ISSN: 0973-7510
E-ISSN: 2581-690X
The use of coal as a raw material for power plants has a good economic impact, but it also has a detrimental environmental impact, particularly due to the presence of Cr and Pb, heavy metals with bioaccumulation and biomagnification qualities. Efforts to control Pb and Cr in liquid coal waste can be achieved by bioremediation. The goal of this study is to screen indigenous bacteria, identify, and test biodegradation on the best bacteria capable of degrading Cr and Pb. Bacterial screening is done experimentally in the lab. Bacterial identification is done using morphological, biochemical, and molecular genetic methods. Using atomic absorption spectroscopy to validate Cr and Pb biodegradation research. Biodegradation experiments revealed that the efficacy of indigenous bacteria reduced Pb by 216% (0.238 ppm to 0.11 ppm) and Cr by 195% (0.34 ppm to 0.174 ppm). The findings of biochemical, morphological, and molecular genetic studies revealed that the top bacterial strains were up to 96% related. using Chromobacterium haemolyticum strain W15. Chromobacterium haemolyticum strain X, an indigenous bacteria capable of degrading Cr and Pb, was successfully isolated from liquid waste.
Bioremediation, Cr and Pb, Molecular Genetics, Biochemical and Morphological Tests, Chromobacterium haemolyticum Strain X Bacteria
Indonesia has one of the greatest coal mining outputs in the world. According to statistics from the Indonesia’s Ministry of Energy and Mineral Resources Geological Agency has potential coal reserves of 186 billion tons, with 52% on Sumatra Island, 47% on Kalimantan Island, and 1% on other islands.1 The result of the high number of mining industries is, of course, producing waste; this has a harmful influence on the environment, particularly because of the heavy metal concentration, which is bioaccumulative and biomagnifying. This can be dangerous for the environment, the organisms that live in it, and the impact on humans through the accumulation of heavy metals in the body and food chain at each trophic level.2
Waste produced by the coal industry is often characterized as liquid waste containing heavy metals that are hazardous and poisonous.3 According to Minister of Environment Decree Number 113 of 2003, liquid coal waste contains heavy elements such as manganese (Mn), copper (Cu), cadmium (Cd), zinc (Zn), lead (Pb), arsenic (As), nickel (Ni), chromium (Cr), cyanide (CN), mercury (Hg), iron (Fe), and manganese (Mn), all of which pose significant risks to the environment and individuals.4
Heavy metal bioremediation is a method for reducing heavy metal pollution by using native bacteria enzymatically, resulting in the decomposition of pollutants from dangerous and complex molecules found in heavy metals into harmless chemical compounds.5 Bioremediation is a process that involves designing microorganisms to thrive on certain heavy metals in order to lower heavy metal levels. When the bioremediation process occurs, enzymes generated by microorganisms change the chemical structure through a process known as biotransformation. Bioremediation of Pb and Cr from coal industry liquid waste using indigenous bacteria is an effort to use indigenous bacteria enzymatically to degrade the heavy metals Cr and Pb from dangerous compounds to non-hazardous, non-toxic, and simple chemicals.6 Bioremediation processes necessitate superior and chosen bacteria; hence, screening procedures comprising isolation, identification, and genetic information are necessary to acquire the best sorts of degrading bacteria. In terms of bioremediation technology, the Indonesian government has established a regulatory framework that governs conventional bioremediation operations to address environmental issues caused by mining, oil, and other kinds of pollution. The Minister of State for the Environment issued Decree No. 128 of 2003, which outlines bioremediation procedures and technical criteria for handling petroleum waste and petroleum-contaminated soil. The laws also indicate that bioremediation is carried out by using indigenous microorganisms.7
The goal of this study is to identify native bacteria that are most effective at breaking down Cr and Pb from coal industry liquid waste, as well as to identify the bacteria detected by morphological-biological and molecular biology analysis. It is envisaged that bioremediation technology would be employed to restore habitats polluted with Pb and Cr by indigenous microorganisms.
Types of research
This is an experimental quantitative study that will investigate the capacity of indigenous bacteria to digest Cr and Pb in liquid coal waste. Atomic absorption spectroscopy is used to validate the decrease in Cr and Pb concentrations caused by the breakdown of indigenous microorganisms. Identification of new indigenous bacteria using morphology-biochemistry and molecular genetics
Research variable
The independent variable is indigenous bacteria, the fixed variable is time, and the dependent variable is variations in Cr and Pb levels in liquid coal waste.
Population and Sample:
- Population: the population in this study is coal liquid waste.
- Sample: Selected indigenous bacteria as the best degraders in liquid coal waste
Data collection
This research’s data gathering process is separated into two categories, which are:
Primary information data
Primary information data was taken from observations in the laboratory
- a) Screening data for specific indigenous microorganisms.
- b) Information on the capacity to extract the finest indigenous bacterial strains during the breakdown of liquid coal waste
- c) Results of morphological-biochemical and molecular genetic bacterial tests on the chosen and best bacteria for degrading Cr and Pb in liquid coal waste..
Secondary information data
Secondary information data used to complete research obtained by researchers from various sources.
