ISSN: 0973-7510
E-ISSN: 2581-690X
Banana is one of the primary fruits cultivated in Malaysia and currently decimated by the emergence of a disease, known as banana blood disease (BBD) which caused by Ralstonia syzygii subsp. celebesensis (Rsc). The BBD has significantly affected the yield and profits of the worldwide banana industry. To date, various approaches including chemical and biological controls have been attempted to manage this disease but none of them succeed in controlling the disease. The uses of lactic acid bacteria (LAB) in managing plant diseases have been reported earlier but little information is available. Therefore, this project is designed to identify and investigate potential plant-associated LAB as biological control agent (BCA) against Rsc by using agar well diffusion method. The inhibition zones of each well were observed after 72h and the isolated LAB that showed inhibition zones were proceed for molecular characterization using PCR amplification followed by gel electrophoresis. The sequences were used for phylogenetic analysis. In addition, each of the potential LAB were used to identify their morphological characterizations and biochemical testing. Throughout the study, the highest inhibition zones of LAB from kimchi and fermented milk achieved a diameter of 21.30 mm and 28.70 mm, respectively. Kimchi isolates showed the highest similarity which is 97% as Lactiplantibacillus plantarum species. Among the fermented milk isolates, the highest similarity which is 98% identified as Lacticaseibacillus paracasei.
Banana, Banana Blood Disease, Lactic Acid Bacteria, Biological Control Agent, Ralstonia syzygii subsp. celebesensis (Rsc)
Banana (Musa spp.) is one of the premier, popular, commercial food crops that has been regarded among the most significant income source of Malaysia’s economy in agro-based industry sector.1 It was ranked as top 10 most produced commodities in Malaysia from 1994 to 2021, with an average production of approximately 402,000 tonnes in a year.2 However, the banana production in Malaysia was hampered with the spread of banana blood disease (BBD) which contributed a significant economic loss in Malaysia, Southeast Asia producing countries and beyond.3 In Malaysia, the critical epidemic of the bacterial wilt was first detected in Johor in 2007 due to a massive flood and allow it spread to whole Peninsular Malaysia.4 According to Food and Agriculture Organization of United Nations,5 the yield of bananas in Malaysia had declined from 380,000 to 260,911 tonnes and the cultivation areas was decreased from 25,000 ha to 22,719 ha in 2007. The main threat to the reduction of banana production is possible due to adverse environmental conditions which allowed pathogens able to spread and eventually destroy the banana farms.6 The disease was then reported in Sabah in 2016 and emerged in Sabah’s different districts between 2012 and 2013.7
Ralstonia syzygii subsp. celebesensis (Rsc), a bacterial pathogen causal of the disease will cause the flesh of bananas to rot and turn brown (discoloration) and the leaves to wither.3 Rsc can be transmitted through water, soil, farm equipment, machinery, and insects.8 The transmission by soil and water and on farm tools allowed the pathogen to enter host roots through natural openings or wounds.9,10 Ralstonia often moves quickly to stem tissues after colonizing the root xylem tissues and infecting the roots of both susceptible and resistant plants even through a minor wound.11 The complex xylem, which aids in transporting the water to the entire plant, is made up of tracheids (the dead cells) and parenchyma (the living cells). With the help of xylem, exopolysaccharide, which is a substance that is produced by Ralstonia, will block the activity of the xylem and eventually lead to the appearance of wilting in plants.11 Exopolysaccharide, a substances that produced by Ralstonia helps in the movement and swimming of T-3 secreted effector, and enzyme secretions such as DNAases, cell wall-degrading enzymes, and enzymes that detoxify reactive oxygen species, eventually caused the blockage of xylem.11,12 Therefore, when plants are starting to wilt, the pathogen will start to move from the roots to the soil and it caused Ralstonia to activate a range of virulent factors to induce illness in different infection phases.12
To control bacterial wilt continuously damaging the field of banana, several strategies were investigated in the aspect of physical, chemical, biological, and cultural methods.13 Pesticides such as algicide and fumigants show positive impacts in controlling bacterial wilt and the combination of chemicals successfully caused a significant loss of the infected banana field from 72% to 100%.13 However, this has not always been the case. According to Edwards-Jones G, the consequences of carelessly or without proper knowledge in applying pesticide may result on soil contamination which are able to persist in the environment for many years and might be hazardous to farmers.14 Although there are several methods that have been applied including applying pesticides, solarization and hot water treatment, soil amendment, etc. but none of them are safe to be applied in order to control the yield of bacteria wilt.13
