ISSN: 0973-7510
E-ISSN: 2581-690X
Penicillium citrinum is one of the most prevalent tobacco spoilage fungi. However, the mechanisms underlying fungal growth on tobacco leaves remain largely unknown. In this study, transcriptomic analyses were performed to reveal the genome-wide expression profiles of P. citrinum growing on tobacco leaves. First, a comparative analysis was conducted between two sets of transcriptomic data from P. citrinum growing on chemically defined media and tobacco leaves. Enrichment analyses showed that differentially regulated genes were mainly associated with carbohydrate degradation (e.g., cellulose, pectin, and xylan) and the catabolism of fatty acids and aromatic compounds. Comparative transcriptomic analyses between different time points indicated that the fungal transcriptome varied dynamically during the spoilage process, and the enriched terms were associated with small-molecule degradation and fungal development. Enrichment analyses indicated that more up-regulated genes appeared in all enriched Gene Ontology terms. Notably, more organelles significantly contributed to further fungal growth on tobacco leaves. In conclusion, P. citrinum activates a comprehensive transcriptome that changes dynamically when causing tobacco mildew.
Mold Contamination, Global Expression, Fungal Degradation, Mildewed Tobacco Leaf
Tobacco, an economically important crop, is extensively grown in the world.1,2 Flue-cured tobacco is prepared by roasting fresh leaves in fire tubes and subjecting the roasted leaves to aging for 2-3 years before they are made into cigarette products.3 Tobacco leaves are rich in carbon and nitrogen sources.4,5 Under optimal conditions (e.g., temperature and humidity), extensive fungal growth can cause the decomposition of tobacco leaves.6 Spoilage is significantly detrimental to the quality of tobacco products.7 In addition, fungal contamination in tobacco products poses both direct (e.g., fungal infection) and indirect risks (e.g., mycotoxins) to consumers.8,9 Therefore, it is essential to control spoilage caused by fungi in the tobacco industry.
Saprotrophic species from Aspergillus and Penicillium have long been considered the dominant mildew-causing fungi.7,10 Dominant species vary in different tobacco-producing regions.11 Spoilage is initiated under optimal environmental conditions, and fungi quickly proliferate in tobacco tissues by assimilating various nutrients. Finally, fungi develop visible mycelia and release an unpleasant odor.7 As heterotrophic organisms, saprotrophic fungi deconstruct plant cell walls and degrade their complex materials into simple molecules.12 However, little is known about the mechanisms underlying fungal growth on tobacco leaves. Transcriptomic analysis has been widely used to clarify the molecular basis for plant degradation by fungi. Based on transcriptomic investigations, a multitude of functional genes associated with substrate degradation could be revealed, including hydrolytic enzyme, transporter, transcription factor, and the like.13,14 Thus, transcriptomic analysis is also suitable for deciphering the mechanisms involved in fungal decomposition of tobacco leaves, and the obtained knowledge can be exploited for spoilage control.
In current investigation, RNA sequencing (RNA-seq) analysis was executed on Penicillium citrinum growing on tobacco leaves to clarify the details of its genome-wide expression during degradation of tobacco leaves. This research represents a comparative transcriptome profiling of the mildew-causing fungus grown tobacco leaves.
Fungal strain and cultivation
Wild-type P. citrinum (CCTCC M 2021161) was purchased from the China Center for Type Culture Collection (Wuhan, Hubei, China) and maintained on potato dextrose agar (PDA) (Catalog number: 213400) (BD Company, MD, USA) at 25°C. Conidia were harvested from the 5-d-old culture and used in the following transcriptomic analyses as the initial fungal inocula. Czapek-Dox plates (CZA: 3% glucose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% MgSO4, 0.05% KCl, and 0.001% FeSO4 plus 1.5% agar) were adopted as the chemically defined culture substrate.
Genome annotation and carbohydrate-active enzyme analysis
The genome sequence of P. citrinum F4_1A_F1_F on the National Center for Biotechnology Information (NCBI) (BioProject: PRJNA723004, submitted by Jet Propulsion Laboratory, CA, USA) was used as a reference genome. The gene structures of this genome were predicted with the AUGUSTUS software,15 then the cDNA sequence and its deduced protein sequence were obtained. The resultant proteins were annotated via a BLAST search against a database consisting of the non-redundant proteins which was downloaded from the website (ftp://ftp.ncbi.nih.gov/blast/db/). The annotations with the best hits were selected. Carbohydrate-active enzymes were functionally classified by a local BLAST searching analysis with a database formed from catalytic enzymes and carbohydrate-binding module proteins (CAZy) (http://www.cazy.org/).
