridiflava were among the bacteria listed in Bergey’s Manual of Systematic Bacteriology.22 Among them are P. aeruginosa, an opportunistic human pathogen, and P. syringaea, a phytopathogen that has been detected in a variety of crop plant species.23,24 Study by Palleroni et al.25 verified that Pseudomonas is multigeneric. They separated the genus into five distantly related groupings known as rRNA groups (rRNA groups I–V) by evaluating rRNA:DNA hybridization. However, the phylogenetic distribution of the Pseudomonads was previously ascertained using 16S rRNA sequence analysis, rRNA-DNA hybridization, and polyphasic taxonomic research (including DNA: DNA hybridization) data. At the moment, multilocus sequence analysis (MLSA), which is based on the sequence analysis of the four housekeeping genes (16S rRNA, gyrB, rpoB, and rpoD), is the most trustworthy technique for identifying and categorizing pseudomonas strains.26 Currently limited to the rRNA group I, the genus pseudomonas has 57 true Pseudomonas species that resemble the type species P. aeruginosa in both genomic and morphological traits.27 Most other species have been reclassified as belonging to the family Comamonodaceae, which include the genera Burkholderia, Ralstonia, Brevundimonas, Sphingomonas, Xanthomonas, and Stenotrophomonas, as well as the genera Acidovorax, Comamonas, and Hydrophagaga.28 See the reviews by Gomila et al 29 and Garrido-Sanz 30 for further information on the phylogenomics and systematics of pseudomonas.
Role of secondary metabolites in biocontrol produced by fluorescent pseudomonas
Effective BCA need to possess the capacity to generate secondary metabolites possessing antimicrobial properties against an array of phytopathogens. This is one of their desired characteristics. It has been demonstrated that this bacterium has antibiotic activity against oomycetes, fungi, and pathogenic bacteria.31 According to Budziliewicz,32 secondary metabolites are naturally occurring chemicals that are created as byproducts of primary metabolism and are not useful as reserves or sources of energy. Despite having a less evident function in the organism’s internal economy, their importance for survival functions cannot be overstated.33 The prolonged effectiveness of micro-volatile compounds is significantly impacted by their high volatility and poor water solubility. High concentrations of particular volatiles can be synthesized by microbial strains through the use of engineered strategies.34 The identification of particular micro-volatile compounds may offer a novel diagnostic tool to detect the presence of a given species in infected wounds, as evidenced by the identification of species-specific molecules from both mixed and pure cultures of single species.35 Numerous volatile chemicals that are released by bacteria contribute to interactions across different kingdoms of fungus, plants, and animals. Secondary metabolites are the principal method of antagonism that fluorescent pseudomonas uses against plant diseases, same like other powerful biocontrol PGPR (Table 1 and Figure 1). Phloroglucinol, HCN, PHZ, PLT, PRN, and CLPs are examples of secondary metabolites that drive the antagonistic relationship between fluorescent pseudomonas and phytopathogens. Although the impact of various bacterial volatile chemicals on population dynamics in polymicrobial communities is poorly understood, they do change the physiology and stress responses of bacteria. It has found that volatile hydrogen cyanide (HCN), which inhibits the growth of a wide range of pathogens under in vitro.36
Table (1):
Fluorescent pseudomonads released secondary metabolites against phytopathogens
| S. No. | Secondary metabolites | Effective in | Fluorescent pseudomonads | Plant pathogens | Ref. |
|---|---|---|---|---|---|
| 1 | Hydrogen Cyanamide (HCN) | Many cultivated crops | Pseudomonas sp. P76, P124 | Sclerotium rolfsii | 37 |
| Under laboratory condition only | P. mediterranea and P. corrugata | Botrytis cinerea | 38 | ||
| Under laboratory condition only | Pseudomonas CF1 and CF5 | Macrophomina phaseolina | 39 | ||
| Tomato | Pseudomonas sp. | Clavibacter michiganensis | 40 | ||
| 2 | Hydrogen Cyanamide and volatile compounds | Under laboratory condition only | Pseudomonas donguensis | Rhizoctonia solani and Pythium ultimum | 41 |
| 3 | Pyrrolnitrin (PRN) | Tomato | Pseudomonas chloraphis | Rhizoctonia solani | 42 |
| Soyabean | Pseudomonas fluorescens | Pythium ultimum | 43 | ||
| Under laboratory condition | Pseudomonas cepacia | Colletotrichum truncatum | 44 | ||
| Sugarbeet | Pseudomonas cepacia | Aphanomyces | 45 | ||
| 4 | Phenazine-1-carboxylic acid (PCA) | Under laboratory condition | Pseudomonas sp. | Fusarium oxysporum | 46 |
| Wheat | Pseudomonas sp. | Rhizoctonia solani | 47 | ||
| Pigeon pea and chickpea | Pseudomonas aeruginosa | Fusarium udum, F. ciceri | 48 | ||
| Wheat | Pseudomonas fluorescens | Gaeumannomyces graminis var. tritici | 49 | ||
| 5 | 2,4-Diacetylphloroglucinol | Groundnut | Pseudomonas fluorescens | Aspergillus niger | 50,51 |
| Wheat | Pseudomonas fluorescens VUPf5 | G. graminis var. tritici | 52 | ||
| Tomato | Pseudomonas sp. LBUM300 | C. michiganensis subsp. michiganensis | 40 | ||
| Under laboratory condition only | P. brassicacearum J12 | Ralstonia solanacearum | 53 | ||
| Banana | P. aeruginosa | Fusarium cubens | 54 | ||
| Rice | Pseudomonas sp. | Xanthomonas oryzae pv. oryze | 55 | ||
| Wheat | P. aurantiaca | F. oxysporum | 56 | ||
| 6 | Cyclic lipopeptides (CLPs) | Groundnut | Pseudomonas SH-C52 | Sclerotium rolfsii | 57 |
| Tomato | Pseudomonas fluorescens | Phytophthora infestans | 58 | ||
| Sugarbeet | Pseudomonas fluorescens | R. solani and P. ultimum | 59-60 |
A diverse range of plant-beneficial bacteria may colonize the rhizosphere of various plant species and create biofilms, thanks to the fluorescent Pseudomonas group.61
The rhizosphere and soil microbiomes play a major role in suppressing plant disease by synthesising antagonistic secondary compounds. However, the processes governing the degree of pathogen control remain poorly understood. Numerous Pseudomonas species are linked to the microbiomes of soil and rhizosphere, and there is ample evidence of their capacity to inhibit pathogens.62
HCN (Hydrogen Cyanamide)
Among the inorganic volatile compounds that inhibit the growth of insects, nematodes, and plant pathogens is HCN. In many Pseudomonas species, HCN synthase converts glycine into it.63 Most fluorescent pseudomonads, certain fluorescent pseudomonads, and a small number of species of the genus Chromobacterium, Burkholderia, certain Rhizobia, and Cyanobacteria all report that cyanide synthesis in bacteria occurs frequently.64 Fluorescent pseudomonas produce varying amounts of hydrogen cyanide in the rhizosphere, depending on environmental variables.65 According to Nandi et al,66 glycine is the direct metabolic precursor of cyanide and is decarboxylated into HCN and CO2 when HCN synthase is present.67 HCN synthase as an oxygen-sensitive, membrane-associated enzyme involved in cainogenesis that is the result of the HCN ABC synthase gene cluster. Because it inhibits cytochrome c oxidase, HCN generated by fluorescent pseudomonads has demonstrated toxicity against phytopathogens.68 However, RhdA, a thiosulfate known as cyanide sulfur transferase (rhodanese), which changes cyanide into a less lethal thiocyanate, makes fluorescent pseudomonads immune to cyanide. According to estimates, several Pseudomonas species may produce up to 300 µM cyanide through the oxidative decarboxylation of glycine. According to reports, in microaerophilic settings, cyanide production peaks between 34 and 37°C.69 While the majority of previous research has indicated a clear function for HCN in phytopathogen biocontrol, more recent investigations have cast doubt on this idea. According to Rijavec and Lapanje,70 HCN has less to do with the biological control of phytopathogens and more to do with the chelation of metals, which makes more phosphate available to the plant.