Instead, safer and security treatments are required in controlling the disease. The key to this study is using lactic acid bacteria (LAB) which potentially act as a biological control agent (BCA) in order to create a sustainable treatment against BDB. The ability of LAB act as microbial biopesticides that are beneficial microorganisms provides the benefits by competing with pathogens.15 Numerous antimicrobial compounds, including organic acids (lactic and phenyllactic acids), diacetyl, hydrogen peroxide and bacteriocins that produced by LAB strain create a relatively harsh environment for any pathogens to survive.16,17 However, acids have a major impact on biocontrol abilities. As stated in the previous studies,18,19 LAB cultures or their supernatants have been applied as BCAs in controlling on the plant diseases in chili, tomato and cucumber caused by Colletotrichum capsica, Fusarium oxysporum and Pythium ultimum respectively. In addition, LAB strain that isolated from fresh fruits and vegetables is proven that can reduce the growth infection of phytopathogenic and spoilage bacteria and fungi as Xanthomonas campestris, Erwinia carotovora, Monilinia laxa, and Botrytis cinerea on apple fruits.20 Furthermore, Hamed HA reported LAB has its potential and shows significant impacts in enhancing overall plant development as a BCA for protection of tomato plants.21 Lactic acid bacteria are a promising bacterial group that has the potential to suppress soil-borne diseases and are able to promote plant growth development.19 Therefore, the present investigation was designed to identify and characterize phenotypically and genotypically of the potential LAB. Also, this study investigated the in vitro activity of the potential plant-associated LAB as BCAs against Rsc of the banana blood disease.
Sampling and isolation of lactic acid bacteria
LAB were sampled and isolated from kimchi (a type of Korean fermented vegetables), grape and fermented dairy products. Samples were taken aseptically and analyzed within 24 hours. de Man, Rogosa and Sharpe agar (MRS) were used for the isolation of LAB. Each sample was cleaned before being crushed using a homogenizer. The crushed samples were mixed with 0.75% NaCl solution. Serial dilutions of the mixed solution (10 to 108-fold) were spread directly onto the surface of MRS-agar plates.22 The samples were incubated for 3 to 5 days at 28°C (28 ± 2°C). Colonies of acid-producing bacteria were identified morphologically using Gram staining and catalase test and were selected randomly to purify the colonies by replating them on the MRS agar plates. This procedure was repeated several times using streaking method until pure cultures were obtained. The pure cultures of the LAB isolates were maintained in MRS agar for subsequent use.
Morphological and biochemical characterization of LAB
The biochemical tests of potential LAB used for characterizations including pH tolerance test, NaCl tolerance test and temperature sensitivity test followed the methods as described by Ayo-Omogie and Okorie.23
Gram staining
LAB colony was randomly selected from the cultures and smeared on the slide. The smear was then flooded with crystal violet for 60s and washed off with distilled water and air-dried. Iodine was applied on the smear for 60s and again washed with distilled water and allowed to air dry. 95% alcohol was slowly added on the smear until the staining run clear. The smear was washed with distilled water and air-dried. Counterstains with safranin was spilled slowly for 45s and washed off and air-dried. The bacteria smear was then observed under light microscope (Nikon Model Eclipse E100 LED MIV R) with 1000x magnification to observe the shape and color. A purple staining indicates Gram-positive while pink staining indicates Gram-negative.24
Catalase test
One drop of 3% hydrogen peroxide (H2O2) was dripped to LAB cultures on a glass slide to perform catalase test. The test was evaluated by the formation of oxygen bubbles which indicate the bacteria able to produce catalase enzyme to convert H2O2 to water and oxygen.25
pH tolerance test
Each LAB culture was sub-cultured into sterile MRS broth with the volume ratio 1 to 10 and incubated at 28°C ± 2°C overnight. 1 mL of overnight LAB cultures were then sub-cultured into 9 mL of sterile MRS broth with different pH: pH2, pH3 and pH4 and incubated at 28°C ± 2°C for 24h. MRS broth with different pH were prepared using HCl and NaOH before autoclaved. After 24h incubation, pH tolerance test was evaluated by inoculating 10 µL of inoculum from each tube into MRS agar using track plate method and incubated at 28°C ± 2°C for 48h. The presence and absent growth of LAB on MRS agar indicated the ability of the LAB to sustain in extreme pH conditions as pH tolerant.23