RNA extraction and sequencing
Flue-cured tobacco was prepared and stored in a warehouse of our company (China Tobacco Zhejiang Industrial Co. Ltd, Hangzhou, China). To obtain the in-situ transcriptome of P. citrinum, conidial suspension (10 µl, 106 conidia/ml) was added dropwise on the tobacco leaf surface (TL) and incubated at 25°C. The leaf parts covered with mycelia were taken at 2 and 3 days after cultivation. The mycelia grown on CZA plates were used as the control of chemically defined media (CK). Thus, three samples were prepared for RNA-seq analyses: (i) P. citrinum grown on CZA plates for 2 days (CK2D), (ii) P. citrinum grown on tobacco leaves for 2 days (TL2D), and (iii) P. citrinum grown on tobacco leaves for 3 days (TL3D).
Fungal mycelia were put into a ceramic mortar and frozen with liquid nitrogen, and then ground into powder with a pestle. The total RNA was prepared with extraction reagent RNAisoTM Plus (Cat.: 9108, TaKaRa, Dalian, China), using 1 mL reagent per 100 mg fungal sample. The resultant total RNA was purified with a Qiagen RNeasy Mini kit (Cat.: 74134, Qiagen, Shanghai, China) which was coupled with on-column treatment with DNase I. The quality of total RNA (amount and purity) was determined on NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). Then, the RNA integrity was evaluated on Bioanalyzer 2100 (Agilent, CA, USA). The downstream experiments were conducted when the amount of total RNA was greater than 1 µg, the concentration was not less than 50 ng/µL, and the RNA integrity value was over 7.0. The polyA-containing mRNA was purified through two rounds of affinity adsorption with Dynabeads Oligo (dT) magnetic beads (Cat.: 25-61005, Thermo Fisher, USA). The fragmentation of mRNA was achieved at 94°C for 5 min with the NEBNext® RNA Fragmentation Module (Cat.: E6150S, NEB, USA). cDNA was synthesized with reverse transcriptase SuperScriptTM II (Cat.:1896649, Invitrogen, USA), using the fragmented RNA as template. Then, these complex double strands were converted into the double-stranded DNA molecules with Escherichia coli DNA polymerase I (Cat.: M0209, NEB, USA) and RNase H (Cat.: M0297, NEB, USA). During this conversion reaction, dUTP (Thermo Fisher, cat.R0133, CA, USA) was integrated into the DNA fragments with blunt termini. Then, a base of ‘A’ was added to each end of the double-stranded DNA. Dual-index adapters for Illumina® (Cat.: E7395, NEB, USA) were connected to the fragments, and selection of fragments with expected size was completed with AMPure XP beads (Cat.: E6260, NEB, USA). After treatment with Uracil-DNA Glycosylase (Cat.: M0280, NEB, USA), the collected products were augmented via polymerase chain reaction (PCR) with Index Primer (Cat.: E7335, NEB, USA). The PCR program was launched with initial denaturation at 95°C for 3 min. Amplification was achieved through eight cycles in which each cycle included denaturation at 98°C for 15 sec, annealing at 60°C for 15 sec, and extension at 72°C for 30 sec. Finally, extension reaction was completed at 72°C for 5 min. The final cDNA library was constructed with fragments with average size of 300 ± 50 bp. The resultant library was sequenced on the platform of Illumina NovaseqTM 6000 (Illumina, Inc., CA, USA) in LC Bio Technology Co. Ltd. (Hangzhou, China). All manipulations, including the experimental temperature, reagent concentration, and parameter setting, were conducted according to the detailed instructions provided by manufacturers.
The data for RNA-seq analyses have been submitted to NCBI’s Gene Expression Omnibus under accession no. GSE202277. All treatments were conducted with two independent replicates.
Assay for differentially expressed genes (DEGs)
Transcripts were determined by mapping clean reads onto the database of the annotated genes with the HISAT program.16 The identified genes were normalized as the expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FKPM) with Cufflinks software.17 The Cuffdiff method18 was used to search the DEGs between the paired comparative transcriptomes (i.e., TL2D/CK2D and TL3D/TL2D). Genes were considered to be differentially expressed when an absolute value of log2Ratio (fold change) was greater than 1 and the q-value was less than 0.05. The obtained DEGs were subjected to functional sortation via enrichment analysis. Firstly, each DEG was mapped to the functional term in the systems of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Then, the enriched categories and pathways were revealed with hypergeometric tests, when its corrected P-value was less than 0.05.
