ISSN: 0973-7510
E-ISSN: 2581-690X
The increased prevalence of Helicobacter pylori infection and inadequate genetic research on the same, demands a genetic study among Tamil population of South India to unravel the association of NLRP3 (NLR family pyrin domain containing 3) variants with persistent H. pylori infection. This research was aimed to study the correlation between persistent H. pylori infection and influence of such genetic variants in the development of disease progression. In this study, 200 healthy volunteers and 120 H. pylori-positive cases were screened for two NLRP3 variants, rs74163773 and rs10754558, using allele-specific PCR and TaqManTM SNP genotyping assay, respectively. Based on our genotype and allelic distribution, rs74163773 variant did not associate with the risk of developing the persistent infection. However, a significant association of heterozygous CG variant of rs10754558 with patient showing clinical symptoms of gastritis, PUD and persistent infection in the over-dominant, co-dominant and recessive models was found. Our findings suggest that persistent H. pylori infection susceptibility was influenced by genetic variant rs10754558 and its heterozygous CG variant can serve as an independent risk factor in the manifestation of chronic gastritis and PUD in the South Indian Tamils.
NLRP3, SNV, PUD, Gastritis, H. pylori
Gastric cancer (GC) ranks third in contributing to cancer-related deaths globally and remains the most fatal with a higher incidence rate reported from parts of Central and Southern America, Eastern Europe, and Central and East Asia. About 1.1 million new GC cases were reported in 2020 and it is anticipated to increase by 76% globally by 2030.1,2 GC is rapidly increasing in epidemic proportions across the Indian subcontinent with a projected incidence of approximately 12,000 new cases by 2025 and with a 27% survival rate over five years.3,4 According to the data provided by the National Cancer Registry Program 2020, the north-eastern states of India have experienced a higher incidence of GC, and the incidence rate (age-adjusted) was higher in men than women, owing to the poor molecular understanding of GC progression and lack of early detection and diagnosis.5
H. pylori, a helical-shaped, Gram-negative flagellated bacterium found in the pyloric antrum, is the most recognized etiological risk factor of GC. Its prevalence rate lies between 50-83% and infects the susceptible in their early twenties. Various epidemiological, clinical and preclinical studies strongly suggest that H. pylori cause gastric carcinoma,6 and thus this bacterium is recognized as a class I carcinogenic pathogen in humans, while the genetic and environmental factors were established to be other key risk factors.7
NLR (NOD like receptor) belongs to the family of pattern recognition receptors and are among the important entities of the non-specific immune system in identifying the PAMPs (Pathogen-associated molecular pattern). NLRP3 (NLRP3; genetic locus: 1q44), a subfamily of NLRs, takes part in inflammatory diseases. It forms a multimeric protein complex (inflammasome) that recruits an intracellular cysteine protease known as caspase-1 (CASP1) which results in IL-1b (Interleukin-1b) and IL-18 (Interleukin-18) production, along with the initiation of pyronecrosis and pyroptosis to protect against infection.8 Activation of IL-1ג and IL-18 expression by the NLRP3-CASP1 signalling pathway during persistent H. pylori infection, promotes gastric inflammation and carcinogenesis. Genetic variations that compromise the functions of NLRP3, alter the secretion of these cytokines and are said to affect the immune homeostasis, thereby favouring the H. pylori to thrive, including the augmentation of downstream processes in GC development. Also, persistent infection and immune tolerance lead to GC by over-expression of cyclooxygenase-2 (COX-2) and repression of gastric acid secretion.9
Several studies on NLRP3 polymorphisms have contributed valuable insight into its role in inflammatory disorders like type-1 diabetes mellitus, celiac disease,10 Alzheimer’s disease (AD),11 multiple sclerosis12 and cervical cancer13 other than GC.14 Though less genetic studies are available concerning the relationship between H. pylori and NLR signalling cascade, studies on H. pylori-infected human gastric cells have confirmed that lipopolysaccharides of the bacteria enhance CASP1, IL-1b and IL-18 expression in-vitro.15,16 Moreover, an in-vivo experiment with the NLRP3-knockout mice model showed reduced release of TNF-a, IL-1b and IL-10 due to down-regulated immune response, which made the animals prone to infection leading to ulcerative colitis.17 Conversely, the genetic association between a novel intron variant of NLRP3 (rs74163773) and the vaginal infection caused by Candida sp., namely recurrent vulvovaginal candidiasis (RVVC) leads to over-activation of IL-1b,18 and progression towards GC.19 Also, recent findings have linked the development of H. pylori-induced GC to NLRP3 variants, rs10754558 and rs4612666 in the Chinese Han population.20
Though NLRP3 showed a stronger correlation with immune filtration and serves as prognostic biomarker of GC immune response against H. pylori,21,22 the underlying mechanism remains obscure in the South Indian population. Therefore, we aimed to analyze the influence of NLRP3 genetic variants (rs10754558 and rs74163773) in developing persistent infection caused by H. pylori in the South Indian population. We believe that this study may be a prime approach to throw light on establishing a population-specific biomarker that can be extrapolated to one’s increased risk of persistent H. pylori infection, chronic gastritis, and/or PUD.