PHZ (Phenazines)
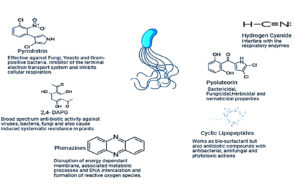
It is a broad class of heterocyclic rings consisting nitrogen that have been found in the archaeal phylum Euryarcheotic, the bacterial phyla Actinobacteria, and Proteobacteria.71 More than 6000 compounds with phenazine as a key component have been synthesized, and more than 100 unique natural PHZ structural variations have been identified for their antibacterial properties.72 The role of secondary metabolites in management of plant pathogen showed in Figure 2.
According to Guttenberger et al,73 there are actually more than 180 PHZ-based compounds that have anti-tumor, anti-fungal, insecticidal and other anti-pathogenic properties. According to Briard et al,74 Phenazine-1-carboxylic acid (PCA), phenazine-1-carboxamide (PCN), and pyocyanin (PYO) all have blue coloration in PHZ isolated from Pseudomonas species, while 1-hydroxyphenazine (1-HP) has an orange coloration. Substitutes at different locations also affect a substance’s redox potential, solubility, and biological activity.75 It is occasionally stated that a single species has many derivatives.76 All other PHZ begin with phenazine-1-carboxylic acid (PCA), which is derived from chorismic acid.77 The seven-gene operon phzABCDEFGen encodes the structural proteins needed for fluorescent pseudomonas to convert chorismic acid into PCA.78 Two different enzymes (PhzM and PhzS) that alter PCA produce PYO, while PhzH converts PCA into PCN. According to the majority of research, reactive oxygen species (ROS) produced by PHZ are mostly responsible for their antibacterial properties.79,80 Pseudomonas strain PCL1391 produces PCA and PCN, which are known to stimulate several ABC transporters in Botrytis cinerea.81 It has been discovered that PCA and PYO phenazines work incredibly well to treat fungal infections. While most PHZ compounds are efficient in biocontrol of many bacterial and fungal infections, they frequently failed to stop the growth of co-occurring immediate competitor microorganisms.82,83 Additionally, PHZ are crucial for the uptake of iron, cell communication, gene expression modulation, biofilm formation, and bacterial survival. Enzymes participating in major metabolic pathways may change PHZ as a result of its reaction with primary metabolites. These characteristics imply a potential involvement for them in primary cell metabolism, namely in the cell’s ability to survive under stressful situations. PHZ function as an electron shuttle and are involved in energy metabolism and transmission.84 There have been recent reports of new developments on the manufacturing of substituted PHZ.85 It is unknown if and how the secondary metabolites of the host plant affect the relationship between the bacteria and their host plant. Numerous investigations into how various biotic variables alter the phenotypic of plants have been documented.86,87 Secondary metabolites (SMs) produced by bacteria are essential to microbial interactions. Even while many natural products’ structures, biosynthesis, and roles have been clarified with great progress, little is still known about what happens to SMs when they are released into a particular niche.88
Phloroglucinol
According to Loper et al,87 Phenologlucinol (1,3,5-benzenetriol or 1,3,5-trihydroxybenzene) and its derivatives are phenolic compounds with a wide spectrum of antiviral, antibacterial, antifungal, antihelminthic, and phytotoxic properties. More than 700 naturally occurring phloroglucinol compounds have been identified from a range of natural sources, including microorganisms, plants, and marine life.88 Nonetheless, fluorescent pseudomonas is a highly conserved microorganism with the capacity to make phloroglucinol and its derivatives.14 According to Troppens et al.,89 DAPG is a well-known kind of phloroglucinol derivative that fluorescent pseudomonads manufacture. Numerous experimental investigations have confirmed that DAPG is a significant antibacterial metabolite that suppress the growth of plant pathogens.90 Studies show that the antibiotic DAPG is only produced by pseudomonas found worldwide. According to reports, pseudomonas isolated from a variety of geographic regions share the same DAPG biosynthesis locus.91 Polyketide synthases (PKSs) catalyse the decarboxylative condensation of monomers such as acetyl coenzyme A (acetyl-CoA), propionyl-CoA, malonyl-CoA, and methylmalonyl-CoA to produce phenolglucinol and its derivatives, which are categorised as secondary metabolites in the polyketides class.92 There are three different types of PKSs that have been approved.93 Three regulatory genes (phlF, phlG, and phlH) and six structural genes (phlA, phlC, phlB, phlD, phlE, and phlI) regulate the synthesis of DAPG.94
While the majority of fluorescent pseudomonas retained the DAPG biosynthesis gene cluster, evolution has caused many strains to lose this ability.95,96 It was demonstrated by Landa et al.96 that genotypes that produce DAPG may be crop-specific. According to Picard and Bosco,97 the biosynthetic gene phlD is essential for the synthesis of the precursor of DAPG and a trustworthy marker for DAPG producers. The selection of a growth medium, stressors brought on by a high concentration of salt, or heat shock can all increase the production of DAPG.98 It has been observed that employing metabolically modified Escherichia coli improves the production of phloroglucinol. Recently, attempts have also been made to express the bacterial phlD gene in plants in an attempt to boost the commercial production of phloroglucinol and its specific derivatives, such as DAPG.99
Pyoluteorin (PLT)
PLT is a phenolic polyketide that was initially identified in P. aeruginosa and subsequently in other fluorescent pseudomonads. It is made up of a substance connected to bichlorinated pyrrole.100
PLT has antifungal, antibacterial, and herbicidal properties.101 PLT biosynthesis requires the pltLABCDEFG gene cluster.102 Reportedly, PLT functions as an auto-inducer and an intercellular signalling mechanism among multiple co-occurring bacterial cell types in the rhizosphere.103 According to a recent study, phloroglucinol affects PLT production in P. protegens and the expression of the genes that make PLT in a concentration-dependent manner.104 Phloroglucinol has been shown to induce two unique metabolites (DAPG and PLT) with separate processes and targets that are phytopathogens at various concentrations.