Gene prediction and annotation for the reference genome
P. citrinum F4_1A_F1_F (Jet Propulsion Laboratory, CA, USA) was used as a reference strain. The predicted database for P. citrium genome consisted of 9,498 genes (Supplementary Table S1). Based on BLAST searching, 7,802 protein genes were annotated (Supplementary Table S1). CAZy annotation revealed that the P. citrinum genome contained numerous carbohydrate-degrading enzymes (572 proteins), which were sorted into 166 groups (Supplementary Table S2). P. citrinum processed several families of cellulases (cellobiohydrolases GH6 and GH7 and endo-b-1,4-glucanses GH12), hemicellulases (endo-b-1,4-xylanases GH11 and GH30, a-arabinofuranosidase GH51, endo-b-1,4-galactanase GH53, a-arabinofuranosidase GH62, and a-glucuronidases GH67). It is well known that tobacco leaves consist of large amounts of cellulose and lignin.19 This finding suggests that P. citrinum has an ability to degrade tobacco leaves. In addition, P. citrinum possessed several enzymes that catalyzed the degrading process of xylan oligomers and polymers, including xyloglucan-specific endo-b-1,4-glucanase A (GH12), endo-1,4- b-xylanase D (GH10), xylan 1,4-b-xylosidase (GH3). In addition, a family of proteins was sorted into the carbohydrate esterases (CEs) (e.g., carboxylesterase (CE10) and esterase (CE12)), which suggests that this fungus has potential in the degradation of esters and lipids.
Summary of global expression analysis
To identify global expression profiles in P. citrinum during the degradation of tobacco leaves, a comparative transcriptomic analysis was performed. Three samples were prepared for RNA-seq analyses: (i) P. citrinum grown on CZA plates for 2 days (CK2D), (ii) P. citrinum grown on tobacco leaves for 2 days (TL2D), and (iii) P. citrinum grown on tobacco leaves for 3 days (TL3D). Each sample contained two independent replicates.
Deep RNA-seq produced a large quantity of valid reads. The summary for quality features is shown for each library (Table). For example, valid reads for two replicates of CK2D were 5.28 and 4.96 × 107. On the whole, the percentage of valid reads to total ones exceeded 97% (97.88 – 98.20%). The Q20 value for each library was up to 99.9%, and all Q30 values were not less than 97.9%. These quality indexes indicated that the obtained high-quality transcriptome was suitable for further analyses.
Table:
Quality control of transcriptomic analyses
| Sample ID | Raw data | Valid data | Ratio (valid reads/raw reads) | Q20 (%) | Q30 (%) | GC content (%) | ||
|---|---|---|---|---|---|---|---|---|
| Reads number | Base number | Reads number | Base number | |||||
| CK2D_R1 | 53772270 | 8.07G | 52805190 | 7.92G | 98.20 | 99.94 | 98.18 | 51.5 |
| CK2D_R2 | 50530882 | 7.58G | 49564364 | 7.43G | 98.09 | 99.97 | 97.96 | 51.5 |
| TL2D_R1 | 53949402 | 8.09G | 52820444 | 7.92G | 97.91 | 99.94 | 97.99 | 52.0 |
| TL2D_R2 | 54691580 | 8.20G | 53739298 | 8.06G | 98.26 | 99.94 | 98.02 | 52.5 |
| TL3D_R1 | 53461890 | 8.02G | 52472228 | 7.87G | 98.15 | 99.94 | 97.98 | 52.0 |
| TL3D_R2 | 54531200 | 8.18G | 53374966 | 8.01G | 97.88 | 99.94 | 98.28 | 52.0 |
Fungal DEGs during fungal growth on tobacco leaves
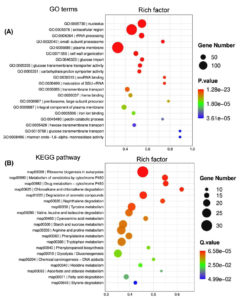
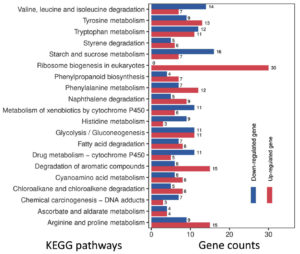
Under optimal temperature and humidity conditions, molds on tobacco leaves cause spoilage during long-term storage.6 To explore the tobacco leaf-induced genes in P. citrinum, transcriptomes for CK2D and TL2D libraries were comparatively analyzed, in which CK2D was used as the control. In TL2D library, genes with q < 0.05 and |log2Ratio| ≥1 were considered to be the DEGs when compared with those in the control library. The results showed that 1,780 and 3,283 genes were significantly up- and down-regulated, respectively (Supplementary Table S3). After annotation of DEGs with GO terms, hypergeometric analyses revealed the over-presented categories were associated with biological process (BP), cellular component (CC), and molecular function (MF) (Figure 1A). Among seven BP terms, the gene counts of up-regulated DEGs were greater than those of down-regulated ones. These terms were mainly associated with catabolic processes for pectin (GO: 