Data processing
Indigenous bacteria isolated from coal mining liquid waste are bacteria that thrive in their environment and can use the substrate as an energy source. Indigenous bacteria were then cultured on solid sodium agar media loaded with coal industry liquid waste, which served as the main energy source. Gradually increase the concentration of liquid coal waste from 1 ppm to 10 ppm qualitatively on solid media. Bromothymol blue was used as an indicator to test bacteria’s breakdown ability. Bacteria that can decompose coal waste will be yellow in color. The Bromothymol Blue indicator is a color change indicator that detects bacterial decomposition in coal waste. The medium turns yellow due to the aerobic catabolism of liquid coal waste, which produces acid compounds that turn the bromothymol blue indicator yellow. Bacteria capable of digesting liquid coal waste on solid medium are next tested on liquid media (natrium broth with brom thymol blue at a concentration of 25-100 ppm) to choose the optimum bacterial degradation capabilities.
The degradation test using liquid media is more real and faster than using solid media. In qualitative observations and testing, yellow color changes were only visible up to a concentration of 75 ppm for three days; However, no color change was seen at a concentration of 100 ppm during three 24 hour observations. As a result, researchers determined that 75 ppm was the ideal amount. Based on the screening results, 5 bacterial isolates were selected that were able to digest liquid coal waste at a concentration of 75 ppm and were evaluated for 3 days with the color changing from blue to yellow.
Data analysis
- Analysis of data on the identification of the best and selected Indigenous bacteria that degrade liquid coal waste quantitatively using morphological-biochemical and molecular genetic methods
- The Atomic Absorption Spectrophotometry (AAS) technique is employed to identify the presence of chromium (Cr) and lead (Pb) heavy metals in liquid coal waste test samples. Refer to SNI 6989.84:2019 for information on the Ex situ Laboratory (Table 1).
- Data processing is carried out by calculating the decrease in Cr and Pb concentrations using linear trend option, forecast display equation on chart using Microsoft Excel software.
Table (1):
Reference Standards and Measurement Methods
Variable |
Parameter |
Reference |
Method |
Measurement |
|---|---|---|---|---|
Main Variables |
Chromium (Cr) and Lead (Pb) |
SNI 6989.84:2019 |
Atomic Absorption Spectrophotometry (AAS) |
Ex situ Laboratorium |
The Atomic Absorption Spectrophotometry (AAS) technique is used to detect heavy metals in liquid coal waste test samples, including chromium (Cr) and lead (Pb). The analysis was conducted at Diponegoro University’s Testing Laboratory, Faculty of Engineering, Environmental Engineering Study Program. The Genetics Laboratory of PT. Genetic Science Indonesia, indigenous bacteria were identified experimentally from liquid coal waste using morphology, biochemistry, and molecular genetics to collect phylogenetic information from chosen indigenous bacteria.
Identification of indigenous bacteria degrading coal liquid waste
Liquid waste samples were diluted with 9 ml physiological NaCl up to 105 dilutions, distributed uniformly over Nutrient Agar substrate using a triangel, then incubated at 37°C for 24 hours.8 The planting technique on nutrient agar media was done three times, yielding about 114 bacterial species. The screening test is then performed on Natrium agar medium that has been combined with liquid waste and incubated with the Bromine Thymol Blue indicator. A formerly blue zone became yellow during this test. Isolates that developed yellow zones were re-cultured on nutrient agar and incubated for 24 hours at 37°C. The yellow zone formed by the deterioration of indigenous bacteria with the greatest zone will be used as the test bacterium in the study.9
Macroscopic observations include the form, color, edges, and elevation of bacterial colonies, and the colony’s surface may be viewed from the side, while its edge can be seen from the top of the dish. Gram staining allows for the observation of bacterial cell shape.10 The slides used for Gram staining were sterilized with 70% alcohol. One bacterial isolate was taken and smeared on a glass slide, then fixed several times in a Bunsen flame. The bacterial isolate was dripped with two drops of crystal violet and left for one minute. The bacterial isolate was with distilled water, and then dried.