Pyrrolnitrin (PRN)
Burkholderia pyrrocinia is the source of PRN, which is halogenated aryl pyrrole.105 A small variety of Gram-negative bacteria, including pseudomonas species, are known to make PRN.106-108 According to Jani et al.,109 PRN generated by fluorescent pseudomonas exhibits antagonistic activity against yeast, fungus, and Gram-positive bacteria. Inhibition of glycerol kinase, which results in glycerol buildup and leakage in the cell membrane, is the mechanism of PRN activity.110 The oxidation of amino-pyrrolnitrin to PRN is a crucial step that is catalyzed by the prnD gene.111 According to Steinberg et al.,112 there have been numerous reports of fluorescent pseudomonad-containing genes from suppressive soils. These genes are specifically recognized for their ability to inhibit Rhizoctonia solani. Wide spread distribution of PRN-producing, highly genotypically linked Pseudomonas from European soils with widespread anti-fungal action was described by Costa et al.113 Phenylpyrroles, chemical derivatives of PRN, are successfully employed as seed and foliar treatments to guard against fungal phytopathogens and are produced on a commercial scale. An efficient phenylpyrrole analog, fludioxonil, is employed against a variety of fungal phytopathogens. Since fludioxonil has been on the market for more than 25 years, there hasn’t been any resistance noted. There hasn’t been a developed analog of PRN other than these two derivatives, which are both successful in the field and on the commercial front.114
Cyclic lipopeptides (CLPs)
According to Raaijmakers et al,115 the short oligopeptides known as CLPs are produced by a range of bacteria and fungi and have a linked fatty acid tail. A brief oligopeptide is cycled by the formation of a lactone ring between two amino acids. However, variability in CLPs is caused by differences in the amount of fatty acids, the manner in which amino acids are changed, and the arrangement of the lactone ring.115,116 Mycoplasmas, enveloped viruses, and Gram-positive bacteria are only a few of the pathogenic microorganisms against which CLPs have demonstrated antibacterial activity in the last few years since their recognition as biosurfactants.115,117,118 Records state that CLPs from fluorescent pseudomonads actively participate in seed and root colonisation, swarming motility, biofilm formation, pathogenicity, and Many CLPs are produced by fluorescent Pseudomonads, some of which require thorough characterization.119 The viscosin, amphisin, tolaasin, and syringomycin families contain the most investigated CLPs.120 Non-ribosomal peptide synthetases (NRPSs) are responsible for the biosynthesis of CLPs. NRPSs are large multienzymes that produce linear or cyclic peptides by sequentially coupling amino acids in an assembly line fashion.121 According to Olorunleke et al.,122 CLPs derived from pseudomonas are presently classified into eight distinct structural groupings based on the length and makeup of the oligopeptide and fatty acid tail. The capacity of CLPs to disrupt cellular membranes is thought to be the mechanism behind their antibacterial activity. The tolaasin and syringomycin groups of CLPs function as cellular poisons by forming pores or tunnels. It was recently found that the supposedly generated CLP orfamide by P. protegens has insecticidal effects.123 In fluorescent pseudomonas, the two-component GacS/GacA regulatory mechanism is crucial for controlling the production of CLP. The possible roles of new regulatory genes required for the synthesis of CLPs in a number of fluorescent pseudomonas species and strains were emphasised.124 The generation and regulation of CLPs are directly influenced by the quorum sensing (QS) system; further research is necessary in this area to fully use the biocontrol potential of fluorescent pseudomonads that produce CLPs. It has been noted that the QS system mediates the synthesis of CLPs such as putisolvin, viscosin, cormycin, and corpeptins. Cell density is required for P. corrugata and P. mediterranea to make cormycin and corpeptin CLPs, and it is controlled by the QS regulatory system.125-127 The QS mechanism plays a crucial role in P. fluorescence producing the biosurfactant viscosin only when the cell density is high enough to overwhelm the host.128 provided evidence of the connection between QS and P. putida putisolvin synthesis. Thus, in coordinating the expression of CLPs throughout the fluorescent Pseudomonads population, the QS system is extremely important. This can also be used to allow CLPs to produce fluorescent pseudomonas and colonize plant roots.
Effective use of secondary metabolites in agriculture
It is commonly recognized that certain soils have antimicrobial properties that prevent phytopathogen growth even in the presence of host plants. Numerous studies have demonstrated the important impact fluorescent pseudomonads play in suppressing illness in these types of soils.129,130 Such suppressive soil mixtures were used in fields with high disease incidence, especially those caused by fungal phytopathogens. Given that it helped spread biocontrol agents to sensitive soil and resulted in phytopathogen control, this could be considered one of the earliest uses of bioformulations for phytopathogen suppression. Over the past few decades, research has demonstrated the critical function that fluorescent pseudomonad secondary metabolites play in suppressing phytopathogens in soils that are susceptible to them (Table 1). Fluorescent pseudomonads, possessing a plethora of secondary metabolites, are currently frequently utilized as biopesticides.131 They are marketed as bioinoculants over the world to treat various plant diseases, and they are thought to be quite successful.132,133 The commercial usage of fluorescent pseudomonad bioformulations is relatively young, having begun in the 1970s, when compared to the use of biopesticides produced from Bacillus thuringiensis or biofertilizers made of other microbes like cyanobacteria and rhizobia.134 Previous studies135 have confirmed that bioformulations containing antagonistic fluorescent pseudomonas were effective in suppressing take-all disease in wheat and barley. Packing houses are also using fluorescent pseudomonas-based bioformulations, Bio-Save®10LP and Bio-Save®11LP, registered with the U.S. Environmental Protection Agency and containing P. syringae ESC-10 and ESC-11 strains, to prevent postharvest fungal diseases during the storage of citrus, pome, and stone fruits.136 P. fluorescens A506 strain is present in a separate formulation called Blight Ban A506, which is used to control fire blight caused by Erwinia amylovora in pear and apple.137 In addition to Cedemon, other biocontrol agents for other phytopathogens include FROSTBAN and AtEze, primarily utilized in the United States. In a similar way, fluorescent pseudomonads have been used in the development of other formulations that are currently in commercial use worldwide. Shenqinmycin, a biopesticide with PCA as its primary ingredient that is generated from P. aeruginosa PA1201, has been registered in China to suppress Xanthomonas oryzae pv. oryzae and R. solani K hn, the agents responsible for bacterial blight and rice sheath blight, respectively. Fluorescent pseudomonads that produce numerous metabolites and have broad activity against phytopathogens are generally preferred in agriculture. Recent research has shown that the treatment of wheat plants with phenazine-1-carboxylic acid (PCA), cyclic lipopeptide (CLP), and lahorenoic acid A, which generates in variable amount from the strains of P. chlororaphis and P. aurantia, dramatically enhanced the growth and development.138 According to a study by Sharifazizi et al,139 fluorescent pseudomonad strain Ps170 has the ability to regulate E. amylovora, which causes fire blight in pears, and can also produce PCA, DAPG, PRN, and PLT. The application of fluorescent pseudomonas metabolites is currently expanding its scope to inhibit protozoa and diseases.140-142 Additionally, recent studies have suggested the use of formulations based on metabolite(s) from fluorescent pseudomonad, either in conjunction with or independently of cells, for biocontrol and a number of other purposes, such as cell signaling, cell protection, gene regulation. In this regard, field testing has demonstrated the extraordinary success of bioformulations based on rhamnolipid and exopolysaccharides (from fluorescent pseudomonas) (Table 2).142,143 According to Yan et al,144 phloroglucinol at nanomolar concentrations is sufficient to initiate PLT synthesis by P. protegens, which effectively inhibits E. amylovora. While the use of luminous pseudomonad metabolites in bioformulations rather than synthetic compounds is an emerging trend that is showing great promise, more extensive field trials are needed to ensure the wider applicability of these techniques.