0045490), cellulose (GO: 0030245), and xylan (GO: 0045493). For example, the significantly inducible genes involved a plenty of genes that catalyzed carbohydrate degradation (e.g., endo-1,4-b-xylanase A, pectin lyase A, fungal cellulose binding protein, endopolygalacturonase A, and a-L-arabinofuranosidase). In the CC category, five terms had more upregulated DEGs than downregulated DEGs, which were mainly associated with nucleus (GO: 0005634) and nucleolus (GO: 0005829). In the MF category, four terms had more up-regulated DEGs than down-regulated DEGs, which were mainly associated with iron ion binding (GO: 0005506) and heme binding (GO: 0020037). After annotation of DEGs with KEGG terms, enrichment analyses revealed that DEGs were over-presented in various pathways (Figure 1B). Among the top 20 enriched pathways (Figure 3), most up-regulated genes were associated with metabolism (e.g., carbohydrates and amino acids) and degradation (e.g., fatty acids and aromatic compounds) processes. Among the physiological terms, the pectin, cellulose, and xylan catabolic processes contributed to fungal growth on the tobacco leaves.
Figure 1. Functional distribution of differentially expressed genes (DEGs) of P. citrinum growing on tobacco leaves. P. citrinum conidia were inoculated onto tobacco leaves and cultured for 2 days. Czapek-Dox plates (CZA) were used as the control for the chemically defined medium. A comparative analysis was conducted between two sets of transcriptomic data to generate a set of DEGs. GO (A) and Kyoto KEGG analyses (B) were used to functionally sort all DEGs, and the top 20 enriched categories are shown in the bubble chart
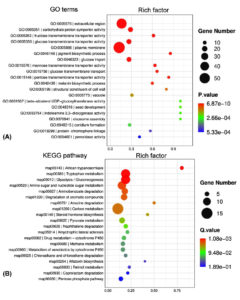
Figure 2. Functional distribution of dynamic transcriptome of P. citrinum during tobacco spoilage. P. citrinum conidia were inoculated onto tobacco leaves and cultured for 2 and 3 days. A comparative analysis was conducted between the transcriptomic data from 2 days to generate a set of DEGs. All DEGs were functionally classified with terms of GO (A) and KEGG (B), and the top 20 enriched categories are shown in the bubble chart
Fungal dynamic transcriptome during tobacco decay
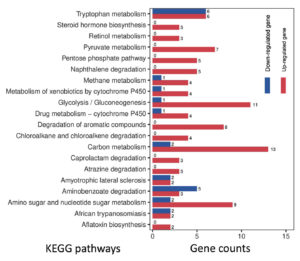
To reveal the global expression dynamics in P. citrinum during the degradation of tobacco leaves, transcriptomic analysis comparing TL2D and TL3D libraries was conducted, in which TL2D was used as the control. The results showed that 1,780 and 3,283 genes were significantly up- and down-regulated, respectively (Supplementary Table S4). On the basis of GO annotation of DEGs, the functional distribution of the annotated terms was revealed via enrichment analyses. The results indicated the DEGs were functionally related to a series of BP, CC, and MF categories (Figure 2A). Among all enriched terms, the amount of up-regulated DEGs was significantly greater than those of down-regulated ones. In the BP category, the terms related to metabolism were mainly associated with processes including carbohydrate metabolism (GO: 0005975), glucose metabolism (GO: 0006006), and polysaccharide catabolism (GO: 0000272). The terms related to transportation of substrate included glucose (GO: 0046323) and hexose (GO: 0035428). In the CC category, the enriched terms were associated with organelles, including peroxisome (GO: 0005777), Golgi apparatus (GO: 0005794), and mitochondrion (GO: 0005769). In the MF category, the enriched categories were in connection with binding activity [e.g., heme binding (GO: 0020037) and zinc ion binding] and transporter activity [e.g., glucose and fructose transmembrane transporter activity (GO: 0005355 and 0005353)]. Based on KEGG classification, DEGs were sorted into different pathways (Figure 2B). Among the top 20 enriched pathways, l6 pathways had more upregulated DEGs than downregulated DEGs (Figure 4). These pathways were associated with carbohydrate metabolism (e.g., pentose phosphate pathway and pyruvate metabolism) and degradation processes (e.g., degradation of xenobiotic and aromatic compounds). These results indicate that the transcriptome of P. citrinum changes dynamically during growth on tobacco leaves, which involves comprehensive metabolic pathways.