The isolate was then dipped in iodine, left for one minute, rinsed with distilled water, and finally dried. The bacterial isolate was progressively dripped with 95% alcohol for 30 seconds before being rinsed with distilled water and dried. The isolate was gradually drizzled with safranin for thirty seconds, rinsed with distilled water, and dried. The dyed bacterial isolates were then examined under a microscope at a magnification of 100 x. Observations include the color and form of bacterial cells. Bacteria gram positive are the colored purple because they can bind crystal violet. Gram-negative bacteria produce a pink hue because they cannot bind crystal violet and are only stained by safranin.11
A biochemical test for lactic acid bacteria. Triple Sugar Iron Agar (TSI) Test: A single colony of bacterial isolates is injected into the Triple Sugar Iron Agar medium by piercing the back perpendicularly and obliquely in a zigzag pattern. After 24 hours of incubation at 37°C, the medium’s color shift became noticeable, If the slanted agar media is red while the backdrop is yellow, it means that the bacteria can ferment glucose. The growth of bacteria on the agar media produces a yellow color indicating that the bacteria are able to digest glucose, lactose and sucrose.12 Indole test for further bacterial identification tests. Bacterial isolates were injected into SIM medium (sulfide, indole, and motility) and incubated at 37°C for 24 hours. Then, 10-12 drops of Kovac’s reagent were used to observe the indole test results.13 The motility test involved putting a colony of bacterial isolates into SIM medium (sulfide, Indole, and motility) and incubation for 48 hours at 37°C. Bacterial growth around the puncture suggests a negative test result. The development of bacteria that spread over the medium demonstrates a favorable test outcome. Methyl red and the Voges-Proskauer (MR-VP) tests. One colony of bacterial isolates was added to MR-VP Medium (Glucose Phosphate Broth) and incubated for 48 hours at 37°C. The methyl red (MR) test was performed by adding three drops of the reagent to the medium. A positive test results in a shift in the color of the medium to red, indicating the formation of acid. The Voges-Proskauer (VP) test involved adding three drops of KOH 3% and five drops of alpha-naphthol shaking for 30 seconds.14 Fermentation test: The test is carried out in a test tube with a Durham tube, then one colony of bacterial isolates is cultured in De Man Rogosa and Sharpe (MRS) broth media. The media culture was then incubated for 48 hours at 37°C. Bacteria that store gas in Durham tubes are categorized as heterofermentative, and those that do not store gas are classified as homofermentative.15 The catalase test was performed by pouring two drops of 3% H2O2 onto 24 hour-old bacterial isolates on a glass slide. The development of air bubbles indicates a favorable catalase response. The development of air bubbles suggests that the bacteria create catalase, an enzyme that converts H2O2 into H2O and O2.16 Temperature Resistance Test One colony of bacterial isolates was added to De Man Rogosa and Sharpe (MRS) broth medium broth medium and incubated at 14°C and 37°C. Turbidity in the medium indicates bacterial proliferation. Temperature resistance test One bacterial isolate colony was put into De Man Rogosa and Sharpe (MRS) broth medium, which was then incubated at 14°C and 37°C. The turbidity in the media suggests bacterial growth.17 Data is presented descriptively in the form of tables and figures, with lactic acid bacteria isolates identified based on colony morphology, cell morphology, and physiological properties of bacteria indicated by biochemical test findings. To isolate molecular genetic genome DNA, bacterial isolates were placed in a microtube with 100 µl dd H2O and 1 ml 0.5% saponin in PBS 1x. The culture was incubated for 24 hours at 4°C, then centrifuged at 12000 rpm for 10 minutes. The supernatant was discarded, and 1 ml of phosphate lysis buffer was added to the mixture. After centrifugation for 5 minutes at 12000 rpm, the supernat Add 100 µl of dd H2O and 50 µl of 20% (b/v) Chelex 100. Place it in a water bath for 10 minutes. The mixture was vortexed every 5 minutes and centrifuged for 15 minutes, at a speed of 12000 rpm, then transfer the pellet from the supernatant to a microtube and add 100 µl of TE solution. Store the DNA pellets at -20°C to produce DNA stock for the quantification test step. Next, use a NanoDrop spectrophotometer to determine the amount of DNA extract. Pipette 1 µl of DNA extract and count at wavelengths of 260 and 280 nm. To amplify DNA using 16S rRNA gene primers (27F and 1491R), add 6 µl ddH2O, 12.5 µl Promega Master Mix, 2 µl forward primer, 2 µl reverse primer, and 2.5 µl DNA template to a microtube. Place the microtube on the PCR equipment and wait approximately 2 hours.18 After the PCR process, the amplification results are continued to the electrophoresis process. 4 µl of loading dye is pipetted, 6 µl of the sample is added, the solution is suspended, the solution is put into the wells using a micropipette, and 5 µl of marker is pipetted then put into the agarose gel wells. The electrophoresis device is then connected to a 100 volt power supply for ± 1 hour. The findings of electrophoresis are shown on the UV Transilluminator.19 The sequencing findings were examined bioinformatically and matched to the Gene bank (www.ncbi.nih.nim.gov) using the BLAST tool.20
Sample at the coal industry waste processing facility in Kalimantan is done using an instantaneous approach (grab sample) at a specific place in the liquid waste disposal channel before it enters the receiving waters, or the river where the effluent is disposed of. Before utilizing the sample container for sampling, the following procedure is performed: Wash the plastic container, including the cap, using a metal and phosphate-free detergent. Rinse with clean water, then wash the bottle with acid by adding 1:1 HCI into the container, twisting the top until tight, and shaking. Then, rinse the bottle with clean water before washing it again with HNO3 1:1. Finally, rinse the bottle with analyte-free water three times and let it to dry. Once dried, firmly seal the bottle and mark it “ready to use.”21 The sample procedure is carried out in accordance with SNI 6989.59:2008 for Water and Wastewater, Section 59: Wastewater sample Methods. Equipment for taking samples must fulfill numerous standards, including being built of materials that will not alter the sample’s qualities, being easy to clean, and being safe to transport. The container that will be used to keep samples is constructed of glass or plastic, can be firmly sealed, is free of pollutants, and is not readily damaged.22 Test samples were collected on August 25, 2023, between 7:00 and 8:00 WIB. Test samples were carried in containers that satisfied the 5 L sampling standards, sealed firmly, and stored in the laboratory downstairs, away from direct sunlight.