Table (2):
Effects of secondary metabolites released by Fluorescent pseudomonas in crop plants
| No. | Metabolites | Observed in | |||
|---|---|---|---|---|---|
| Mechanism | References | Other | Ref. | ||
| 1 | VOC (Volatile compounds) | Having antibacterial and antifungal property | 145,146 | Helps in plant growth promotion | 147,148 |
| 2 | Phenazines | Inhibit conidial germination and mycelial suppression | 74,149 | Signaling and promote plant growth | 150-156 |
| 3 | CLPs | Disruption of cell membrane and cell wall | 157,158 | Having biosurfactant property | 159,160 |
| 4 | 2,4-DAPG | Suppress disease symptoms and disruption of cell membrane | 161,162 | Signaling and co-regulation | 144,163,164 |
| 5 | HCN | Inhibits cytochrome c oxidase | 68,165 | Signaling and chelation of metals | 166,167 |
| 6 | PLT | Disruption of cell membrane | 110,168 | Auto-induction and as signal molecule | 140,169 |
| 7 | PRN | Interfere to electron transport system | 170 | Interferes with osmotic Signal transduction | 171 |
Future scope
Despite the fact that bioformulations and biopesticides have numerous benefits, especially for environmentally friendly agriculture, they still fall well short of chemical pesticides in terms of quality, shelf life, and other concerns as well as uneven performance and narrow spectrum. Future success will hinge on resolving these problems and understanding how biocontrol microorganisms, like fluorescent pseudomonads, function and how their metabolites are regulated in soil environments. Molecular screening techniques can significantly speed up the basic steps involved in choosing strong antagonistic fluorescent pseudomonads. Additionally, it is critical to look into novel secondary metabolites from fluorescent pseudomonas that are either already known to exist or have not yet been discovered, as genomic sequencing of even the most studied microbes has confirmed that we have not yet been able to identify products of most of the gene clusters responsible for secondary metabolites synthesis.172,173 This implies that we still need to comprehend the chemical, environmental, and biological factors that lead to gene clusters expressing secondary biomolecules that have already been identified. This will support the creation of more effective bioformulations as well as the large-scale synthesis of secondary metabolites. In this regard, metabolic approaches can be highly helpful in identifying new compounds as well as figuring out how stimuli or gene expression affect the production of metabolites that are already known. Metabolomics can therefore be used to support molecular methods like gene sequencing and identification, opening the door for more research on secondary metabolites and the development of reliable bioformulations made from them. These are combined with bioinformatics technologies, which could greatly advance secondary metabolite research in the future. Finding biosynthetic gene clusters for secondary metabolites in bacterial genomes and metagenomes is the aim of the Integrated Microbial Genomes Atlas of Biosynthetic Gene Clusters (IMG-ABC) data mart.174 The NCBI’s verified gene clusters and their secondary metabolites provide even more value to this resource. IMG-ABC can be very helpful in the investigation of both known and new metabolites from fluorescent pseudomonads and other PGPR. Numerous fluorescent pseudomonads are known to produce a range of metabolites with the capacity to inhibit one or more phytopathogens. Further research is required on these multi-secondary metabolites-producing fluorescent pseudomonas because these strains will be more effective in treating soils damaged by numerous diseases and will be able to demonstrate a larger host range. It has previously been documented that multifaceted strains of fluorescent pseudomonas can be created via recombinant DNA technology, and this technique can be applied even more successfully in the future. When Yang et al.175 found that the recombinant strain that produced the combination of metabolites (CLP and PCA) inhibited G. graminis far more efficiently than the wild strain after introducing the gene for PCA synthesis from P. synxantha into P. fluorescens. The creation of recombinant fluorescent pseudomonas strains with appropriate metabolite genes can be aided using the IMG-ABC data base. Additionally, precursors and intermediates might be included in formulations to increase secondary metabolite synthesis.175 Phloroglucinol, an intermediary of DAPG biosynthesis, can also be employed to control PLT production in the instance of fluorescent pseudomonas. The use of such intermediates at very low concentrations for the large-scale synthesis of secondary metabolites may prove advantageous for the development of low-cost metabolite-based bioformulations in the future.176 Formulations based on metabolites will be especially helpful in the case of fluorescent pseudomonas, such P. aeruginosa, which is known to produce a range of anti-fungal metabolites and is a potential human pathogen. Because of its potential therapeutic repercussions, P. aeruginosa should not be used as a biocontrol agent. Additionally, authorities and experts are very concerned about the use of this bacteria in biopesticides. In the future, though, we should be able to extract the needed metabolite from fluorescent pseudomonads by using more advanced methods, understanding how to stimulate the creation of secondary metabolites through the ecology of gene expression, and access to inexpensive nutritional sources. To employ fluorescent pseudomonads in large-scale secondary metabolite production for agricultural bioformulations in the future, production technology needs to be carefully examined. Fluorescent pseudomonad secondary metabolites are today regarded as highly significant bioproducts with a variety of applications in the medical (anti-microbial, anti-cancer) and other industries.177 But further investigation is needed to find out how they could be applied to sustainable agriculture, particularly as strong natural anti-phytopathogenic agents.
Even after much research, the proportion of microbial dependent biocontrol products in global plant health management is still quite small when compared to synthetic chemicals. Additionally, there are extremely few registered biopesticide products that contain fluorescent Pseudomonas or their metabolites. The development of bioformulations using PGPR metabolites is still in its early stages. Without a question, fluorescent Pseudomonas are an effective biological control agent for phytopathogens because of their ability to produce dependable bioproducts. Nevertheless, there is still room for improvement in the way these amazing creatures are used. This can be accomplished by employing a comprehensive strategy that combines improved or innovative production techniques with metabolomic, molecular, and bioinformatics tools to investigate and use fluorescent Pseudomonas and their metabolites for the creation of new bioformulations.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Morales-Cedeno LR, Carmen Orozco-Mosqueda M, Loeza-Lara PD, Parra Cota FI, Santos-Villalobos S, Santoyo G. Plant growth promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: fundamentals, methods of application and future perspectives. Microbiol. 2021;242:126612.
Crossref - Gribben. The Great Famine and the Irish Diaspora in America. University of Massachusetts Press, Amherst. 1999.
- Strange RN, Scott PR. Plant disease a threat to global food security. Annu Rev Phytopathol. 2005;43:83-116.
Crossref - FAO, Global Forest Resources Assessment 2015. Desk Reference FAO. 2015:252.
- Dubey NK, Kumar A, Singh P, Shukla R. Exploitation of natural compounds in eco-friendly management of plant pest. In Gisi U, Chet I, Gullino ML (Eds.). Recent Developments in Managements of Plant Diseases. 2010;1:181-198.
Crossref - Aktar W, Sengupta D, Chowdhury A. Impact of pesticides use in agriculture their benefits and hazards. Interdiscip Toxicol. 2009;2(1):1-12.
Crossref - Huang W, Long C, Lam E. Roles of plant-associated microbiota in traditional herbal medicine. Trends Plant Sci. 2018;23(7):559-562.
Crossref - Hiltner L. Uber neue erfahrungen und probleme auf dem gebiete der bodenbakteriologie. Arbeiten der DLG. 1904;98:59-78.
- Kloepper JW, Schroth MN. Plant growth-promoting rhizobacteria on radishes INRA, Angers, France. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria. Station de Pathologie Vegetale et de Phytobacteriologie. 1978;2:879-882.
- Antoun H, Kloepper JW. Plant growth-promoting rhizobacteria (PGPR). In Brenner S, Miller JH (Eds.), Encyclopedia of Genetics Academic Press, New York. 2001:1477-1480.
Crossref - Arora NK. Plant Microbes Symbiosis Applied Facets. Springer. 2015:383.
Crossref - Goswami D, Thakker JN, Dhandhukia PC.Portraying mechanics of plant growth promoting rhizobacteria (PGPR) a review. Cogent Food Agric. 2016;2(1):1127500.
Crossref - Lucy M, Reed E, Glick BR. Applications of free living plant growth promoting rhizobacteria. Anton van Leeuwenhoek. 2004;86(1):1-25.
Crossref - Weller DM. Pseudomonas biocontrol agents of soilborne pathogens looking back over 30 years. Phytopathology. 2007;97(2):250-256.
Crossref - Ciancio A, Pieterse CMJ, Mercado-Blanco J. Editorial harnessing useful rhizosphere microorganisms for pathogen and pest biocontrol. Front Microbiol. 2016;7:1620.
Crossref - Finkel OM, Salas-Gonzalez I, Castrillo G, et al. A single bacterial genus maintains root growth in a complex microbiome. Nature.2020;587(7832):103-108.
Crossref - Zhang J, Sun YS, Xue YH, Chen L. Effects of storage temperature on SOD and POD activities and MDA contents in Chinese cabbage leaves. J Northwest A F Univ Nat Sci. 2019:113-119.
- Palleroni NJ. The Pseudomonas story. J Environ Microbiol. 2010;12(6):1377-1383.
Crossref - Migula W. System Der Bakterien Vol 2 Gustav Fischer, Jena, Germany. 1900.
- Aida Raio. Diverse roles played by “Pseudomonas fluorescens complex” volatile compounds in their interaction with phytopathogenic microrganims, pests and plants. World J Microbiol Biotechnol. 2024;40(3):80.
Crossref - Anjum A, Zuber M, Zia KM, Noreen A, Anjum MN, Tabasum S.Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers a review of recent advancements. Int J Biol Macromol. 2016;89:161-174.