Sodium chloride (NaCl) tolerance test
The potential LAB culture suspensions were inoculated into 10ml sterile MRS broth with 4%, 5%, and 6% NaCl concentrations. The suspensions were incubated at 28°C ± 2°C for 48h and the suspensions were observed by visual inspection and compared with the control. The broth with LAB cultures without NaCl acted as positive control while the broth with additional of NaCl but without LAB cultures acted as negative control. The growth of LAB in varying concentration of NaCl was examined using track plate and incubated at 28°C ± 2°C for 48h.23
Temperature sensitivity test
A single colony of each LAB cultures were inoculated into 10ml sterile MRS broth and incubated in a range of temperature 25°C ± 2°C to 40°C ± 2°C to observe the ability of LAB in different temperatures. The growth of LAB cultures was observed by repeating track plate method as mentioned beforehand to appoint isolates as temperature tolerant.23
In vitro screening of antagonistic activity of isolated lactic acid bacteria against Rsc
Pathogenic bacteria (Rsc) were acquired through the collection of microorganisms from Plant Pathology Laboratory, Horticulture Research Centre, MARDI. Rsc were cultured on the Kelman’s tetrazolium chloride (TZC) medium until the exponential phase was reached.7 Antagonistic activities between potential LAB and Rsc were tested through the agar well diffusion method. A maximum of eight wells with a diameter of 7mm were cut per plate using a 1mL sterile pipette tip to ensure there are enough spaces to observe the inhibition zone that produced by the LAB isolates. Antagonistic bacteria (LAB isolates) were cultured in MRS broth for overnight and 80 µL of the bacteria suspensions (approximately 109 CFU/mL) was added into each well.26 Sterile MRS broth and kanamycin (0.128 mg/mL) were used as negative and positive control, respectively. Each treatment was performed in three replicates. The plates were incubated at 28°C ± 2°C for 72h, after which the diameter of the growth inhibition zones surrounding the wells were measured. Colonies with inhibition zones and distinct morphologies were randomly selected and sub-cultured on MRS agar to acquire pure cultures for subsequent investigations.
Molecular identification of colony
16S rRNA gene sequencing analysis was performed by colony polymerase chain reaction (PCR) and the potential LAB isolates were amplified using the prokaryotic 16S ribosomal DNA universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1525R (5’- AAGGAGGTGATCCAGCCGCA-3’). Colony amplification was performed in a total volume 20 µL containing 10 µL of master mix (KOD ONETM PCR Master Mix- Blue), 0.6 µL of 10 mM of each primer, 1 µL of genomic DNA and 7.8 µL of distilled water. The PCR process was conducted as described by Khoo et al.27 with the initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 10s, annealing at 55°C for 5s, elongation at 72°C for 2s. A final extension at 72°C for 5min and 10°C for storage. The PCR products were then analyzed using 2% of agarose gel. Gel electrophoresis was conducted at 100 V for 30 min with the addition of Sybrsafe. The PCR products that were obtained in gel electrophoresis and compared with 1kbp plus DNA Ladder (ThermoFisher Scientific®). The products were then sent for DNA sequencing. Sequence data were edited and analyzed by Bioedit and sequence similarity searches were performed using the Basic Local Alignment Search Tool (BLAST) programs in the GenBank (NCBI). Phylogenetic tree was constructed by Neighbor-Joining method using ClustalW and MEGA Software as described by Ennahar et al.28 Grouping stability was calculated by 1000 bootstrap.
Sampling and isolation of lactic acid bacteria
Three isolates from kimchi (KC-1, KC-2, KC-3), grape (G-1, G-2, G-3) and fermented milk (FM-1, FM-2, FM-3) were obtained from serial dilution. Dilution of 10 to 108-fold were grown on MRS agar which is the selective media for LAB.
Morphological and biochemical characterization of LAB
Gram staining and catalase test acted as preliminary tests for the characterization of isolates to validate the identities of isolates as a LAB. pH tolerance test, NaCl tolerance test, and temperature sensitivity test served as tests for the properties of LAB isolates. Morphological characterizations including colony morphology and cell morphology were observed using naked eyes and under light microscope, respectively. Table 1 shows all the isolated colony morphologies were consistent with the features of round shape, slippery edge, convex elevation and yellowish white in colour. In respect to the cell morphology observation, LAB from kimchi and fermented milk isolates were bacilli with rod shape while from grape isolates cocci with spherical shape. Isolates from kimchi and fermented milk revealed there was no production of bubbles when added with H2O2 while, there was oxygen produced in all isolates from grape.