Figure 3. Gene numbers in the differentially regulated pathways for P. citrinum grown on tobacco leaves. On the basis of KEGG analyses, the top 20 enriched pathways are shown. The numbers near the right side of the bar indicate the gene numbers for the up- and down-regulated DEGs in the corresponding pathway
Figure 4. Gene numbers in the differentially regulated pathways for P. citrinum during further growth. On the basis of KEGG analyses, the top 20 enriched pathways are illustrated. The numbers near the right side of the bar indicate the gene counts for the up- and down-regulated DEGs in the enriched pathway
P. citrinum is one of the most important tobacco spoilage fungi, causing concerns about product quality and economic losses in tobacco industry. In this study, physiological basis of P. citrinum growing tobacco leaves was revealed through comparative transcriptomic analyses.
Tobacco leaves are rich in carbon (e.g., cellulose) and nitrogen (e.g., amino acid) sources.4,5 Among the physiological terms, the metabolism of carbohydrate and amino acids was found to be involved in fungal growth on the tobacco leaves. These findings suggest that molds might be beneficial for tobacco aging by changing the chemical components in leaves at the early stage of growth, but cause breakdown of tobacco leaves owing to overgrowth under optimal conditions. Similar results have been observed in the brown rot fungus Gloeophyllum trabeum growing on lignocelluloses.14 Significantly, a large quantity of genes associated with carbohydrate degradation (e.g., endo-1,4-b-xylanase A, pectin lyase A, fungal cellulose binding protein, endopolygalacturonase A, and a-L-arabinofuranosidase) were induced or enhanced when P. citrinum was cultured on tobacco leaves. For example, pectin lyase and endo polygalacturonase are required for pectin degradation.20 Fungal cellulose binding protein acts as a non-catalytic module binding substrate during cellulose degradation.21 Arabinofuranosidase is induced by xylan, which suggests that this enzyme functions as a xylan-degrading enzyme.22 These results reinforced that, in the same way as other plant-degrading fungi, P. citrinum breaches tobacco issues (e.g., the cell wall) and degrades the complex materials (e.g., cellulose and polysaccharide) into simple molecules. In addition, on tobacco leaves, P. citrinum activates a group of genes responsible for the metabolism of fatty acids as well as aromatic compounds. These results are well explained by the fact that tobacco leaves contain these two types of chemicals.23,24 These findings suggest that the significantly induced genes in fungi growing on tobacco leaves might be used to evaluate the spoilage potential of fungi.
The transcriptome of P. citrium dynamically changes during growth on tobacco leaves. This dynamic change in the transcriptome has also been observed in the brown rot fungus G. trabeum growing on lignocelluloses.14 Among the physiological terms, the processes of glucose metabolism, hexose transport, conidium formation, and pigment biosynthesis were significantly enhanced in the 3-day-old mycelia. This result suggests that after degrading the complex carbohydrates, P. citrinum actively assimilates simple molecules, such as hexose, to support fungal development and secondary metabolism. Notably, degradation processes [e.g., degradation of xenobiotic and aromatic compounds) were significantly enhanced with further fungal growth. These significantly induced genes could be used to evaluate the spoilage degree during tobacco storage.
In conclusion, the tobacco-spoilage fungus P. citrinum activates a comprehensive transcriptome during the degradation of tobacco leaves, which dynamically changes as the spoilage process proceeds. The significantly induced genes could be used as markers to indicate the fungal capacity to cause tobacco spoilage; in particular, might be applied in evaluation of mildew potential of newly emerged molds. The results provide an initial framework for exploring the mechanisms involved in tobacco spoilage caused by molds. This knowledge will foster the rational design of anti-mold strategies, e.g., fungicide, biocontrol agent, and control of storage condition. This study improves our understanding of mildew events in stored tobacco and provides clues for mildew control in the tobacco industry.