Liquid waste samples are collected and screened in the laboratory to identify the best (selected) indigenous bacteria capable of biodegrading the Cr and Pb concentrations in liquid coal waste. The screening procedure involves cultivating indigenous bacteria from liquid coal waste on solid Natrium Agar media and using Brom thymol blue as an indicator medium for the degradation ability of liquid coal waste. Change the tint from green to yellow to obtain biodegradation capabilities. The faster the color change, the greater the degrading ability of liquid coal waste (Figure 1).
Based on the color change speed test with the Brom Thymol Blue indicator, 5 isolates were identified as capable of decomposing liquid coal waste. A brighter color change suggests that the bacteria may diminish the indication. Visually assess the five bacteria that are capable of degrading; strain 5 has the quickest color change rate as shown in Table 2.
Table (2):
Screening for selected bacteria based on color changes
Natrium Broth |
Day 1 |
Day 2 |
Day 3 |
Day 4 |
|---|---|---|---|---|
The bacterial isolate no. 1 |
– |
++ |
++ |
++++ |
The bacterial isolate no. 2 |
– |
+ |
++ |
++ |
The bacterial isolate no. 3 |
– |
+ |
++ |
++ |
The bacterial isolate no. 4 |
– |
+ |
++ |
+++ |
The bacterial isolate no. 5 |
– |
++ |
+++ |
+++++ |
Based on the screening results, test bacterium No. 5 has the greatest capacity to breakdown coal waste, as evidenced by the quickest color shift from green to yellow on visual inspection using the bromine thymol blue indicator. Test bacterium No. 5 was then utilized as a degrading bacteria in a study on the cleansing of coal liquid waste with Pb and Cr characteristics.
Measurement of Pb and Cr concentration in coal liquid waste
Coal liquid waste contains the dangerous heavy elements Cr and Pb. As a result, efforts must be made to reduce Pb and Cr levels using bioremediation techniques that involve indigenous microorganisms.23 Bioremediation techniques can reduce lead and chromium concentrations in coal waste, allowing it to be safely disposed of in water systems.24
The concentrations of Pb (lead) and Cr (chromium) in this study were determined laboratory test method,25 at the Environmental Engineering testing laboratory, Diponegoro University using the atomic absorption spectrometry (AAS).
Table (3):
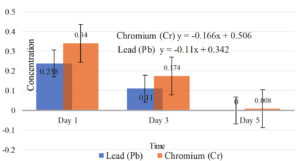
Results of Measurement of Lead (Pb) and Chromium (Cr)25
Parameter |
Day 1 |
Day 3 |
Day 5 |
Indonesian Government Regulation No. 22/2021 |
|---|---|---|---|---|
Lead (Pb) |
0.238 |
0.110 |
0.018 |
0.03 mg/L (class 3) and 0.5 mg/L (class 4) |
Chromium (Cr) |
0.340 |
0.174 |
0.008 |
0.05 mg/L (class 3 ) and 1 mg/L (class 4) |
Note: Appendix VI. Indonesian Government Regulation No 22/2021: Implementation of Environmental Protection and Management: Water Quality: Classification and Criteria26
Table 3 displays the findings of the degradation of heavy metal concentrations of Pb (lead) and Cr (chromium) in liquid coal waste during the research period by the selected and best indigenous bacteria. The success of lowering heavy metal concentrations Cr (Chromium) and Pb quality criteria before being discharged into waterways in line with Appendix VI. The Indonesian Government Regulation Number 22 of 2021: implementation of environmental protection and management. Morphology, biochemistry, and molecular biology are used to identify the finest indigenousbacteria.27,28
Table (4):
Selected Bacteria Identification Result
| Morphological and biochemical tests Bacillus sp. | Morphological and biochemical tests Chromobacterium sp. | |||||||
|---|---|---|---|---|---|---|---|---|
| Morphological form | Stem (rod) | Acid fast | – | Morphological form | Stem (rod) | Acid fast | – | |
| Gram staining | + | Tween 20 Hydrolysis | x | Gram staining | + | Tween 20 Hydrolysis | x | |
| Motility | + | Acid Form ASS Medium | Motility | + | Acid Form ASS Medium | |||
| Length 3>mm | – | a) Glucose | + | Length 3>mm | V | a) Glucose | v | |
| Spore Position and Length | VX | b) Celibiose | + | Spore Position and Length | VX | b) Celibiose | v | |
| Spores | + | c) Galactose | + | Spores | V | c) Galactose | v | |
| Growth on 50o | + | d) Mannose | + | Growth on 50o | + | d) Mannose | v | |
| Growth on 37o | + | e) Melibiose | + | Growth on 37o | + | e) Melibiose | v | |