Table (1):
Morphological and biochemical characterization of LAB isolated from kimchi, grape and fermented milk
| Sample source | Isolate | Colony Morphology | Cell Morphology | Catalase Test | ||||
|---|---|---|---|---|---|---|---|---|
| Type | Color | Edge | Elevation | Cell Shape | Gram Staining | |||
| Kimchi | KC-1 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – |
| KC-2 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – | |
| KC-3 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – | |
| Grape | G-1 | Circle | Yellowish white | Smooth | Convex | Cocci (spherical) | Positive and negative | + |
| G-2 | Circle | Yellowish white | Smooth | Convex | Cocci (spherical) | Positive and negative | + | |
| G-3 | Circle | Yellowish white | Smooth | Convex | Cocci (spherical) | Positive and negative | + | |
| Fermented Milk | FM-1 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – |
| FM-2 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – | |
| FM-3 | Circle | Yellowish white | Smooth | Convex | Bacilli (rod) | Positive | – | |
+ production of oxygen; – without production of oxygen.
According to the preliminary tests, the results proved that isolates from kimchi and fermented milk have the characteristics of LAB while isolates from grape showed absence of LAB properties. Hence, only isolates from kimchi and fermented milk were further investigated. Table 2 shows the growth of the isolates in varying of growth conditions. At extreme pH, only one out of three kimchi isolates were able to withstand acidic conditions with low pH level. Instead, there were more fermented milk isolates (two out of three) were able to grow in the extreme pH level. Most of the isolates can withstand pH3.0 and pH4.0 but only KC-2 cannot withstand in all acidic conditions. With respect to salt tolerance, all isolates tolerated and grew at NaCl concentration of 4-6%. As for temperature sensitivity, fermented milk isolates (FM-1, FM-2, and FM3) were a-ble to tolerate at temperatures ranging from 25°C ± 2°C to 40°C ± 2°C, however kimchi isolates (KC-1, KC-2, and KC-3) were not able to tolerate in 35°C ± 2°C and 40°C ± 2°C.
Table (2):
pH tolerance, sodium chloride tolerance and temperature tolerance tests of potential LAB isolates
| Sample | Isolate | pH tolerance | Growth in (NaCl) | Growth at | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 3.0 | 4.0 | 4% | 5% | 6% | 25°C | 30°C | 35°C | 40°C | ||
| Kimchi | KC-1 | – | + | + | + | + | + | + | + | – | – |
| KC-2 | – | – | – | + | + | + | + | + | – | – | |
| KC-3 | + | + | + | + | + | + | + | + | – | – | |
| Fermented Milk | FM-1 | + | + | + | + | + | + | + | + | + | + |
| FM-2 | – | + | + | + | + | + | + | + | + | + | |
| FM-3 | + | + | + | + | + | + | + | + | + | + | |
+ indicates presence of growth, – indicates absence of growth.
In vitro screening of antagonistic activity of LAB against Rsc
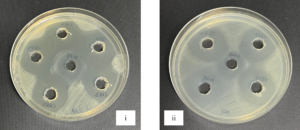
Antagonistic activities between LAB isolates and Rsc were observed through the inhibition zones that produced by LAB isolates as shown in Figure 1. From the normality test (p < 0.001), data was found not normally distributed. Kruskal-Wallis Test (N=35) was applied to test whether kimchi and fermented milk isolates were came from the same distribution. The result showed p < 0.001 among the samples and among the isolates which indicates kimchi isolates and fermented milk isolates were not originated from the same distribution. Table 3 shows the mean of triplicates and followed by standard deviation. The data were then analyzed using Kruskal-Wallis test as the data were not normally distributed. Kruskal-Wallis test showed the result of p < 0.001.
Table (3):
In-vitro antagonistic test between Rsc and isolates LAB via agar well diffusion method after 72 h
| Sample | Isolates | Diameter of inhibition zone (mm) | Diameter of control (mm) |
|---|---|---|---|
| Kimchi | KC-1 | 15.60 ± 0.40 | 23.40 ± 0.93 |
| KC-2 | 21.30 ± 0.24 | ||
| KC-3 | 15.00 ± 0.00 | ||
| Fermented Milk | FM-1 | 25.70 ± 0.37 | |
| FM-2 | 26.30 ± 0.49 | ||
| FM-3 | 28.70 ± 0.68 |
aThe diameter of the inhibition zone (mm) was measured as twice from the middle point of wells to the inhibition zone (the measurement of diameter = 2 times of radius). The values are the means of five replications. Data presented in the table indicates mean ± standard deviation.