Study subjects and screening for H. pylori infection
All the study subjects have given their consent for sample collection and the institutional Ethical Committee of Rajiv Gandhi Medical College and Hospital Chennai, India, has approved the study (EC Reg. No. ECR/270/Inst./TN/2013). Healthy and willing volunteers of South Indian Tamil ethnicity (n = 200) who were devoid of gastrointestinal complications, autoimmune diseases and diabetes mellitus were included. The study subjects were screened for H. pylori infection using GastroPanel® H. pylori ELISA kit (#606040; Biohit Healthcare, Finland) to check for IgG level against the bacteria (HpIgG).23 The cut-off value of >30 U/mL was considered positive for active H. pylori infection.24
Patients with the complaint of discomfort or pain in the upper abdomen were recruited from the Department of Medical Gastroenterology, MMC, Chennai, from December 2018 to October 2019. The gastric tissue samples from the pyloric antrum region were collected and rapid urease test kit (RUT DRY Test Kit, Gastro Cure Systems, Kolkata, India) was used to screen the samples for H. pylori infection. The samples tested positive for RUT and also the subjects who had not been administered any anti-inflammatory drugs, non-steroidal drugs, proton pump inhibitors, and antimicrobial drugs were considered as H. pylori-positive cases (n=120).25 The relevant pathological data and clinical hallmarks of all the participants in this study were obtained along with informed written consent.
Genomic DNA isolation and quantification
Salting-out method was used for genomic DNA isolation.26 DNA quality and purity were confirmed by gel electrophoresis using 0.8% agarose gel and the UV absorbance 260/280 ratio has ranged consistently between 1.6 and 1.8, indicating excellent deproteinization. The resultant DNA was sufficient to carry out further PCR reactions and stored at -20°C until use.
Genotyping of NLRP3 variants
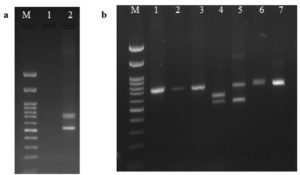
Genotyping of VNTR rs74163773 was performed using the allele-specific PCR method. The region harbouring the 42 bp VNTR in the intron of NLRP3 was amplified using the primers described by Omi et al. (Table 1).27 Template genomic DNA of 1 µL (50 ng) was used in a reaction volume of 25 µL with 0.2 µL of 5 U/µL Taq DNA polymerase (#MBT060; HiMedia Labs, India), 0.5 µL of each primer (5 pmol/µL; Bioserve, India), 1 µL of dNTP mix (25 mM each) (#MBT058; HiMedia Labs, India), 2.5 µL of 10X reaction buffer (20 mM MgCl2) (#MBT060; HiMedia Labs, India) and rest nuclease-free water.27 The thermocycler was set at 95°C for 5 minutes for initial denaturation, followed by denaturation at 95°C for 30 seconds, annealing/extension at 49°C for 30 seconds for 35 cycles, and the final extension at 72°C for 10 minutes.27 The genotype patterns 12/13, 12/12, 12/9, 12/7, 7/9, and 7/7 as reported by Omi et al were followed,27 and the PCR products of the major allele homozygote (12-12 genotype: 720 bp – 12 repeats in both alleles), minor allele homozygote (7-9 genotype: 510 bp – 7 repeats in allele 7 and 594 bp – 9 repeats in allele 9; 7-7 genotype: 510 bp – 7 repeats in both alleles) and heterozygotes with both major and minor alleles (12-13 genotype: 720 bp – 12 repeats in allele 12 and 760 bp – 13 repeats in allele 13; 12-9 genotype: 720 bp – 12 repeats in allele 12 and 594 bp – 9 repeats in allele 9; 12-7 genotype: 720 bp – 12 repeats in allele 12 and 510 bp – 7 repeats in allele 7) were analyzed. The amplicons of the genotypes of rs74163773 were separated on a 1.3% ethidium bromide-stained agarose gel and then photographed and analyzed (Figure 1a and 1b).
Figure 1(a-b). Electrophoresis pattern of rs74163773 genotyping by allele-specific PCR in 1.3% agarose gel
M: 100 bp ladder (#N3231S; New England Biolabs® Inc., India); (a) Lane 2: Novel genotype (X-X) (510 bp/720 bp/762 bp); (b) Lane 1-3,7: 12/12 genotype (720 bp/720 bp); Lane 4: 7/9 genotype (510 bp/594 bp); Lane 5:12/7 genotype (720 bp/510 bp); Lane 6: 12/13 genotype (720 bp/762 bp)
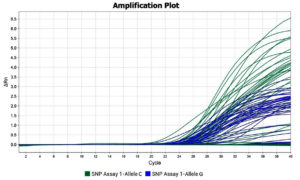
The rs10754558 (C > G) in the 3’-UTR region was screened using the TaqManTM allelic discrimination assay (assay ID: C_26052028_10) (#4351379; Applied Biosystems, Carlsbad, USA). The TaqManTM Genotyping Master Mix (#4371353; Applied Biosystems, Carlsbad, USA), containing the AmpliTaqGoldTM DNA Polymerase, was used to amplify the genomic DNA in a real-time PCR machine (Applied BiosystemsQuantStudio 3) with the help of sequence-specific primers. The reaction mixture consisted of 20 ng of diluted genomic DNA with 5 µL of TaqManTM Genotyping Master Mix, 0.5 µL of TaqManTM Genotyping Assay Mix and the reaction volume was set to 10 µL with nuclease-free water. The thermocycler conditions were set at 95°C for 10 minutes for initial denaturation, followed by denaturation step at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute for 40 cycles in 96 wells in the real-time PCR machine. The amplification of each allele by TaqManTM probes resulted in the production of fluorescent signals which were detected by the sequence detection system and their normalized differences (ΔRn) were plotted against the amplification cycle for both the alleles (Figure 2). The representative amplification plot indicating the allele-specific distribution is shown in Figure 3. The fluorescent probe used for the detection of allele C was fluorescein amidite (FAMTM)-labelled probes and that for allele G was the Victoria (VIC®)-labelled probe (Table 1).