Crossref - Palleroni NJ.Pseudomonadaceae. In Krieg NR, Holt JG (Eds.), In Bergey’s Manual of Systematic Bacteriology Vol 1. Williams and Wilkins, Baltimore.1984, pp. 141–199.
- de Bentzmann S, Plesiat P. The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ Microbiol. 2011;13(7):1655-1665.
Crossref - Morris CE, Kinkel LL, Xiao K, Prior P, Sands DC. Surprising niche for the plant pathogen Pseudomonas syringae. Infect Genet Evol. 2007;7(1):84-92.
Crossref - Palleroni NJ, Kunisawa R, Contopoulou R, Doudoroff M. Nucleic acid homologies in the genus Pseudomonas. Int J Syst Evol Microbiol. 1973;23(4):333-339.
Crossref - Mulet M, Gomila M, Lemaitre B, Lalucat J, Garcia-Valdes E. Taxonomic characterization of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst Appl Microbiol. 2012;35(3):145-149.
Crossref - Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. Phylogenetic affiliation of the Pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50(Pt 4):1563-1589.
Crossref - Kersters K, Ludwig W, Vancanneyt M, DeVos P, Gillis M, Schleifer KH. Recent changes in the classification of the Pseudomonads an overview. Syst Appl Microbiol. 1996;19(4):465-477.
Crossref - Gomila M, Pena A, Mulet M, Lalucat J, Garcia-Valdes E. Phylogenomics and systematics in Pseudomonas. Front Microbiol. 2015;6:214.
Crossref - Garrido-Sanz D, Meier-Kolthoff JP, Goker M, Martin M, Rivilla R, Redondo-Nieto M. Correction genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS One. 2016;11(4):e0153733.
Crossref - Murata K, Suenaga M, Kai K. Genome Mining Discovery of Protegenins A-D, Bacterial Polyynes Involved in the Antioomycete and Biocontrol Activities of Pseudomonas protegens. ACS Chem Biol. 2021;17(12):3313-3320.
Crossref - Budzikiewicz H. Secondary metabolites from fluorescent Pseudomonads. FEMS Microbiol Rev. 1993;10(3-4):209-228.
Crossref - Arnold AE, Mejia LC, Kyllo D, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci U S A. 2003;100(26):15649-15654.
Crossref - Lu Y, Yang Q, Lin Z, Yang X. A modular pathway engineering strategy for the high-level production of b-ionone in Yarrowia Lipolytica. Microb Cell Factories. 2010;19(1):1-13.
Crossref - Lu Y, Zeng L, Li M, et al. Use of GC-IMS for detection of volatile organic compounds to identify mixed bacterial culture medium. AMB Express. 2022;12(1):1-11.
Crossref - Letoffe S, Wu Y, Darch SE, Beloin C, Whiteley M, Touqui L, Ghigo JM. Pseudomonas aeruginosa Production of Hydrogen Cyanide Leads to Airborne Control of Staphylococcus aureus Growth in Biofilm and In Vivo Lung Environments. mBio. 2022;13(5):e0215422.
Crossref - Priyanka AT, Kotasthane AS, Kosharia A, Kushwah R, Zaidi NW, Singh US. Crop specific plant growth promoting effects of ACCd enzyme and siderophore producing and cynogenic fluorescent Pseudomonas. Biotechnology. 2017;7(1):27.
Crossref - Strano CP, Bella P, Licciardello G, Caruso A, Catara V. Role of secondary metabolites in the biocontrol activity of Pseudomonas corrugata and Pseudomonas mediterranea. Eur J Plant Pathol. 2017;149(1):103-115.
Crossref - Reetha AK, Pavani SL, Mohan S. Hydrogen cyanide production ability by bacterial antagonist and their antibiotics inhibition potential on Macrophomina phaseolina (Tassi) goid. Int J Curr Microbiol Appl Sci. 2014;3(5):172-178.
- Lanteigne C, Gadkar VJ, Wallon T, Novinscak A, Filion M. Production of DAPG and HCN by Pseudomonas sp lbum300 contributes to the biological control of bacterial canker of tomato. Am Phytopathol Soc. 2012;102(10):967-973.
Crossref - Ossowicki A, Jafra S, Garbeva P. The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS One. 2017;12(3):e0174362.
Crossref - Park JY, Oh SA, Anderson AJ, Neiswender J, Kim JC, Kim YC. Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett Appl Microbiol. 2011;52(5):532-537.
Crossref - Leon M, Yaryura PM, Montecchia MS, et al. Antifungal activity of selected indigenous Pseudomonas and Bacillus from the soybean rhizosphere. Int J Microbiol. 2009;2009:572049.
Crossref - Burkhead KD, Schisler DA, Slininger PA. Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl Environ Microbiol. 1994;60(6):2031-2039.
Crossref - Homma Y. Mechanisms in biological control focused on the antibiotic pyrrolnitrin. In Ryder MH, Stephens PM, Bowen GD (Eds.), Improving Plant Productivity With Rhizobacteria. CSIRO Division of Soils, Adelaide, Australia. 1994:100-103.
- Patil S, Nikama M, Anokhinab T, Kochetkovb V, Chaudhari A. Multi-stress tolerant plant growth promoting Pseudomonas spp. MCC 3145 producing cytostatic and fungicidal pigment. Biocatal Agric Biotechnol. 2017;10:53-63.
Crossref - Jaaffar AK, Parejko JA, Paulitz TC, Weller DM, Thomashow LS. Sensitivity of rhizoctonia isolates to phenazine-1-carboxylic acid and biological control by phenazine-producing Pseudomonas spp. Phytopathology. 2017;107(6):692-703.
Crossref - Anjaiah V, Cornelis P, Koedam N. Effect of genotype and root colonization in biological control of fusarium wilts in pigeonpea and chickpea by Pseudomonas aeruginosa PNA1. Can J Microbiol. 2003;49(2):85-91.
Crossref - Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var tritici. J Bacteriol. 1988;170(8):3499-3508.
Crossref - Thomashow LS, Weller DM, Bonsall RF, Pierson LS. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56(4):908-991.
Crossref - Sherathia D, Dey R, Thomas M, Dalsania T, Savsani K, Pal KK. Biochemical and molecular characterization of DAPG-producing plant growth promoting rhizobacteria (PGPR) of groundnut (Arachis hypogaea L). Legume Res. 2016;39(OF):614-622.
Crossref - Lagzian A, Sabari Riseh R, Khodaygan P, Sedaghati E, Dashti H. Introduced Pseudomonas fluorescens VUPf5 as an important biocontrol agent for controlling Gaeumannomyces graminis var tritici the causal agent of take-all disease in wheat. Arch Phytopathol Plant Prot. 2013;46(17):2104-2116.
Crossref - Zhou T, Chen D, Li C, et al. Isolation and characterization of Pseudomonas brassicacearum J12 as an antagonist against Ralstonia solanacearum and identification of its antimicrobial components. Microbiol Res. 2012;167(7):388-394.
Crossref - Ayyadurai N, Ravindra Naik P, Rao MS, et al. Isolation and characterization of a novel banana rhizosphere bacterium as fungal antagonist and microbial adjuvant in micropropagation of banana. J Appl Microbiol. 2006;100(5):926-937.
Crossref - Velusamy P, Gnanamanickam SS. Identification of 2,4-diacetylphloroglucinol production by plant-associated bacteria and its role in suppression of rice bacterial blight in India. Curr Sci. 2003;85(9):1270-1273.
- Garagulya AD, Kiprianova EA, Boiko OI. Antibiotic effect of bacteria from the genus Pseudomonas on phytopathogenic fungi. Mikrobiologischez (Kiev).1974;36(2):197-202.
- Le CN, Kruijt M, Raaijmakers JM. Involvement of phenazines and lipopeptides in interactions between Pseudomonas species and Sclerotium rolfsii, causal agent of stem rot disease on groundnut. J Appl Microbiol. 2012;112(2):390-403.
Crossref - Tran H, Ficke A, Asiimwe T, Hofte M, Raaijmakers JM. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 2007;175(4):731-742.