Additional file: Additional Table S1-S4.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This research was supported by China Tobacco Zhejiang Industrial Co. Ltd (No.: ZJZY2021B003).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript. Sequence data have been deposited in NCBI’s Gene Expression Omnibus with accession no. GSE202277.
ETHICS STATEMENT
Not applicable.
- Eickholt DP, Lewis RS. Effect of an introgressed Nicotiana tomentosa leaf number QTL on yield and quality characteristics in flue-cured tobacco. Crop Sci. 2014;54(2):586-594.
Crossref - Tong Z, Xiao B, Jiao F, et al. Large-scale development of SSR markers in tobacco and construction of a linkage map in flue-cured tobacco. Breed Sci. 2016;66(3):381-390.
Crossref - Zhou J, Yu L, Zhang J, et al. Characterization of the core microbiome in tobacco leaves during aging. Microbiologyopen. 2020;9(3):e984.
Crossref - Ding L, Xie F, Zhao M, Wang S, Xie J, Xu G. Rapid quantification of sucrose esters in oriental tobacco by liquid chromatography-ion trap mass spectrometry. J Sep Sci. 2007;30(1):35-41.
Crossref - Sun JG, He JW, Wu FG, et al. Comparative analysis on chemical components and sensory quality of aging flue-cured tobacco from four main tobacco areas of China. Agric Sci China. 2011;10(8):1222-1231.
Crossref - Lu J, Zhang H, Su X, Ma D, Bai X, Du Y. The influence of different storage conditions on the cured tobacco leaves mildew. J Pure Appl Microbiol. 2014;8(3):2135-2139.
- Zhou J, Cheng Y, Yu L, Zhang J, Zou X. Characteristics of fungal communities and the sources of mold contamination in mildewed tobacco leaves stored under different climatic conditions. Appl Microbiol Biotechnol. 2022;106:131-144.
Crossref - Verweij PE, Kerremans JJ, Voss A, Meis J. Fungal contamination of tobacco and marijuana. JAMA. 2000;284(22):2875.
- Ogundero VW. Studies on thermophilic fungi associated with the spoilage of flue-cured tobacco leaves during storage. Mycopathologia. 1983;82:153-158.
Crossref - Welty RE. Fungi isolated from flue-cured tobacco inoculated in the field with storage fungi. Appl Microbiol. 1971;21(3):552-554.
Crossref - Peng Q, Yi T. Analysis of tobacco mildew and its control measures. Chinese Agric Sci Bull. 2007;23(11):146-150.
- Crowther TW, Boddy L, Jones TH. Functional and ecological consequences ofsaprotrophic fungus-grazer interactions. ISME J. 2012;6(11):1992-2001.
Crossref - Miyauchi S, Navarro D, Grisel S, Chevret D, Berrin JG, Rosso MN. The integrative omics of white-rot fungus Pycnoporus coccineus reveals co-regulated CAZymes for orchestrated lignocellulose breakdown. PLoS One. 2017;12(4):e0175528.
Crossref - Umezawa K, Niikura M, Kojima Y, Goodell B, Yoshida M. Transcriptome analysis of the brown rot fungus Gloeophyllum trabeum during lignocellulose degradation. PLoS One. 2020;15(12):e0243984.
Crossref - Mario S, Stephan W. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215-ii225.
Crossref - Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357-360.
Crossref - Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511-515.
Crossref - Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46-53.
Crossref - Best M, Koenig K, McDonald K, Schueller M, Rogers A, Ferrieri RA. Inhibition of trehalose breakdown increases new carbon partitioning into cellulosic biomass in Nicotiana tabacum. Carbohydr Res. 2011;346(5):595-601.
Crossref - Mata-Gomez MA, Heerd D, Oyanguren-Garcia I, Barbero F, Rito-Palomares M, Fernandez-Lahore M. A novel pectin-degrading enzyme complex from Aspergillus sojae ATCC 20235 mutants. J Sci Food Agric. 2015;95(7):1554-1561.
Crossref - Blackman LM, Cullerne DP, Hardham AR. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics. 2014;15:785.
Crossref - Dobozi MS, Szakacs G, Bruschi CV. Xylanase activity of Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58(11):3466-3471.
Crossref - Chu H, Tso TC. Fatty acid composition in tobacco. Plant Physiol. 1968;43(3):428-433.
https://doi.org/10.1104/pp.43.3.428 - Popova VT, Ivanova TA, Stoyanova AS, et al. Chemical constituents in leaves and aroma products of Nicotiana rustica L. tobacco. Int J Food Stud. 2020;9(1):146-159.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.