Table (1):
Primer and probe sequences of NLRP3 polymorphisms
Gene |
Genotyping Method |
Primer/Probe sequence |
|---|---|---|
NLRP3 (rs74163773) |
Allele-specific PCR |
Forward primer (5’→3’): CTGACCTCCCAATGTGCCTT Reverse primer (5’→3’): CAGAGCTTCTTCAGATTGCA |
NLRP3 (rs10754558) |
TaqManTM allelic discrimination assay |
VIC®-labelled probe: GACAATGACAGCATCGGGTGTTGTT[C]TCATCACAGCGCCTCAGTTAGAGGA FAMTM-labelled probe: GACAATGACAGCATCGGGTGTTGTT[G]TCATCACAGCGCCTCAGTTAGAGGA |
Figure 2. Representative amplification plot for rs10754558 SNV in control and H. pylori cases using real-time PCR
Probe for allele G was labelled with VIC® and that for allele C with FAMTM
DRn -Normalized difference
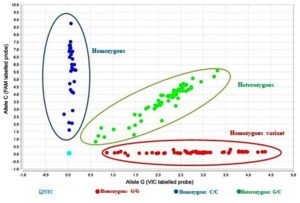
Figure 3. Scatter plot of TaqManTM allelic discrimination of rs10754558
The homozygous wild genotype (CC) is shown in blue. The homozygous variant genotype (GG) is shown in red. The heterozygous genotype (CG) is shown in green. NTC: No template control
Statistical analysis
t-test and chi-square (χ2) test was performed for the analysis and the association between genotypes of NLRP3 and H. pylori cases using SPSS® software. Allelic and genotype frequencies were calculated. Any deviation from HWE (p < 0.05 was found to be statistically significant) for genotype distribution between the study subjects was determined.
STRING analysis
STRING analysis was done with human NLPR3 with database version 11 on standard settings.28 Gene-Gene interaction was obtained with String analysis. NLPR3 gene served as an input, and PPI network was obtained and analyzed based on the scientific evidence linked with STRING.
Our control group included 115 males and 85 females, and the H. pylori-positive cases comprised 78 male subjects and 42 female subjects. The population statistical data and the clinical features of the study subjects are shown in Table 2. All of them were genotyped for rs74163773 and rs10754558. Table 3 depicts the genotypic distribution and allele frequencies of these variants. The screening of 42 bp VNTR of rs74163773 had resulted in six genotypes (12/12, 12/13, 12/9, 12/7, 7/7, and 7/9), where 12/12 genotype was found to be the most prevalent genotype among others. Interestingly, we observed a novel genotype (X-X) in our study population and found that it has not been reported in any other population. No further validation of this novel genotype was done as it is less frequent in our population. The genotype frequency of rs74163773 showed deviation from HWE in both control and H. pylori cases. This disequilibrium could be due to the rare occurrence of the majority of the genotypes of NLRP3 (rs74163773) in our population. Also, rs74163773 has failed to show significant risk in the development of persistent infection and its associated clinical manifestations (data not shown).
Table (2):
Clinical Characteristics of Healthy control and H. pylori positive cases
| Particulars | Healthy Controls (n=200) (%) | H. pylori +ve Cases (n=120) (%) |
|---|---|---|
| Mean age (years) ± SD Median age (years) >50 <50 |
46.62 ± 15.26 49 95 (47.5) 105 (52.5) |
53.45 ± 18.04 57 77 (64.2) 43 (35.8) |
| Gender | ||
| Male Female |
115 (57.5) 85 (42.5) |
78 (65) 42 (35) |
| Persistent gastritis Peptic Ulcer Disease |
Nil Nil |
80 (66.7) 40 (33.3) |
| Smoking | ||
| Yes No |
43 (21.5) 157 (78.5) |
31 (25.8) 89 (74.2) |
| History of alcohol consumption | ||
| Yes No |
11 (5.5) 189 (94.5) |
18 (15) 102 (85) |
The minor allele frequency (MAF) reported for rs10754558 was found to be 0.54, which was in agreement with the reported number for the Asian population (MAFAsia=0.5).29 No deviation from HWE was observed for this variant in our control and case population. Also, we discovered a significant association between rs10754558 with the risk of persistent H. pylori infection in the co-dominant model (p = 0.006) and recessive model (p = 0.008) (Table 3). Further, we observed that the H. pylori-infected individuals harbouring the heterozygous CG variant have a decreased risk of developing persistent gastritis (p = 0.048) and PUD (p = 0.039) (Table 4).