Crossref - Nielsen TH, Thrane C, Christophersen C, Anthoni U, Sorensen J. Structure, production characteristics and fungal antagonism of tensin – a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J Appl Microbiol. 2000;89(6):992-1001.
Crossref - Nielsen TH, Sorensen D, Tobiasen C, et al. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl Environ Microbiol. 2002;68(7):3416-3423.
Crossref - Noirot Gros MF, Shinde S, Larsen PE, et al. Dynamics of aspen roots colonization by Pseudomonads reveals strain specific and mycorrhizal specific patterns of biofilm formation. Front Microbiol. 2018;9:853.
Crossref - Hansen ML, Wibowo M, Jarmusch SA, Larsen TO, Jelsbak L. Sequential interspecies interactions affect production of antimicrobial secondary metabolites in Pseudomonas protegens. DTU9.1. ISME J. 2022;16(12):2680-2690.
Crossref - Veselova MA, Plyuta VA, Khmel IA. Volatile compounds of bacterial origin structure, biosynthesis, and biological activity. Microbiology. 2018;88(3):261-274.
Crossref - Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173(3):170-177.
Crossref - Schippers B, Bakker AW, Bakker PAHM, Van Peer R. Beneficial and deleterious effects of HCN-producing Pseudomonads on rhizosphere interactions. Plant Soil. 1990;129:75-83.
Crossref - Nandi M, Selin C, Brawerman G, Fernando WGD, Kievit T. Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol Control. 2017;108:47-54.
Crossref - Devi KK, Deepika S, Bhaduri A, Kothamasi D. Polymorphism in hcnAB gene in Pseudomonas species reveals ecologically distinct hydrogen cyanide-producing populations. Geomicrobiol J. 2013;30(2):131-139.
Crossref - Devi KK, Kothamasi D. Pseudomonas fluorescens CHA0 can kill subterranean termite Odontotermes obesus by inhibiting cytochrome c oxidase of the termite respiratory chain. FEMS Microbiol Lett. 2009;300(2):195-200.
Crossref - Pessi G, Haas D. Transcriptional control of hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quoram- sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182(24):6940-6949.
Crossref - Rijavec T, Lapanje A. Hydrogen cyanide in the rhizosphere not suppressing plant pathogens, but rather regulating availability of phosphate. Front Microbiol. 2016;7:1785.
Crossref - Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1- carboxamide from Pseudomonas aeruginosa PA01. J Bacteriol. 2001;183(21):6454-6465.
Crossref - Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417-445.
Crossref - Guttenberger N, Blankenfeldt W, Breinbauer R. Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorg Med Chem. 2017;25(22):6149-6166.
Crossref - Briard B, Bomme P, Lechner BE, et al. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep. 2015;5:8220.
Crossref - Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2(2):71-78.
Crossref - Pierson LS, Pierson EA. Metabolism and function of phenazines in bacteria in environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86(6):1659-1670.
Crossref - Laursen JB, Nielsen J. Phenazine natural products biosynthesis, synthetic analogues, and biological activity. Chem Rev. 2004;104(3):1663-1686.
Crossref - Tupe SG, Kulkarni RR, Shirazi F, Sant DG, Joshi SP, Deshpande MV. Possible mechanism of antifungal phenazine-1-carboxamide from Pseudomonas sp. against dimorphic fungi Benjaminiella poitrasii and human pathogen Candida albicans. J Appl Microbiol. 2015;118(1):39-48.
Crossref - Schoonbeek H, Raaijmakers JM, De Waard MA. Fungal ABC transporters and microbial interactions in natural environments. Mol Plant Microbe Interact. 2002;15(11)1165-1172.
Crossref - Chincholkar SB, Thomashow L. Microbial Phenazines. Springer-Verlag, Berlin, Heidelberg. 2013.
Crossref - Beifuss U, Tietze M. Methanophenzine and other natural biologically active phenazines. Top Curr Chem. 2005;244(37):77-113.
Crossref - Chen JJ, Chen W, He H, et al. Manipulation of microbial extracellular electron transfer by changing molecular structure of phenazine-type redox mediators. Environ Sci Technol. 2012;47(2):1033-1039.
Crossref - Zhou L, Jiang H, Jin K, et al. Isolation, identification and characterization of rice rhizobacterium Pseudomonas aeruginosa PA1201 producing high level of biopesticide “Shenqinmycin” and phenazine-1-carboxamide. Wei Sheng Wu Xue Bao. 2015;55(4):401-411.
- Namwongsa J, Jogloy S, Vorasoot N, Boonlue S, Riddech N, Mongkolthanaruk W. Endophytic bacteria Improve root traits, biomass and yield of Helianthus tuberosus L under normal and deficit water conditions. J Microbiol Biotechnol. 2019;29(11):1777-1789.
Crossref - Berendsen RL, Vismans G, Yu K, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12(6):1496-1507.
Crossref - Lozano Andrade CN, Hansen ML, Jarmusch SA, Jelsbak L. On the biotransformation of Pseudomonads secondary metabolites. The Danish Microbiological Society Annual Congress. 2023:29-29.
- Loper JE, Hassan KA, Mavrodi DV, et al. Comparative genomics of plant-associated Pseudomonas spp. insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784.
Crossref - Singh PI, Bharate SB. Phloroglucinol compounds of natural origin. Nat Prod Rep. 2006;23(4):558-591.
Crossref - Troppens DM, Moynihan JA, Barret M, O’Gara F, Morrissey JP. Genetics and evolution of 2, 4-diacetylphloroglucinol synthesis in Pseudomonas fluorescens. Molecular Microbial Ecology of the Rhizosphere. 2013;1-2:593-605.
Crossref - Sonnleitner E, Haas D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol. 2011;91(1):63-79.
Crossref - Keel C, Weller DM, Natsch A, Defago G, Cook RJ, Thomashow LS. Conservation of the 2, 4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62(2):552-563.
Crossref - Achkar J, Xian M, Zhao H, Frost JW. Biosynthesis of phloroglucinol. J Am Chem Soc. 2005;127(15):5332-5333.
Crossref - Mandryk-Litvinkovich MN, Muratova AA, Nosonova TL, Evdokimova OV, Valentovich LN, Titok MA, Kolomiets EI. Molecular genetic analysis of determinants defining synthesis of 2,4-diacetylphloroglucinol by Pseudomonas brassicacearum BIM B-446 bacteria. Appl Biochem Microbiol. 2017;53(1):31-39.
Crossref - De La Fuente L, Landa BB, Weller DM. Host crop affects rhizosphere colonization and competitiveness of 2, 4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology. 2006;96(7):751-762.
Crossref - Moynihan JA, Morrissey JP, Coppoolse ER, Stiekema WJ, O’Gara F, Boyd EF. Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Appl Environ Microbiol. 2009;75(7):2122-2131.
Crossref - Landa BB, Mavrodi OV, Schroeder KL, Allende-Molar R, Weller DM. Enrichment and genotypic diversity of phlD-containing fluorescent Pseudomonas spp in two soils after a century of wheat and flax monoculture. FEMS Microbiol Ecol. 2006;55(3):351-368.
Crossref - Picard C, Bosco M. Genetic diversity of phi D gene from 2,4-diacetylphloroglucinol- producing Pseudomonas spp. strains from the maize rhizosphere. FEMS Microbiol Lett. 2003;219(2):167-172.
Crossref - Hultberg M, Alsanius B. Influence of nitrogen source on 2, 4-diacetylphloroglucinol production by the biocontrol strain Pf-5. Open Microbiol J. 2008;2(1):74-78.
Crossref - Cao Y, Jiang X, Zhang R, Xian M. Improved phloroglucinol production by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2011;91(6):1545-1552.
Crossref - Abdel-Ghany SE, Day I, Heuberger AL, Broeckling CD, Reddy ASN. Production of phloroglucinol, a platform chemical, in Arabidopsis using a bacterial gene. Sci Rep. 2016;6:38483.