Figure 1. Antagonistic activities between Rsc and isolates LAB from kimchi (i) and fermented milk (ii) through agar well diffusion method
Molecular characterization of the potential LAB isolates
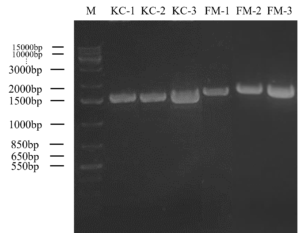
Gel electrophoresis in Figure 2 shows the result of the bands of each DNA fragment which indicates the molecular size of the DNA isolates. The DNA marker that used in this experiment was 1kbp plus DNA Ladder (ThermoFisher Scientific®) and act as a marker of molecular size of DNA fragments. The amplified genomic DNA of kimchi isolates (KC) were approximately 1500 bp, while fermented milk isolates (FM) were slightly higher than kimchi isolates (KC). The gene sequencing was then processed using BLAST. Kimchi isolates showed a similarity of 95% to 97% of identity with Lactiplantibacillus plantarum, whereas fermented milk isolates showed the similarity of 96% to 98% identity with the species of Lacticaseibacillus paracasei. The phylogenetic analysis conducted using neighbour joining method is shown in Figure 3. According to the phylogenetic tree that shows in Figure 3, each of the potential LAB were classified in different nodes and specifically in different clades. KC-1, KC-2, and KC-3 showed that three of the species are having the same clade as L. plantarum, whereas FM-1, FM-2 and FM-3 showed that three of the species are having the same clades as L. paracasei.
Figure 2. PCR amplification of kimchi (KC) and fermented milk (FM) isolates that produced inhibition zone against Rsc with 27F and 1525R primers. M: 1 kb plus DNA ladder
Figure 3. Phylogenetic trees based on Neighbour Joining method showing the relationship between isolates of L. plantarum and L. paracasei identified in the present study with other retrieved sequences from NCBI. The species details and NCBI accession number of each sequence were provided at the end of the node. Numbers above branches indicate the bootstrap values. The information about the species and the NCBI accession number of each sequence were presented at the end of the node. The bootstrap values are shown as numbers above the branches
In this study, a total of six LAB isolates obtained from different sources were characterized morphologically and tested with several biochemical assays. Morphological identification was conducted based on colony morphology and characteristics of cytology (including cell type and Gram staining). All isolates showed the same colony morphology which are circle, yellowish white, smooth edge and convex. In the respect of cell morphology, the shape of kimchi and fermented milk were observed as bacillus while grape isolates were observed as coccus. The shape of LAB consists of cocci or bacillus form which are circles and rods, respectively.29 For the Gram staining, kimchi and fermented milk isolates were stained in purple as Gram-positive bacteria while grape isolates stained with purple and pink. According to Tripathi & Sapra, this situation, called gram variable cells, might happen due to evaluation using old cultures as degradation of the peptidoglycan cell walls are susceptible to older culture.30 Consequently, the gram-positive cells show the characteristics as gram-negative or gram variable cells.
LAB is a group of heterogenous bacteria that share common characteristics such as gram-positive, non-spore-forming, non-respiring, and catalase negative bacteria.31, 32 By referring to Table 1, kimchi and fermented milk isolates do not produce air bubbles while grape isolates produce air bubbles when the colony was added with H2O2. The formation of air bubbles indicates the ability of LAB to produce catalase enzyme which is able to break down H2O2 to water (H2O) and oxygen (O2).25 In another word, LAB do not produce cytochromes or catalase enzyme as they are obligate anaerobes and thrive in the absence of oxygen.33 Thus, kimchi and fermented milk isolates were displayed as LAB characteristics while grape isolates do not. According to Khushboo et al., the findings in a study in isolating LAB from pickle, milk, curd and wheat dough found out that there are 18 isolates showed negative response to catalase among 60 isolates and all of these isolates were Gram-positive and did not produce spores.33
Biochemical test is an important test in order to identify the ability of LAB and its effectiveness. A low pH of medium is crucial in ascertaining LAB capability to survive in extremely high pH conditions. Referring to Table 2, KC-3, FM-1, and FM-3 were thrive in extreme condition even at pH 2, but KC-2 was not able to survive even at pH 4. KC-1 and FM-2 can only withstand up to pH 3 acidic conditions. G-Alegria et al. highlighted that these two LAB, L. plantarum and Oenococcus oeni from wine, showed a growth performance at pH 3.2 with the presence of 13% of ethanol at 18°C.34
According to Schillinger et al., the temperature range of LAB is highly varied and categories as mesophiles.31 They typically grow best between 20 and 40°C, reaching a maximum temperature of 50°C, while certain lactobacilli, such L. delbrueckii subsp. delbrueckii, may even thrive at 55°C. In the present study, KC-1, KC-2, and KC-3 were not able to grow well at 35°C and above while FM-1, FM-2 and FM-3 can survive. These results are supported by research which proved there is possibility of LAB such as Leuconostoc cremoris that is not able to grow at 40°C. L. cremoris which is a fermentative lactic acid bacterium that is predominantly found in industrial dairy fermentations.23,35