| Growth on 42o | X | f) Rafinose | + | Growth on 42o | X | f) Rafinose | v | |
| Growth on 48o | X | g) Sucrose | + | Growth on 48o | X | g) Sucrose | v | |
| Growth with 10% NaCl | + | h) Xylose Acid Form | – | Growth with 10% NaCl | + | h) Xylose Acid Form | v | |
| Anaerobic | + | Medium | Anaerobic | + | Medium | |||
| Aerobics | + | a) Glucose | X | Aerobics | + | 1. Glucose | v | |
| ONPG | + | b) Celibiose | X | ONPG | + | 2. Celibiose | v | |
| Utilization of Citrate | + | c) Galactose | X | Utilization of Citrate | + | 3. Galactose | v | |
| Urease | – | d) Mannose | X | Urease | – | 4. Mannose | v | |
| Indol | – | e) Melibiose | X | Indol | – | 5. Melibiose | v | |
| VP (Acetoin detection) | – | f) Rafinose | X | VP (Acetoin detection) | – | 6. Rafinose | v | |
| Nitrate Reduced | – | g) Sucrose | X | Nitrate Reduced | – | 7. Sucrose | v | |
| Scratch hyde barasis stone | + | h) Xylose | X | Scratch hyde barasis stone | + | 8. Xylose | v | |
| Oxidase | + | i) Growth on Cetrimide Agar | X | Oxidase | + | 9. Growth on Cetrimide Agar | v | |
| (O/F) test | – | j) Lactose | – | (O/F) test | – | 10. Lactose | – | |
| H2S | – | k) Urease | – | H2S | – | 11. Urease | – | |
| Catalase | + | l) Yellow Pigment | – | Catalase | + | 12. Yellow Pigment | – | |
| Arginine | – | m) Lysine | – | Arginine | – | 13. Lysine | – | |
| Methyl red | – | n) Simon citrate | + | Methyl red | – | 14. Simon citrate | + | |
| Vogus proskauer | + | o) Saccharose | + | Vogus proskauer | + | 15. Saccharose | + | |
+ = Facultative anaerobic; Vx = Central oval; X = Not tested
Table (5):
Morphological and Biochemical Analysis of Test Bacteria
No. |
Parameter |
Results |
Method |
|---|---|---|---|
1 |
Bacillus sp. atau Chromobacterium sp. |
Positif |
Dinkes/ Balabkes PAK/ P/ SPO/ 03/ MB/ PK/105. Morphological and Biochemical Analysis of Test Bacteria |
Table 4 and 5 displays the morphological and biochemical analyzes carried out on the test bacteria. The results of morphological and biochemical tests were identified as Bacillus sp. or Chomobacterium sp, therefore biomolecular genetic testing is necessary.
Table (6):
Nucleic Acid (Genomic DNA) Quantification (Nanodrop) Bacteria
No. |
Nama Sample |
Sample Code |
Conc. (ng/µl) |
A260/280 |
A260/230 |
Volume (µl) |
|---|---|---|---|---|---|---|
1 |
ISW Udinus 2023 |
2776 |
158.8 |
1.92 |
2.08 |
30 |
Table 6 reveals that the DNA purity value, the ratio 260/230, is a value of 2.08 (standard 2.0 to 2.2), implying that the DNA quality is good based on the quality standards for the DNA purity value at the 260/230 ratio, namely in the range of 2.0 to 2.2.
Identification of the best selected indigenous bacteria
Morphological and biochemical analysis
Morphological observations were made on chosen indigenous bacterial isolates, including colony shape, colony edge shape, colony size and color, gram-positive and negative observations, and microscopic shape of the bacteria.29 Biochemical tests include testing the physiological characteristics of bacteria based on Bergey’s Manual of Determinative Bacteriology, as follows.30
When visual and biochemical tests on chosen indigenous bacteria using the Bacillus sp. and Chorobacterium sp. identification techniques are compared, the results are almost identical, indicating the need for molecular biological testing to validate the kind and name of the bacteria.
Molecular biology testing of the best selected indigenous bacteria
Bacterial genetic analysis-based samples Example name: ISW-BAC 1 PCR Primer: 16 seconds (27 F/1492 R) PCR Results: Bacteria Species Barcoding (~1400 bp).
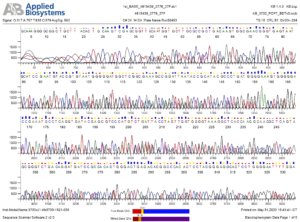
Genetic analysis approaches employ genomic DNA extraction with the Quick-DNA Bacterial Miniprep Kit (Zymo Research, D6005), PCR amplification with (2x) My Taq HS Red Mix (Bioline, BIO-25048), and bidirectional sequencing (Figure 2).
*Sequence 27F Primer:5’–AGAGTTTGATCMTGG CTCA G– 3’
**Sequence 1492R Primer: 5’–TACGGYTACCTTG TTACGACTT– 3’
Table 5 shows the quantification of nucleic acid (genomic DNA) (nanodrop), as follows:
Microorganisms examined based on Sequence Assembly Results – PCR Products, Primer Sequence 1492 R, 867bp, as follows:
Microorganisms examined based on Sequence Assembly Results – PCR Products, Primer
Sequence 1492 R, 867bp, as shown in Figure 3.