Table (3):
Genotype and allele frequencies of NLRP3 variants (rs74163773 and rs10754558) in H. pylori-positive cases
| Genes | Genotypes/allele frequency | Controls (n=200) (%) | H. pylori-positive cases (n=120) (%) | ap-value | #OR | 95% #CI |
|---|---|---|---|---|---|---|
| NLRP3 (rs74163773) | 12/12 genotype | 154 (77) | 83 (69) | – | Reference | – |
| 12/13 genotype | 27 (14) | 15 (12.5) | 0.93 | 1.03 | 0.52-2.04 | |
| 12/9 genotype | 8 (4) | 3 (2.5) | 0.83 | 0.69 | 0.17-2.69 | |
| 7/7 genotype | 2 (1) | 5 (4) | 0.11 | 4.63 | 0.88-24.44 | |
| Novel genotype (X-X) | 6 (3) | 8 (7) | 0.16 | 2.47 | 0.83-7.37 | |
| 12/7 genotype | 3 (1) | 5 (4) | 0.22 | 3.09 | 0.72-13.26 | |
| 7/9 genotype | 0 | 1 (1) | 0.75 | 5.55 | 0.22-137.88 | |
| NLRP3 (rs10754558) (C>G) | CC (Homozygote reference) | 38 (19) | 22 (18) | – | – | – |
| CG (Heterozygote) | 107 (53.5) | 47 (39) | 0.48 | 0.76 | 0.41-1.42 | |
| GG (Homozygote variant) | 55 (27.5) | 51 (43) | 0.20 | 1.6 | 0.84-3.06 | |
| C | 183 (46) | 91 (38) | – | – | – | |
| G | 217 (54) | 149 (62) | 0.06 | 1.38 | 1.00-1.91 | |
| Dominant model | CC vs. CG+GG | – | – | 1.00 | 1.04 | 0.58-1.87 |
| Co-dominant model | CG vs. GG | – | – | 0.006 | 2.11 | 1.26-3.52 |
| CC vs. GG | – | – | 0.20 | 1.60 | 0.84-3.06 | |
| CC vs. CG | – | – | 0.48 | 0.76 | 0.41-1.42 | |
| Recessive model | CG+CC vs. GG | – | – | 0.008 | 1.95 | 1.21-3.14 |
| G vs. C | – | – | – | 0.06 | 0.72 | 0.52-1.00 |
*Significant results are in bold font.
aχ2 analysis, p-value<0.05 was considered to be significant.
HWE for controls and cases for rs74163773 are in disequilibrium. p<0.05.
χ2=7.03, df =2, p =0.02 for genotype frequencies in cases vs. control.
HWE for controls and cases for rs10754558 are in equilibrium. p>0.05.
χ2=8.31, df =2, p =0.015 for genotype frequencies in cases vs. control.
χ2=3.45, df =1, p =0.06 for allele frequencies in cases vs. control.
# OR-Odds Ratio; CI- Confidence Interval
Table (4):
Influence of NLRP3 variant (rs10754558) with clinical manifestations of H. pylori infection
| Gene | Genotypes/ allele frequency | Controls (n=200) (%) | Chronic gastritis (n= 80) (%) | #OR (95% #CI)ap-value | PUD (n=40) (%) | #OR (95% #CI)ap-value |
|---|---|---|---|---|---|---|
| NLRP3 (rs10754558) (C>G) | CC | 38 (19) | 23 (28.3) | Reference | 13 (32.5) | Reference |
| CG | 107 (53.5) | 32 (40.5) | 0.49 (0.25-0.94) 0.048 | 14 (35) | 0.38 (0.16-0.88) 0.039 | |
| GG | 55 (27.5) | 25 (31.2) | 0.75 (0.37-1.51) 0.53 | 13 (32.5) | 0.69 (0.28-0.65) 0.54 | |
| C | 183 (46) | 78 (48.7) | – | 40 (50) | – | |
| G | 217 (54) | 82 (51.3) | 1.12 (0.78-1.62) 0.58 | 40 (50) | 1.18 (0.73-1.91) 0.56 |
*Significant results are in bold font.
aχ2 analysis, p <0.05 was considered to be significant.
HWE for controls and chronic gastritis for rs10754558 are in equilibrium. p>0.05.
χ2=4.87, df =2, p=0.09 for genotype frequencies in control vs. chronic gastritis.
χ2=0.30, df =1, p=0.58 for allele frequencies in control vs. chronic gastritis.
HWE for controls and PUD for rs10754558 are in equilibrium. p>0.05.
χ2=5.42, df =2, p=0.07 for genotype frequencies in control vs. PUD.
χ2=0.33, df =1, p=0.57 for allele frequencies in control vs. PUD.
# OR-Odds Ratio; CI- Confidence Interval
Among the cases, rs10754558 was observed to show significant risk in the progression of clinical phenotypes such as chronic gastritis and PUD in addition to a predisposition to persistent H. pylori infection. The following different models of inheritance of rs10754558 were associated significantly with the persistent H. pylori infection development [over-dominant model(p = 0.01)], chronic gastritis [co-dominant model (p = 0.03); over-dominant model (p = 0.04)] and PUD [co-dominant model (p = 0.02); over-dominant model (p = 0.03)] (Table 5). The above data shows that the heterozygous variant of NLRP3 (rs10754558) to be strongly associated as an independent risk factor in the progression of clinical manifestations like chronic gastritis and PUD. Thus, our findings suggest that rs10754558 influences significant development of clinical manifestations of H. pylori infection, chronic gastritis, and PUD in our population.