All the six isolates revealed remarkable salt tolerance even exposed to 6% of NaCl. The tolerance of all isolates to high NaCl concentration (4% to 6%) further supports the idea that they can endure challenging circumstances. These results are consistent with a previous study on screening potential LAB as probiotic using antagonistic.36 The authors demonstrated LAB isolates from curd could withstand 1-6% NaCl. LAB is also categorized as halophiles bacteria in which it is an extremophile that can survive in high salt concentrations. Yacinnkaya and Kilic reported that halophilic lactic acid bacteria (HLAB), particularly in fish products, which need sodium chloride to develop and can withstand high sodium chloride concentrations up to 32% of NaCl.37
One of the most important screenings criteria for lactic acid bacteria that are efficient and unique is their antimicrobial activity.36 The six LAB potential isolates were examined to consider their antimicrobial activity by modified agar-well diffusion method. The outcome of antagonistic effects of isolates were subjected against the pathogen Rsc. All six LAB potential isolates showed inhibition zone against the Rsc which is an indication that they possessed varying abilities to exert antimicrobial effects on pathogens.23 This result revealed that all six isolates exhibited the inhibition zones from the lowest 15.6 mm to the highest 28.7 mm (Table 3). The current result showed a consistent result in the previous study as they assessed the antimicrobial reaction of L. acidophilus and L. delbrueckii strains compared to other isolated bacteria including Escherichia coli, Staphylococcus aureus, Bacillus cereus, and Salmonella typhimurium.33 Their findings according to the inhibition zones that produced by CM1 and OS1 stains indicated that these strains had more antibacterial activity when compared to other bacteria, suggesting that they could be useful as probiotics. Kruskal-Wallis test’s result shown has rejected the null hypothesis which are “the distribution of diameter of inhibition zone (mm) is the same across categories of sample” and “the distribution of diameter of inhibition zone (mm) is the same across categories of isolates” as both tests showed p < 0.001 which is not greater than 0.05, thus there is different between the different categories. Sample indicates six different isolates while categories of isolates indicate kimchi and fermented milk. In comparison to kimchi isolates, fermented milk isolates had a greater inhibition against Rsc.
Amplified DNA size of 1500bp (Figure 2) was observed in all 6 isolates and those isolates were proceeded to nucleotide sequencing. Six isolates were edited using Bioedit software packaging, and the consensus sequences obtained were blasted in NCBI. By using the reference strains in NCBI, the BLAST findings showed that kimchi isolates were matched with L. plantarum while fermented milk isolates were matched with L. paracasei and recognized as LAB. The analysis of the genomes of the six isolates showed that all of them have at least 95% and above similarities to Lactiplantibacillus and Lacticaseibacillus, respectively. These results are consistent with the study conducted by Tilahun et al. in which the tested 10 LAB were obtained only 92% to 98% similarity to GenBank of NCBI. 10 bacterial isolates were found to belong to the genera Lactobacillus, Enterococcus, and genus Bacillus.38
A phylogenetic tree constructed from the alignment of the six isolates sequences and the representative sequences from NCBI database were used to create a Neighbour Joining phylogenetic tree in MegaX. According to the phylogenetic categorization, strains with comparable sequences were clustered together and probably considered of as close relatives.38 Neighbour Joining phylogenetic tree based on the isolated strains and their closest possible and related species. Bootstrap values calculated for 100 replications.
This study has potential limitations. The issue happened on grape isolates as observed as Gram-variable can be troubleshooting as grape has huge diversity of LAB as compared to other fruits.39 PCR amplification can be done after a proper DNA extraction and purification to avoid unconfirmed sequence bases in International Union of Pure and Applied Chemistry (IUPAC) codes such as K as Keto group indicates either G or T/U, M as aMino group indicates A or C, R as purine indicates A or G, and Y as pYrimidine indicates C or T/U.40 This study is also extendable to obtain the antimicrobial substance of LAB isolates which shows the compatibility of the isolates against Rsc.
Gram-positive and catalase-negative bacteria were indicates in the preliminary screening of unknown isolates. Biochemical testings were conducted for each isolate to examine the ability of the LAB to grow in different extreme conditions. Throughout the study, KC-1, KC-2 and KC-3 were identified as L. plantarum while FM-1, FM-2 and FM3 were identified as L. paracasei. All isolates showed at least 95% of the percentage of identities to the species. Based on the antagonistic activity between isolated LAB and Rsc in agar well diffusion, both L. plantarum and L. paracasei were able to inhibit Rsc by showing the highest inhibition zones which are 21.30 mm and 28.70 mm, respectively. Therefore, this study concludes that L. plantarum and L. paracasei were able to control the in vitro growth of Rsc.