1JACCCTCTGTACCGACCATTGTATGACGTGTGAAGCCCTGGTCATAAGGGCCATGAGGACT61TGACGTCATCCCCACCTTCCTCCGGTTTGTCACCGGCAGTCCCATTAGAGTGCCCAACTT121AATGATGGCAACTAATGGCAAGGGTTGCGCTCGTTGCGGGACTTAACCCACATCTCACG181ACACGAGCTGACGACAGCCATGCAGCACCTGTGTTAACGCTCCCTTTCGGCACCGCCAC241ATCTCTGCGGTCGTCTCATTGACATGTCAAGACCAGGAAAGGTTTTTCGCGTTGCATCAA301ATTAATCCACATCATCCACCGCTTGTGCGGGTCCCCGTCAATTCCTTTGATTTTTAACCT361TGCGGCCGAACTCCCCAGGCGGTCGATTTCTGGCGTTAGCTTCGCTACCAAGGATTCAAA421CCCCCAACAGCTAGTTGACATCGTTTAAGGCGTGGACTACCAGGGTATCTAATCCTGTTT481GCTCCCCACGCTTTCGTGCATGAGCGTCAGTGTCATCCCAGGGGGCTGCCTTCGCCATCG541GTATTCCTCCGCATCTCTACGCATTTCACTGCTACACACGGAATTCTACCCCCCTCTGAC601GCACTCTAGTCTTGCAGTCTCCAATGACGCTCCCAGGTTAAGCCCGGGGCTTTCACATCA661GACTTGCAAAACCGCCTGCGCACGCTTTACGCCCAATAATTCCGAATAACGCTTGCACCC721TACGTATTACCGCGGCTGCTGGCACGTAGTTAGCCGGGGCTTATTGTTCGGTACTCTCAA781CCCCCTAGGTATTAACAATGGGATTTGCTCCCGTACAAAGTACTTTACAACCCGAAGCCT841TCTTCAACACCCGGCTGGCTGCATCAG
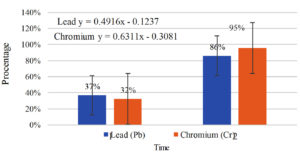
Figures 4 and 5 show the effectiveness and percentage of degradation of selected test bacteria on chromium and lead parameters in liquid coal waste.
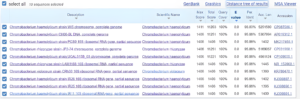
Based on Figure 6, the bacteria that has the closest relationship to the test bacteria is Chromobacterium haemolyticum strain W15 with a relationship level of 96%.
Figure 6. Shows the Phylogenetic Tree Construction of the best selected indigenous bacteria that degrade Chromium (Cr) and Lead (Pb) in liquid coal waste
Figure 7 depicts the phylogenetic tree construction of Chromobacterium haemolyticum strain W15.
Table 7 is the result of identifying selected bacteria and the best degraders of Cr and Pb based on bacterial nucleotide base sequences (molecular genetics).
Table (7):
Identification results based on bacterial nucleotide base sequences
No. Isolate ISW |
Nucleotide Length (bp) |
Bacteria Name |
Homology % |
Code |
|---|---|---|---|---|
ISW Dinus |
2776 |
Chromobacterium haemolyticum |
96% |
Strain 5 ISW |
Explanation note
a) Description is the title of the information consisting of genus, species, type of strain, DNA fragment, and the type of completeness of the DNA sequence displayed per information (partial sequence means only part of the DNA sequence of the entire gene/fragment, while full Coding Sequence (CDS) means the complete DNA sequence coding for the gene).
b) The Max Score is the total score acquired by aligning the input/query sequence with the database sequence. This value is derived via matrix computations, specifically the substitution matrix.
c) The overall score is calculated by adding all alignments between the input, query, and parallel database sequences. If there are several alignments, this value may differ from the highest score.
d) The query coverage is a percentage that reflects how large/long the input sequence is in relation to the target DNA sequence. If the DNA sequence entered corresponds to the entire target DNA sequence in the NCBI database, the percentage is 100%.
e) The E value estimates the chances that this alignment will occur by chance. A reasonable E value is around zero. As a result, it is improbable that the alignment occurred by chance.
f) Percentage identity measures how closely the input and target DNA sequences match.
g) Microorganism samples evaluated with the 16S rRNA marker are deemed identical at the species level if the “percentage identity” value exceeds 97.5%, and at the genus level if the value exceeds 95%.31
Bioremediation is an attempt to transform poor environmental circumstances or heavy metal-polluted conditions into improved environmental conditions or heavy metal-free conditions by the use of enzymatic biological agents. Chromium (Cr) and lead (Pb) are heavy metals that pollute water due to industrial activities, especially liquid coal waste. Chromium (Cr) and lead (Pb) are non-biodegradable and can accumulate in living tissue; therefore, it is very important to treat coal wastewater before it is discharged into the environment. The bioremediation technique aims to reduce Pb and Cr levels in coal-fired liquid waste.
The research results reveal that indigenous bacteria picked from coal waste may decrease Chromium (Cr) and Lead (Pb) concentrations (Figure 4 and Figure 5).