Crossref - Nowak-Thompson B, Gould SJ, Loper JE. Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene. 1997;204(1-2):17-24.
Crossref - Takeda R. Pseudomonas pigments iii derivatives of pyoluteorin. J Bull Agric Chem Soc Jpn. 1958;23(2):126-130.
Crossref - Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J Bacteriol. 1999;181(1):2166-2174.
Crossref - Brodhagen M, Henkels MD, Loper JE. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 2004;70(3):1758-1766.
Crossref - Clifford JC, Buchanan A, Vining O, et al. Phloroglucinol functions as an intracellular and intercellular chemical messenger influencing gene expression in Pseudomonas protegens. Environ Microbiol. 2016;18(10):3296-3308.
Crossref - Arima K, Imanaka H, Kousaka M, Fukuta A, Tamura G. Pyrrolnitrin, a new antibiotic substance, produced by Pseudomonas. Agric Biol Chem. 1964;28(8):575-576.
Crossref - Mujumdar SS, Bashetti SP, Chopade BA. Plasmid pUPI126-encoded pyrrolnitrin production by Acinetobacter haemolyticus A19 isolated from the rhizosphere of wheat. World J Microbiol Biotechnol. 2014;30(2):495-505.
Crossref - Weller DM, Thomashow LS, Mavrodi DV, Yang M, Zhang J. Pseudomonas fluorescens 2-79 with genes for biosynthesis of pyrrolnitrin improves biocontrol activity US. 2016.
- Jani J, Parvez N, Mehta D. Metabolites of Pseudomonads a new avenue of plant health management. Chakravarthy AK (Ed.). Horizons In Insect Science Towards Sustainable Pest Management. 2015:61-69.
Crossref - Pillonel C, Meyer T. Effect of phenylpyrroles on glycerol accumulation and protein kinase activity of Neurospora crassa. Pest Sci. 1997;49(3):229-236.
Crossref - Nakatsu CH, Straus NA, Wyndham RC. The nucleotide sequence of the TN6271 3- chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141(2):485-495.
Crossref - Steinberg C, Edel-Hermann V, Alabouvette C, Lemanceau P. Soil suppressiveness to plant diseases. In Van Elsas JD, Trevors JT, Jansson JK(Eds.), Modern Soil Microbiology II. CRC Press, Boca Raton FL, USA. 2006:455-478.
- Costa R, van Aarle IM, Mendes R, van Elsas JD. Genomics of pyrrolnitrin biosynthetic loci evidence for conservation and whole-operon mobility within gramnegative bacteria. Environ Microbiol. 2009;11(1):159-175.
Crossref - Kilani J, Fillinger S. Phenylpyrroles 30 years, two molecules and (nearly) no resistance. Front Microbiol. 2016:7.
Crossref - Raaijmakers JM, de Bruijn I, de Kock MJD. Cyclic lipopeptide production by plant- associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol Plant Microbe Interact. 2006;19(7):699-710.
Crossref - Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M, Natural functions of lipopeptides from Bacillus and Pseudomonas more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34(6):1037-1062.
Crossref - Li W, Rokni-Zadeh H, Vleeschouwer De M, et al. The antimicrobial compound xantholysin defines a new group of Pseudomonades cyclic lipopeptides. PLoS One. 2013;8(5):e62946.
Crossref - Raju R, Kandhasamy S, Nalliappan GK, Natarajan KV, Gandhi K, Chandrasekaran B. Cyclic depsipeptide producing fluorescent Pseudomonads exerts antifungal activity against fungal pathogens of maize (Zea mays). Afric J Microbiol Res. 2016;10(42):1767-1774.
Crossref - Nielsen TH, Nybroe O, Koch B, Hansen M, Sorensen J. Genes involved in cyclic lipopeptide production are important for seed and straw colonization by Pseudomonas sp. strain DSS73. Appl Environ Microbiol. 2005;71(7):4112-4116.
Crossref - Nybroe O, Sorensen J. Production of Cyclic Lipopeptides by Fluorescent Pseudomonads 3. Springer, US. 2014:147-172.
Crossref - Baltz RH. Combinatorial biosynthesis of cyclic lipopeptide antibiotics a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol. 2014;3(10):748-758.
Crossref - Olorunleke FE, Kieu N P, Hofte M. Recent advances in Pseudomonas biocontrol. In Murillo J, Vinatzer BA, Jackson RW, Arnold DL(Eds.), Bacterial-Plant Interactions Advance Research and Future Trends. Caister Academic Press, Norfolk. 2015:167-198.
Crossref - Loper JE, Henkels MD, Rangel LI, et al. Rhizoxin, orfamide A, and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol. 2016;18(10):3509-3521.
Crossref - Song C, Sundqvist G, Malm E, et al. Lipopeptide biosynthesis in Pseudomonas fluorescens is regulated by the protease complex ClpAP. BMC Microbiol. 2015;15:29.
Crossref - Licciardello G, Bertani I, Steindler L, Bella P, Venturi V, Catara V. The transcriptional activator rfiA is quorum-sensing regulated by cotranscription with the luxI homolog pcoI and is essential for plant virulence in Pseudomonas corrugata. Mol Plant Microbe Interact. 2009;22(12):1514-1522.
Crossref - Licciardello G, Strano CP, Bertani I, et al. N-Acyl-homoserine-lactone quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in the regulation of lipodepsipeptide production. J Biotechnol. 2012;159(4):274-282.
Crossref - Cui J, Bahrami AK, Pringle EG, et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA. 2005;102(5):1791-1796.
Crossref - Dubern JF, Lagendijk EL, Lugtenberg BJ, Bloemberg GV. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J Bacteriol. 2006;187(17):187.
Crossref - Mazzola M. Mechanisms of natural soil suppressiveness to soilborne diseases. Anton van Leeuwenhoek. 2002;81(1-4):557-564.
Crossref - Vida C, Bonilla N, de Vicente A, Cazorla FM. Microbial profiling of a suppressiveness-induced agricultural soil amended with composted almond shells. Front Microbiol. 2016;7:4.
Crossref - Haggag WM, Abo El Soud M. Production and optimization of Pseudomonas fluorescens biomass and metabolites for biocontrol of strawberry grey mould. Am J Plant Sci. 2012;03(07):836-845.
Crossref - Kupferschmied P, Maurhofer M, Keel C. Promise for plant pest control: root associated Pseudomonads with insecticidal activities. Front Plant Science. 2013;4:1-17.
Crossref - Singh JS, Kaushal S, Kumar A, Vimal SR, Gupta VK. Agriculturally beneficial microbes in sustainable crop production. Front Microbiol. 2016.
- Arora NK, Mishra J. Prospecting the roles of metabolites and additives in future bioformulations for sustainable agriculture. Appl Soil Ecolo. 2016;107:405-407.
Crossref - Kwak YS, Weller DM. Take-all of wheat and natural disease suppression a review. Plant Pathol J. 2013;29(2):125-135.
Crossref - Stockwell VO, Stack JP. Using Pseudomonas spp. for integrated biological control. Phytopathology. 2007;97(2):244-249.
Crossref - Stockwell VO, Johnson KB, Sugar D, Loper JE. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology. 2010;100(12):1330-1339.
Crossref - Shahid I, Rizwan M, Baig DN, Saleem RS, Malik KA, Mehnaz S. Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis and P. aurantiaca strains isolated from cactus, cotton, and para grass. J Microbiol Biotechnol. 2017;27(3):480-491.
Crossref - Sharifazizi M, Harighi B, Sadeghi A. Evaluation of biological control of Erwinia amylovora, causal agent of fire blight disease of pear by antagonistic bacteria. Biological Control. 2017;104:28-34.
Crossref - Meyer SLF, Halbrendt JM, Carta LK, et al. Toxicity of 2,4-diacetylphloroglucinol (DAPG) to plant-parasitic and bacterial-feeding nematodes. J Nematol. 2009;41(4):274-280.