Table (5):
Odds Ratio (OR) calculated in different models of inheritance of NLRP3 variant (rs10754558) in H. pylori-positive cases and with clinical manifestations of H. pylori infection
| Genetic model | Genotypes/ allele frequency | H. pylori-positive cases | Chronic gastritis | PUD | |||
|---|---|---|---|---|---|---|---|
| ap-value | #OR(95% #CI) | ap-value | #OR(95% #CI) | ap-value | #OR(95% #CI) | ||
| Dominant model | CC | – | Reference | – | Reference | – | Reference |
| CG+GG | 0.88 | 0.96 (0.53-1.71) | 0.07 | 1.72 (0.94-3.13) | 0.06 | 2.05 (0.97-4.35) | |
| Co-dominant model | CC vs. CG | 0.38 | 1.32 (0.70-2.47) | 0.03 | 2.02 (1.06-3.88) | 0.02 | 2.61 (1.13-6.06) |
| CC vs. GG | 0.15 | 0.62 (0.33-1.19) | 0.42 | 1.33 (0.66-2.68) | 0.41 | 1.45 (0.30-3.46) | |
| Recessive model | CG+CC | – | Reference | – | Reference | – | Reference |
| GG | 0.006 | 0.51 (0.32-0.83) | 0.53 | 0.83 (0.47-1.47) | 0.52 | 0.79 (0.38-1.64) | |
| Over-dominant model | CC+GG | – | Reference | – | Reference | – | Reference |
| CG | 0.01 | 1.79 (1.13-2.83) | 0.04 | 1.73 (1.02-2.92) | 0.03 | 2.14 (1.05-4.33) | |
*Significant results are highlighted in bold font.
aχ2 analysis, p<0.05 was considered to be significant.
# OR-Odds Ratio; CI- Confidence Interval
STRING analysis
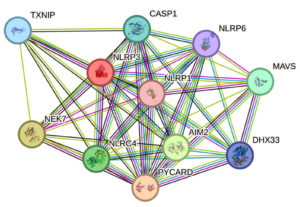
In this study, a functional interaction is defined as a direct interaction between the query protein and other proteins found in the same KEGG pathway. PPI network was obtained with human NLRP3 as a search query (Figure 4). The genes which plays a major role in NLRP3 inflammasome complex are shown in Figure 4. NLPR plays a vital role in innate immunity and inflammation against bacterial toxin, viral DNA, Bacterial DNA or Ca2+. When a pathogen or its associated signals sensed, NLPR initiates the formation of inflammasome complex comprising of NLPR, ASC and pro-CASP1. The pro-CASP1 then activates CASP1 there by activating IL-18 and IL-1b gene. The activated NLPR inflammasome complex leads to the maturation and production of IL-1β and IL-18 cytokines.
Figure 4. STRING Protein-Protein Interaction Network
NLPR3 gene served as an input. Distinct functional clusters were determined. All of the proteins plays an important role in Inflammasome pathway
Evidence suggests that, genetic variants in the NLPR3 gene might increase the expression of components of the NLRP3–IL-1b signalling pathway thereby leading to overexpression of pro-inflammatory cytokines and it’s secretion. The increased serum IL-1b further activates NLPR3, and the latter promotes the infiltration of neutrophils. Gastric acid secretion decreases due to local neutrophil infiltration leading to long-term survival of H. pylori pathogen in the gastric mucosa there by leading to Gastritis and Gastric cancer.30,31
Gastric cancer poses a serious global health problem and challenges researchers due to its high morbidity and mortality rates. A recent study have shown that therapeutic approaches with NLRP3 would be imperative in treating GC by promoting cancer by pyroptosis, a form of antimicrobial response that occur upon infection with intracellular pathogens.32 H. pylori, a gastric pathogen is responsible for the development of mild asymptomatic gastritis to severe PUD, MALT lymphoma, and gastric adenocarcinoma.33 Besides H. pylori infection, the susceptibility to infection and it’s associated disease progression were also determined by host genetic factors. Polymorphisms compromising the functions of NLRs result in recurrent H. pylori infections and increase the risk to develop a GC malignancy over time. Thus, our study was aimed to screen for the variants in NLRP3 in the South Indian population and establish its genetic association with persistent H. pylori infection.
Inflammasomes are multimeric protein complexes that are responsible for the host’s innate immune system to defend against invading pathogens. Microbial activation of inflammasomes is a prerequisite for the activation of CASP1, causing inflammation by the secretion of pro-inflammatory cytokines.34,35 In addition to this, inflammasomes restrict intracellular bacterial growth by pyroptosis and effectively protect the host against bacterial infections.8 However, bacterial toxins modify these effectors and regulate the inflammasomes for their growth and survival.