Based on the results of the effectiveness of laboratory tests, there was a reduction in the parameters Cr (98%) and Pb (108%) in liquid coal waste through a bioremediation process with the best native bacterial strains. Graph of reduction in Pb (Lead) and Chromium (Cr) concentrations in liquid coal waste. The findings of the degradation speed based on the linear regression equation demonstrate that the capacity to degrade indigenous bacteria is selected quicker on Chromium (Cr) with the linear equation y = – 0.166 x + 0.506 (95%) than on Lead (Pb) with the linear equation y = – 0.11 x + 0.342. (86%).
Identification of indigenous bacteria
The best original bacteria (isolate strain no. 5) used in the research were then identified using morphological-biochemical and molecular genetic tests. Bacterial metabolic characteristics are revealed in biochemical tests by the interaction of their metabolites with chemical reagents and their ability to use certain substances as carbon and energy sources.32 Gram staining is the first bacterial identification procedure that determines the gram type of bacteria based on the appearance of bacterial cells.33 The shapes of bacteria are round (singular: coccus, plural: cocci), rods or cylinders (singular: bacillus, plural: bacilli), and spiral, namely curved or circular rods.34 The shape and color of bacterial cells can be determined after gram staining and checking under a microscope with 100X magnification. Bacilli-shaped bacteria are bacteria that have the shape of short sticks or small rods and are cylindrical.35 The gram staining technique demonstrates that the test microorganisms are gram-positive. Gram-positive bacteria’s cell wall is made up of peptidoglycan and special components such as teichoic and teichuronic acids and polysaccharides. Gram-negative bacteria’s cell wall is also made up of peptidoglycan, but the special components are lipoprotein, outer membrane, and lipopolysaccharide.36 Gram-positive bacillus bacteria that form spores with a positive catalase test are bacillus species.37 Bacillus sp. shows positive oxidase test results because it is able to produce the enzyme cytochrome oxidase, an enzyme from the bacterial electron transport chain.38 The carbohydrate fermentation test determines the capacity of the test microorganisms to ferment certain carbohydrates. The test bacteria from the Bacillus genera were capable of fermenting glucose. Celibiose, Galactose, Mannose, Melibiose, Rafinose, and Sucrose are fermentable, although mannitol and lactose cannot be.39 The indole test determines an organism’s capacity to break down the amino acid tryptophan and make indole. The tests carried out showed negative indole test results. This shows that the test bacteria were unable to break down the amino acid tryptophan because they lacked the tryptophan enzyme which can hydrolyze the amino acid tryptophan with the indole side group.40 Triple sugar iron (TSI) agar is a differential medium that measures carbohydrate fermentation and H2S generation. Triple sugar iron (TSI) distinguishes bacteria based on their capacity to ferment lactose, glucose, and sucrose, as well as their generation of hydrogen sulfide. Bacteria from the genus Bacillus cannot manufacture H2S on differential triple sugar iron (TSI) agar media.41 Bacillus is typically catalase positive, oxidase positive, capable of metabolizing carbohydrates by fermentation, unable to make acid from mannitol, and certain Bacillus taxa are anaerobic heterofermentative. Bacillus bacteria can also metabolize carbohydrates, proteins, amino acids, and convert nitrates to nitrites. Bacillus are catalase and oxidase positive, capable of fermenting carbohydrates, unable to produce acid from mannitol, and certain Bacillus taxa are anaerobic heterofermentative. Bacillus can metabolize carbohydrates, proteins and amino acids, and convert nitrates into nitrites.42
Molecular biology tests are required to guarantee that the test microorganism is identified as Bacillus sp, because it still enables other bacteria with the same features as Bacillus.
The molecular biology test
The agarose gel electrophoresis method was used to detect PCR products containing the 16S rDNA/16S rRNA gene (PCR-amplified 16S rRNA) from the tested bacterial species. Agarose gel separation works effectively for DNA fragments ranging in size from 50 to 20,000 bp (Figure 2). The 16S rDNA/16S rRNA gene was investigated because it is conserved and is part of the structural RNA section of the ribosome, which plays a crucial role in protein synthesis. As a result, the 16S rRNA gene is always present and owned by prokaryotic organisms, is conserved, and is almost never transported horizontally, making it ideal for phylogenetic tree reconstruction and prokaryote identification.43 The ISW Udinus 2023 strain 5 bacterial genome isolation technique was marked by the creation of one band for each genome of the test bacterium after being seen using a UV transilluminator with a 1.5 kb 16S rRNA gene coding band and compared with a marker (1 kb DNA ladder). The nucleotide sequences of 16S rRNAs were sequenced and compared for similarity using GenBank and the BLAST-N (Basic Local Alignment Search Toll-Nucleotide) tool to establish the test bacteria’s homology and species. To evaluate the phylogeny and relationship with other organisms, the 16S rDNA
The 16S rDNA sequence data was then aligned with the clustalX version 2.0 program. A phylogenetic tree was generated using the neighbor-joining tree statistical technique in conjunction with a 1000-level bootstrap p-distances model. The PCR results for the 16S rDNA gene were represented by a single band on the electrophoresis gel that was around 1500 bases long.9 The bacterial nucleotide base sequence can be revealed by sequencing using forward and reverse primers, as follows:
The test bacteria with code Strain 5 ISW is a molecule type-nucleic acid with a query length of 2776. with a lineage report as follows
Kingdom : Procaryotae,
Division : Bacteria,
Class : Betaproteobacteria,
Family : Neisseriales,
Ordo : Chromobacteriaceae,
Genus : Chromobacterium
Species : Chromobacterium haemolyticum strain X
Based on phylogenetic tree analysis, strain 5 ISW (test bacteria) has a very tight connection with Chromobacterium haemolyticum strain W15, with a maximum identity of 96%. Table 6 displays ten bacteria that are closely related to the test microorganisms, as follows.