ACKNOWLEDGMENTS
The authors would like to thank Universiti Malaysia Sabah for administrative and technical support throughout the research. The authors are also thankful to the financial support from the Ministry of Higher Education (MoHE), Malaysia, under Fundamental Research Grant Scheme (FRGS) and UMSgreat from Universiti Malaysia Sabah.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was financially supported by the Ministry of Higher Education (MoHE), Malaysia, under Fundamental Research Grant Scheme (FRGS) FRGS/1/2022/WAB04/UMS/01/1 and Universiti Malaysia Sabah under UMSGreat GUG0610-1/2023.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Onsang N, Pebrian D, Anggraini F. Sustainability of Malaysian smallholder banana farming: an energy efficiency use-based audit. AgriEngInt: CIGR Journal Open. 2023;25(1):111-122.
- Food and Agriculture Organization of United Nations.. Production/ yield quantities of bananas in Malaysia from 1994 to 2021. 2022. https://www.fao.org/faostat/en/#data/QCL/visualize].
- Ray JD, Subandiyah S, Rincon-Florez VA, et al. Geographic Expansion of Banana Blood Disease in Southeast Asia. Plant Dis. 2021;105(10):2792-2800.
Crossref - Teng S-K, Aziz NAA, Mustafa M, et al. The Occurrence of Blood Disease of Banana in Selangor, Malaysia. Int J Agric Biol. 2016;18(1):92-97.
Crossref - Food and Agriculture Organization of United Nations (n.d. Crops and livestock products. https://www.fao.org/faostat/en/#data/QCL/visualize
- Tan BC. Can Banana be a Success Story for Malaysia? Journal of Agribusiness Marketing. 2022;9(1):13-22.
Crossref - Talib MAA, Azeme NFN, Shafie KA, et al. Lateral Flow Immunoassay Detection of Ralstonia syzygii subsp. celebensis Causing Banana Blood Disease in Symptomatic and Asymptomatic Banana Plants. J Trop Plant Physiol. 2021;13(1):12.
Crossref - Safni I, Subandiyah S, Fegan M. Ecology, Epidemiology and Disease Management of Ralstonia syzygii in Indonesia. Front Microbiol. 2018;9:419.
Crossref - Gaumann E. Investigations on the blood-disease of bananas in Celebes I. Rev Appl Mycol. 1921;1:225-227.
- Gaumann E. Investigations on the blood disease of banana in Celebes II. Rev. Appl. Mycol. 1924;1:344-346.
Crossref - Ingel B, Caldwell D, Duong F, et al. Revisiting the Source of Wilt Symptoms: X-Ray Microcomputed Tomography Provides Direct Evidence That Ralstonia Biomass Clogs Xylem Vessels. PhytoFrontiersTM. 2022;2(1):41-51.
Crossref - de Pedro-Jove, R, Puigvert M, Sebastia P, Macho AP, et al. Dynamic expression of Ralstonia solanacearum virulence factors and metabolism-controlling genes during plant infection. BMC Genomics. 2021;22(1):170.
Crossref - Yuliar, Nion YA, Toyota K. Recent Trends in Control Methods for Bacterial Wilt Diseases Caused by Ralstonia solanacearum. Microbes Environ. 2015;30(1):1-11.
Crossref - Edwards-Jones G. Do benefits accrue to ‘pest control’ or ‘pesticides?’: A comment on Cooper and Dobson. Crop Protection. 2008;27(6):965-967.
Crossref - Verschuere L, Rombaut G, Sorgeloos P, Verstraete W. Probiotic Bacteria as Biological Control Agents in Aquaculture. Microbiol Mol Biol Rev. 2000;64(4):655-671.
Crossref - Steglinska A, Koltuniak A, Motyl I, et al. Lactic Acid Bacteria as Biocontrol Agents against Potato (Solanum tuberosum L.) Pathogens. Appl Sci. 2022;12(15):7763.
Crossref - Ariyapitipun T, Mustapha A, Clarke AD. Microbial Shelf Life Determination of Vacuum-Packaged Fresh Beef Treated with Polylactic Acid, Lactic Acid, and Nisin Solutions. J Food Protec. 1999;62(8):913-920.
Crossref - El-Mabrok A, Hassan Z, Mokhtar A, Hussain K. Screening of lactic acid bacteria as biocontrol against (Colletotrichum capsica) on chili Bangi. Res J Appl Sci. 2021;7(9):466-473.
- Lutz M, Michel V, Martinez C, Camps C. Lactic acid bacteria as biocontrol agents of soil-borne pathogens. Biological control of fungal and bacterial plant pathogens IOBC-WPRS Bull, 2012;78:285-288.