NLRP3 is a family member of the canonical inflammasomes and is activated either by direct recognition of PAMPs and DAMPs through ASC recruitment domain or by activated TLRs and NOD1/NOD2.36 The activated NLRP3 recruits CASP1 which is involved in release of IL-18 and IL-1b.9 Here, IL-1b helps in the survival of the gastric pathogen H. pylori through suppression of gastric acid secretion by gastric parietal cells and leads to gastric atrophy and metaplasia, which are prerequisites for GC development.37-39 Further, it plays an important part in gastric carcinogenesis by promoting intravasation and metastasis of GC cells.40 Kang et al reported that IL-18 was involved in immune escape and metastasis of GC cells.41 Also, IL-18 was found to be associated with chronic gastritis in patients with H. pylori.42 Myung et al has reported the association between a polymorphism in IL-18 and susceptibility to pathogenic infection.43 Its virulence factors cause cellular damage through lysosomal rupture, mitochondrial damage, endoplasm- and endosome-mediated stress leading to oxidative stress and inflammasome activation that results in pyroptosis.44 Thus, oxidative stress activates inflammasome machinery to secrete pro-inflammatory cytokines resulting in gastric inflammation, a major contributor to gastric tumorigenesis.45
The role of innate immune system in the primary immune response against H. pylori infection is considered to be crucial. Variants in the genes involved in conferring immunity against H. pylori may be important in the pathophysiology of PUD and gastric tumorigenesis.46 Studies have revealed that the polymorphisms in NLRP3 had been associated with the susceptibility to inflammatory diseases such as rheumatoid arthritis,47 and ulcerative colitis.48 Castano-Rodriguez et al has shown that the SNPs involved in the NLR signalling pathway such as NLRP3 had been associated with H. pylori infection and related it to GC in the Chinese Han population.20
Omi et al investigated the influence of a 42 bp VNTR (rs74163773) in the intron of NLRP3 with the development of inflammation-based hypertension. Their findings suggested that the length of this VNTR might affect the expression of NLRP3. Also, they showed that the subjects with 12/12 genotype had significantly higher levels of NLRP3 mRNA in the peripheral leukocytes.27 Another study to analyze the influence of the rs74163773 on vulvar vestibulitis syndrome (VVS) revealed that allele 7 (7/7 genotype) was significantly associated with VVS.49 Similarly, Jaeger et al showed that the 12/7 genotype of rs741633773 was significantly associated with RVVC.18 In our study, we found that the 12/12 genotype of rs74163773 was found to be the most prevalent genotype and a novel genotype (X-X) to be the least prevalent among the South Indian population, moreover failed to show the risk of developing persistent infection and its associated clinical phenotypes. Lack of association could be due to ethnic differences in the population, mechanisms of disease progression, geographical conditions, and environmental modifications. The differences in microbial population and immune homeostasis may be the reason behind the inconclusive relationship between the clinical features and the severity of the disease.
Screening of 3’-UTR variant (C > G) of rs10754558 polymorphism revealed it’s significant association with persistent H.pylori infection. Also, the influence of rs10754558 with its clinical manifestations revealed that this variant was associated with chronic gastritis and PUD as that of the study reported by Castano-Rodriguez et al.20 as well as with HPV infection and cervical cancer.13 We believe there are no studies that have been conducted so far to investigate the association of NLRP3 variants and the risk of persistent H. pylori infection, which contributes for disease progression to GC. rs10754558 was also associatively reported with many inflammatory diseases such as primary gout arthritis,50 aphthous stomatitis,51 juvenile spondyloarthritis,52 anaphylaxis,53 and ulcerative colitis.48 Hitomi et al showed that allele G of rs10754558 could influence NLRP3 mRNA levels and their stability.53 Furthermore, this variant was also associated with upregulated secretion of IL-1b,54 and favours H. pylori to adapt in gastric mucosa by suppressing gastric acid secretion.
The observed MAF of rs10754558 in our control group was found to be 54% whereas MAFs reported in the Chinese Han,54 and Brazilian populations were 43%.10 These differences could be attributed to the differences in the ethnicity of the population, hence displaying genetic heterogeneity. The multiple regression models such as the co-dominant model and recessive model were found to be mainly associated with persistent pathogenic infection in the population. Consequently, we observed a statistical association between GG genotype and chronic pathogenic infection. Our results are concordant with Tan et al findings on the late onset of AD recessive model (p = 0.005) in the Chinese Han population.11 Besides, we observed that the individuals with CG genotype of rs10754558 have a decreased risk of manifesting chronic gastritis and PUD. Our findings on different models of inheritance of rs10754558 have revealed that the over-dominant and co-dominant models were associated with H. pylori released complications. In previous studies, we have reported the association between genetic polymorphisms of TLR4 (rs4986790 and rs4986791) and TLR9 (rs352140) with chronic pathogenic infection in South Indian Tamils,55 whereas genetic polymorphisms of TLR5 (rs2072493, rs5744174, and rs5744168) showed no significant association with persistent H. pylori infection among the subject group.56 This implies that various genetic factors could influence the pathogenesis of disease and its clinical phenotypes. Our study could be further strengthened with larger sample size and the corresponding functional analyses to elucidate the role of NLRP3 in the pathogenesis of persistent H. pylori infection, and its extrapolation to the predisposition to GC.
In conclusion, genotyping of NLRP3 polymorphisms has revealed that rs10754558 was significantly associated with persistent H. pylori infection in the Tamil South Indian population and the heterozygous genotype of NLRP3 (rs10754558) acts as an independent risk factor for developing chronic gastritis and PUD. Further, we observed that VNTR rs74163773 has failed to show significant risk in developing persistent H. pylori infection and its associated clinical manifestations in the Tamil South Indian population. Though, our preliminary study have revealed the association of rs10754558 with chronic gastritis and PUD, further validation of this result using expressional and functional studies may delineate the importance of rs10754558 in the development of these disease.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the DBT-BIOCARE Scheme for Women Scientists, Department of Biotechnology, Ministry of Science and Technology, Government of India, under grant No. BT/PR18428/BIC/101/261/2016.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethical Committee, Madras Medical College (MMC), Chennai (EC Reg No. ECR/270/Inst./TN/2013; IEC No. 31082015).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-249.
Crossref - Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137-2150.
Crossref - Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893-2917.