- Jousset A, Rochat L, Scheu S, Bonkowski M, Keel C. Predator-prey chemical warfare determines the expression of biocontrol genes by rhizosphere-associated Pseudomonas fluorescens. Appl Environ Microbiol. 2010;76(15):5263-5268.
Crossref - Tewari S, Arora NK. Talc based exopolysaccharides formulation enhancing growth and production of Hellianthus annuus under saline conditions. Cell Mol Biol. 2014;60(5):73-81.
- Borah SN, Goswami D, Sarma HK, Cameotra SS, Deka S. Rhamnolipid biosurfactant against Fusarium verticillioides to control stalk and ear rot disease of maize. Front Microbiol. 2016;7:1505.
Crossref - Yan Q, Philmus B, Chang JH, Loper JE. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. eLife. 2017;6:e22835.
Crossref - Zhou JY, Zhao XY, Dai CC. Antagonistic mechanisms of endophytic Pseudomonas fluorescens against Athelia rolfsii. J Appl Microbiol. 2014;117(4):1144-1158.
Crossref - Giorgio A, De Stradis A, Lo Cantore P, Iacobellis NS. Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front Microbiol. 2015;6:1056.
Crossref - Jishma P, Hussain N, Chellappan R, Rajendran R, Mathew J, Radhakrishnan EK. Strain-specific variation in plant growth promoting volatile organic compounds production by five different Pseudomonas spp. as confirmed by response of Vigna radiata seedlings. J Appl Microbiol. 2017;123(1):204-216.
Crossref - Park Y S, Dutta S, Ann M, Raaijmakers JM, Park K. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem Biophys Res Commun. 2015;461(2):361-365.
Crossref - Wallace RL, Hirkala DL, Nelson LM. Postharvest biological control of blue mold of apple by Pseudomonas fluorescens during commercial storage and potential modes of action. Post Harvest Bio Technol. 2017;133:1-11.
Crossref - Simionato AS, Navarro MOP, de Jesus MLA, et al. The effect of phenazine-1-carboxylic acid on mycelial growth of Botrytis cinerea produced by Pseudomonas aeruginosa LV strain. Front Microbiol. 2017;8:1102.
Crossref - Glasser NR, Wang BX, Hoy JA, Newman DK. The pyruvate and a-ketoglutarate dehydrogenase complexes of Pseudomonas aeruginosa catalyze pyocyanin and phenazine-1-carboxylic acid reduction via the subunit dihydrolipoamide dehydrogenase. J Biol Chem. 2017;292(13):5593-5607.
Crossref - Morohoshi T, Yamaguchi T, Xie X, Wang WZ, Takeuchi K, Someya N. Complete genome sequence of Pseudomonas chlororaphis subsp aurantiaca reveals a triplicate quorum-sensing mechanism for regulation of phenazine production. Microbes Environ. 2017;32(1):47-53.
Crossref - Jain R, Pandey A. A phenazine-1-carboxylic acid producing polyextremophilic Pseudomonas chlororaphis (MCC2693) strain, isolated from mountain ecosystem, possesses biocontrol and plant growth promotion abilities. Microbiol Res. 2016;190:63-71.
Crossref - Morohoshi T, Wang WZ, Suto T, et al. Phenazine antibiotic production and antifungal activity are regulated by multiple quorum-sensing systems in Pseudomonas chlororaphis subsp aurantiaca StFRB508. J Biosci Bioeng. 2013;116(5):580-584.
Crossref - Khare E, Arora NK. Dual activity of pyocyanin from Pseudomonas aeruginosa-antibiotic against phytopathogen and signal molecule for biofilm development by rhizobia. Can J Microbiol. 2011;57(9):708-713.
Crossref - Kumar RS, Ayyadurai N, Pandiaraja P, et al. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J Appl Microbiol. 2005;98(1):145-154.
Crossref - D’Aes J, De Maeyer K, Pauwelyn E, Hofte M. Biosurfactants in plant Pseudomonas interactions and their importance to biocontrol. Environ Microbiol Rep. 2010;2(3):359-372.
Crossref - Lee DW, Kim BS. Antimicrobial cyclic peptides for plant disease control. Plant Pathol J. 2015;31(1):1-11.
Crossref - Andersen JB, Koch B, Nielsen TH, et al. Surface motility in Pseudomonas sp DSS73 is required for efficient biological containment of the rootpathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology. 2003;149(Pt 1):37-46.
Crossref - Kuiper I, Lagendijk EL, Pickford R, et al. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol. 2004;51(1):97-113.
Crossref - de Souza JT, Weller DM, Raaijmakers JM. Frequency, diversity, and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch takeall decline soils. Phytopathology. 2003;93(1):54-63.
Crossref - Mishra S, Arora NK. Management of black rot in cabbage by rhizospheric Pseudomonas species and analysis of 2,4-diacetylphloroglucinol by qRT-PCR. Biol Control. 2012;61(1):32-39.
Crossref - Maurhofer M, Baehler E, Notz R, Martinez V, Keel C. Cross talk between 2,4- diacetylphloroglucinol-producing biocontrol Pseudomonads on wheat roots. Appl Environ Microbiol. 2004;70(4):1990-1998.
Crossref - Gutierrez-Garcia, K, Neira-Gonzalez A, Perez-Gutierrez RM, et al. Phylogenomics of 2, 4-diacetylphloroglucinol-producing Pseudomonas and novel antiglycation endophytes from Piper auritum. J Nat Prod. 2017;80(7):1955-1963.
Crossref - Weisskopf L, Schulz S, Garbeva P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat Rev Microbiol. 2021;19(6):391-404.
Crossref - Faramarzi MA, Brandl H. Formation of water-soluble metal cyanide complexes from solid minerals by Pseudomonas plecoglossicida. FEMS Microbiol Lett. 2006;259(1):47-52.
Crossref - Rampioni G, Schuster M, Greenberg EP, et al. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2007;66(6):1557-1565.
Crossref - Salas-Gonzalez I, Reyt G, Flis P, et al. Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science. 2021;371:6525.
Crossref - Tilocca B, Cao A, Migheli Q. Scent of a killer: Microbial volatilome and its role in the biological control of plant pathogens. Front Microbiol. 2020.
Crossref - Lambowitz AM, Slayman CW. Effect of pyrrolnitrin on electron transport and oxidative phosphorylation in mitochondria isolated from Neurospora crassa. J Bacteriol. 1972;112(2):1020-1022.
Crossref - Okada A, Banno S, Ichiishi A, Kimura M, Yamaguchi I, Fujimura M. Pyrrolnitrin interferes with osmotic signal transduction in Neurospora crassa. J Pest Sci. 2005;30(4):378-383.
Crossref - Covington BC, McLean JA, Bachmann BO. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat Prod Rep. 2017;34(1):6-24.
Crossref - Walsh CT, Fischbach MA. Natural products version 2.0 connecting genes to molecules. J Am Chem Soc. 2010;132(8):2469-2493.
Crossref - Markowitz VM, Chen IMA, Chu K, et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42(D1):D568-D573.
Crossref - Yang M, Mavrodi DV, Mavrodi OV, Thomashow LS, Weller DM. Construction of a recombinant strain of Pseudomonas fluorescens producing both phenazine-1-carboxylic acid and cyclic lipopeptide for the biocontrol of take-all disease of wheat. Eur J Plant Pathol. 2017;149(7):683-694.
Crossref - Shelake RM, Waghunde RR, Kim JY. Plant-Microbe-Metal (PMM) interactions and strategies for remediating metal ions. Plant-Metal Interactions. 2019:247-262.
Crossref - Waghunde RR, Shinde CU, Pandey P, Singh C. Fungal biopesticides for agro-environmental sustainability. Industrially Important Fungi for Sustainable Development. Fungal Biology. 2021:479-508.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.