- Axel C, Coffey EZA, Guo J, Waters DM, Arendt EK. Ecofriendly control of potato late blight causative agent and the potential role of lactic acid bacteria: a review. Appl Microbiol Biotechnol. 2012;96(1):37-48.
Crossref - Hamed H, Moustafa Y, Abdel-Aziz S. In vivo efficacy of lactic acid bacteria in biological control against Fusarium oxysporum for protection of tomato plant. Life Sci J. 2011;8(4):462-468.
- Chen Y, Wu H, Yanagida F. Isolation and characteristics of lactic acid bacteria isolated from ripe mulberries in Taiwan. Braz J Microbiol. 2010;41(4):916-921.
Crossref - Ayo-Omogie H, Okorie E. In vitro Probiotic Potential of Autochthonous Lactic Acid Bacteria and Microbiology of Kunu Made from Mixed Grains. Br Microbiol Res J. 2016;14(4):1-10.
Crossref - Resti F, Hartanto I. Isolation and characterization of Lactic Acid Bacteria (Lactobacillus sp) from strawberry (Fragaria vesca). J Phys: Conf Ser. 2019;1317:012086.
Crossref - Ismail YS, Yulvizar C, Mazhitov B. Characterization of lactic acid bacteria from local cow’s milk kefir. IOP Conf Ser: Earth Environ Sci. 2018;130:012019.
Crossref - Taha MDM, Jaini MFM, Saidi NB, Rahim RA, Shah UKM, Hashim AM. Biological control of Erwinia mallotivora, the causal agent of papaya dieback disease by indigenous seed-borne endophytic lactic acid bacteria consortium. PLOS ONE. 2019;14(12):e0224431.
Crossref - Khoo YW, Teng TH, Khaw YS, Li SF, Chong KP. First Report of Lasiodiplodia theobromae Causing Leaf Spot on Aloe vera var. chinensis in Malaysia. Plant Dis. 2022;106 (8):2258.
Crossref - Ennahar S, Cai Y, Fujita Y. Phylogenetic Diversity of Lactic Acid Bacteria Associated with Paddy Rice Silage as Determined by 16S Ribosomal DNA Analysis. Appl Environ Microbiol. 2003;69(1):444-451.
Crossref - Bjorkroth KJ, Koort J. Lactic acid bacteria: Taxonomy and Biodiversity. Encyclopedia of Dairy Sciences. 2011:45-48.
Crossref - Tripathi N, Sapra A. Gram Staining. StatPearls – NCBI Bookshelf. 2023. https://www.ncbi.nlm.nih.gov/books/NBK562156/
- Schillinger U, Holzapfel W, Bjorkroth KJ. Lactic acid bacteria. Food Spoilage Microorganisms. 2006:541-578.
Crossref - Sabatini N. A Comparison of the Volatile Compounds, in Spanish-style, Greek-style and Castelvetrano-style Green Olives of the Nocellara del Belice Cultivar: Alcohols, Aldehydes, Ketones, Esters and Acids. Olives and Olive Oil in Health and Disease Prevention. 2010:219-231.
Crossref - Khushboo, Karnwal A, Malik T. Characterization and selection of probiotic lactic acid bacteria from different dietary sources for development of functional foods. Front Microbiol. 2023;14:1-14.
Crossref - G-Alegrִia E, Lopez I, Ruiz JI, et al. High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett. 2004;230(1):53-61.
Crossref - Ozcan E, Selvi SS, Nikerel E, Teusink B, Oner ET, Cakyr T. A genome-scale metabolic network of the aroma bacterium Leuconostoc mesenteroides subsp. cremoris. Appl Microbiol Biotechnol. 2019;103(7):3153-65.
Crossref - Prabhurajeshwar C, Chandrakanth RK. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed J. 2017;40(5):270-83.
Crossref - Yalcikaya S, Kilic GB. Isolation, identification and determination of technological properties of the halophilic lactic acid bacteria isolated from table olives. J Food Sci Technol. 2019;56(4):2027-37.
Crossref - Tilahun B, Tesfaye A, Muleta D, Bahiru A, Terefework Z, Wessel G. Isolation and Molecular Identification of Lactic Acid Bacteria Using 16s rRNA Genes from Fermented Teff (Eragrostis tef (Zucc.)) Dough. Int J Food Sci. 2018;1-7.
Crossref - Bae S, Fleet GH, Heard GM. Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol. 2006;100(4):712-27.
Crossref - Johnson AD. An extended IUPAC nomenclature code for polymorphic nucleic acids. Bioinformatics. 2010;26(10):1386-89.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.