Crossref - Cancer Tomorrow. Accessed July 28, 2021. https://gco.iarc.fr/tomorrow/en
- Mathur P, Sathishkumar K, Chaturvedi M, et al. Cancer Statistics, 2020: Report From National Cancer Registry Programme, India. JCO Glob Oncol. 2020;(6):1063-1075.
Crossref - Dikshit R, Gupta PC, Ramasundarahettige C, et al. Cancer mortality in India: A nationally representative survey. Lancet. 2012;379(9828):1807-1816.
Crossref - Kumar Pachathundikandi S, Brandt S, Madassery J, Backert S. Induction of TLR-2 and TLR-5 expression by Helicobacter pylori switches cagPAI-dependent signalling leading to the secretion of IL-8 and TNF-a. PLoS One. 2011;6(5):e19614-e19614.
Crossref - Sahoo M, Ceballos-Olvera I, Del Barrio L, Re F. Role of the inflammasome, IL-1b, and IL-18 in bacterial infections. Scientific World Journal. 2011;11:2037-2050.
- He Q, Fu Y, Tian D, Yan W. The contrasting roles of inflammasomes in cancer. Am J Cancer Res. 2018;8(4):566-583.
- Pontillo A, Brandao L, Guimaraes R, Segat L, Araujo J, Crovella S. Two SNPs in NLRP3 gene are involved in the predisposition to type-1 diabetes and celiac disease in a pediatric population from northeast Brazil. Autoimmunity. 2010;43(8):583-589.
Crossref - Tan MS, Yu JT, Jiang T, et al. NLRP3 polymorphisms are associated with late-onset Alzheimer’s disease in Han Chinese. J Neuroimmunol. 2013;265(1-2):91-95.
Crossref - Inoue M, Shinohara ML. NLRP3 inflammasome and MS/EAE. Autoimmune Dis. 2013;2013:859145.
Crossref - Lu Q, Lao X, Gan J, et al. Impact of NLRP3 gene polymorphisms (rs10754558 and rs10733113) on HPV infection and cervical cancer in southern Chinese population. Infect Agent Cancer. 2023;18(1):64.
Crossref - Zhang XY, Li C, Chen D, et al. H. pylori CagA activates the NLRP3 inflammasome to promote gastric cancer cell migration and invasion. Inflamm Res. 2022;71(1):141-155.
Crossref - Basak C, Pathak SK, Bhattacharyya A, Mandal D, Pathak S, Kundu M. NF-kB- and C/EBPb-driven interleukin-1b gene expression and PAK1-mediated caspase-1 activation play essential roles in interleukin-1b release from Helicobacter pylori lipopolysaccharide-stimulated macrophages. J Biol Chem. 2005;280(6):4279-4288.
Crossref - Hitzler I, Sayi A, Kohler E, et al. Caspase-1 Has Both Proinflammatory and Regulatory Properties in Helicobacter Infections, Which Are Differentially Mediated by Its Substrates IL-1b and IL-18 . J Immunol. 2012;188(8):3594-3602.
Crossref - Hanaei S, Sadr M, Rezaei A, et al. Association of NLRP3 single nucleotide polymorphisms with ulcerative colitis: A case-control study. Clin Res Hepatol Gastroenterol. 2018;42(3):269-275.
Crossref - Jaeger M, Carvalho A, Cunha C, et al. Association of a variable number tandem repeat in the NLRP3 gene in women with susceptibility to RVVC. Eur J Clin Microbiol Infect Dis. 2016;35(5):797-801.
Crossref - Yin S, Lan C, Pei H, Zhu Z. Expression of interleukin 1b in gastric cancer tissue and its effects on gastric cancer. Onco Targets Ther. 2015;9:31-35.
Crossref - Castano-Rodriguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM. The NOD-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer: A case-control study and gene expression analyses. PLoS One. 2014;9(6):e98899.
Crossref - Kumar S, Dhiman M. Inflammasome activation and regulation during Helicobacter pylori pathogenesis. Microb Pathog. 2018;125:468-474.
Crossref - Wang P, Gu Y, Yang J, et al. The prognostic value of NLRP1/NLRP3 and its relationship with immune infiltration in human gastric cancer. Aging (Albany NY). 2022;14(24):9980-10008.
Crossref - Ghoshal UC, Kumar S, Krishnani N, Kumari N, Chourasia D, Tripathi S. Serological assessment of gastric intestinal metaplasia and atrophy using pepsinogen-I, pepsinogen-II and gastrin-17 levels in a low incidence area of gastric cancer endemic for H. pylori infection. Trop Gastroenterol. 2011;32(4):292-298.
- Chen TS, Li FY, Chang FY, Lee SD. Immunoglobulin G antibody against Helicobacter pylori: Clinical implications of levels found in serum. Clin Diagn Lab Immunol. 2002;9(5):1044-1048.
Crossref - Ohyauchi M, Imatani A, Yonechi M, et al. The polymorphism interleukin 8 – 251 A/T influences the susceptibility of Helicobacter pylori related gastric diseases in the Japanese population. Gut. 2005;54(3):330-335.
Crossref - Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Crossref - Omi T, Kumada M, Kamesaki T, et al. An intronic variable number of tandem repeat polymorphisms of the cold-induced autoinflammatory syndrome 1 (CIAS1) gene modifies gene expression and is associated with essential hypertension. Eur J Hum Genet. 2006;14(12):1295-1305.
Crossref - Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(1 D):D638-D646.
Crossref - rs10754558 RefSNP Report – dbSNP – NCBI. www.ncbi.nlm.nih.gov. Published online 2022. https://www.ncbi.nlm.nih.gov/snp/rs10754558%0Ahttps://www.ncbi.nlm.nih.gov/snp/rs10754558?vertical_tab=true#frequency_tab
- Israel DA, Peek RM. Chapter 63 – Mechanisms of Helicobacter pylori-Induced Gastric Inflammation. In: Said HMBTP of the GT (Sixth E, ed. Academic Press; 2018:1517-1545.
Crossref - Pachathundikandi SK, Blaser N, Backert S. Mechanisms of Inflammasome Signaling, microRNA Induction and Resolution of Inflammation by Helicobacter pylori. Curr Top Microbiol Immunol. 2019;421:267-302.
Crossref - Liu J, Qi X, Gu P, Wang L, Song S, Shu P. Baicalin Induces Gastric Cancer Cell Pyroptosis through the NF-kB-NLRP3 Signaling Axis. J Cancer. 2024;15(2):494-507.
Crossref - Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori Infection and the Risk of Gastric Carcinoma. N Engl J Med. 1991;325(16):1127-1131.
Crossref - Schroder K, Tschopp J. The Inflammasomes. Cell. 2010;140(6):821-832.
Crossref - Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278-286.
Crossref - Kolb R, Liu GH, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: A double-edged sword. Protein Cell. 2014;5(1):12-20.
Crossref - Apte RN, Dotan S, Elkabets M, et al. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006;25(3):387-408.
Crossref - El-Omar EM. The importance of interleukin 1b in Helicobacter pylori associated disease. Gut. 2001;48(6):743-747.
Crossref - Saperas E, Yang H, Tache Y. Interleukin-1beta acts at hypothalamic sites to inhibit gastric acid secretion in rats. Am J Physiol – Gastrointest Liver Physiol. 1992;263(3):G414-G418.
Crossref - Dinarello CA. Why not treat human cancer with interleukin-1 blockade? Cancer Metastasis Rev. 2010;29(2):317-329.
Crossref - Kang JS, Bae SY, Kim HR, et al. Interleukin-18 increases metastasis and immune escape of stomach cancer via the downregulation of CD70 and maintenance of CD44. Carcinogenesis. 2009;30(12):1987-1996.
Crossref - Fera MT, Carbone M, Buda C, et al. Correlation between Helicobacter pylori Infection and IL-18 mRNA Expression in Human Gastric Biopsy Specimens. Ann N Y Acad Sci. 2002;963(1):326-328.
Crossref - Myung DS, Lee WS, Park YL, et al. Association between interleukin-18 gene polymorphism and Helicobacter pylori infection in the Korean population. Sci Rep. 2015;5(1):11535.
Crossref - Salama NR, Hartung ML, Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11(6):385-399.
Crossref - Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436-444.
Crossref - Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403-414.
Crossref - Kastbom A, Verma D, Eriksson P, Skogh T, Wingren G, Soderkvist P. Genetic variation in proteins of the cryopyrin inflammasome influences susceptibility and severity of rheumatoid arthritis (The Swedish TIRA project). Rheumatology. 2008;47(4):415-417.
Crossref - Zhang HX, Wang ZT, Lu XX, Wang YG, Zhong J, Liu J. NLRP3 gene is associated with ulcerative colitis (UC), but not Crohn’s disease (CD), in Chinese Han population. Inflamm Res. 2014;63(12):979-985.
Crossref - Lev-Sagie A, Prus D, Linhares IM, Lavy Y, Ledger WJ, Witkin SS. Polymorphism in a gene coding for the inflammasome component NALP3 and recurrent vulvovaginal candidiasis in women with vulvar vestibulitis syndrome. Am J Obstet Gynecol. 2009;200(3):P303.E1-303.E6.
Crossref - Zhang QB, Qing YF, He YL, Xie WG, Zhou JG. Association of NLRP3 polymorphisms with susceptibility to primary gouty arthritis in a Chinese han population. Clin Rheumatol. 2018;37(1):235-244.
Crossref - Bidoki AZ, Harsini S, Sadr M, et al. NLRP3 gene polymorphisms in Iranian patients with recurrent aphthous stomatitis. J Oral Pathol Med. 2016;45(2):136-140.
Crossref - Perica M, Vidovic M, Lamot L, et al. Single nucleotide polymorphism of toll-like receptor 4 (TLR4) is associated with juvenile spondyloarthritis in Croatian population. Clin Rheumatol. 2015;34(12):2079-2086.
Crossref - Hitomi Y, Ebisawa M, Tomikawa M, et al. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J Allergy Clin Immunol. 2009;124(4):779-785.e6.
Crossref - Zhou D, Wang X, Chen T, et al. The NLRP3 rs10754558 polymorphism is associated with the occurrence and prognosis of coronary artery disease in the Chinese Han population. Biomed Res Int. 2016;2016:3185397.
Crossref - Loganathan R, Nazeer M, Goda V, et al. Genetic variants of TLR4 and TLR9 are risk factors for chronic Helicobacter pylori infection in South Indian Tamils. Hum Immunol. 2017;78(2):216-220.
Crossref - Goda V, Jayaraman M, Loganathan R, et al. TLR5 Polymorphisms rs2072493, rs5744174, and rs5744168 Are Not Genetic Risk Factors for Chronic Helicobacter pylori Infection in Indian Tamils. Immunol Invest. 2017;46(6):537-543.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.