Figure 6 shows the Phylogenetic Tree Construction of the best selected indigenous bacteriathat degrade Chromium (Cr) and Lead (Pb) in liquid coal waste.
Utilization of Chromobacterium haemolyticum bacteria was successfully isolated from coal wastewater. and has the greatest ability to degrade the heavy metals chromium (Cr) and lead (Pb) contained in coal wastewater. Chromobacterium hemolyticum strain Because this bacterium is not pathogenic to the environment, its use as a bioremediation agent to degrade lead and chromium in coal wastewater is considered quite safe.
Pb (lead) and Cr (chromium) are heavy metals that accumulate in tissues over time and can impair organisms’ metabolic systems. They also have magnifying qualities because their concentration increases with trophic level in the food chain. The accumulation and magnifying process will upset the environmental balance. Bioremediation of chromium (Cr) and lead (Pb) from liquid coal waste is an attempt to remove or minimise chromium and lead pollution in the environment by utilising native (indigenous) microorganisms.
The bioremediation study of liquid coal waste using native bacteria was effective in reducing Pb and Cr levels, as well as identifying native bacteria Chromobacterium haemolyticum strain W15.
ACKNOWLEDGMENTS
The authors express their gratitude to the Health Laboratory team at Dian Nuswantoro University Semarang for their valuable contribution in facilitating the execution of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Yodha A. Sustainable development in Indonesia: holistic assessments and pathways. Massachusetts Institute of Technology. 2018.
- Ali MM, Hossain D, Al-Imran, et al. Environmental pollution with heavy metals: A public health concern. Heavy Met Environ Impacts Mitig. 2021:143-152.
- Kalisz S, Kibort K, Mioduska J, Lieder M, Malachowska A. Waste management in the mining industry of metals ores, coal, oil and natural gas-A review. J Environ Manage. 2022;304:114239.
Crossref - Luo C, Routh J, Dario M, et al. Distribution and mobilization of heavy metals at an acid mine drainage affected region in South China, a post-remediation study. Sci Total Environ. 2020;724:138122.
Crossref - Rathoure AK. Heavy metal pollution and its management: Bioremediation of heavy metal. In: Waste management: concepts, methodologies, tools, and applications. IGI Global. 2020:1013-1036.
Crossref - Tayang A, Songachan LS. Microbial bioremediation of heavy metals. Curr Sci. 2021;120(6):113891.
Crossref - Syakti AD. Marine bioremediation in indonesia: die before blossom. Omni-Akuatika. 2018;14(3):117-127.
Crossref - Thomas L, Ram H, Singh VP. Inducible cellulase production from an organic solvent tolerant Bacillus sp. SV1 and evolutionary divergence of endoglucanase in different species of the genus Bacillus. Braz J Microbiol. 2018;49(2):429-442.
Crossref - Isworo S, Oetari PS, Prabowo D, Cerlyawati H. Bioremediation Tofu Liquid Waste Based on Chemical Oxygen Demand (COD) Parameters. Annu Res Rev Biol. 2022;37(12):94-104.
Crossref - Sousa AM, Machado I, Nicolau A, Pereira MO. Improvements on colony morphology identification towards bacterial profiling. J Microbiol Methods. 2013;95(3):327-335.
Crossref - Badar RAD, Carmona JLR, Collantes JGC, et al. Staining Capability of Plant Extracts for the Identification of Gram-positive and Gram-negative Bacteria: A Systematic Review. Asian J Biol Life Sci. 2022;11(2):277-284.
Crossref - Chauhan A, Jindal T, Chauhan A, Jindal T. Biochemical and molecular methods for bacterial identification. Microbiol Methods Environ Food Pharm Anal. 2020;425-468.
Crossref - Kumar BP, Vineetha M, Satyanagalakshmi K, Reddy NS, Sparjanbabu DS. In vitro screening of indigenous competent bacterial strains for their multiple plant growth promoting attributes. Int J Trop Agric Food Sys. 2022;39(3):247-258.
- Masi C, Gemechu G, Tafesse M. Isolation, screening, characterization, and identification of alkaline protease-producing bacteria from leather industry effluent. Ann Microbiol. 2021;71:1-11.
Crossref - Soraya R. Inhibitory power and probiotic properties of lactic acid bacteria strains isolated from cow’s milk from the chlef region, Algeria. Ashok Yakkaldevi. 2022.
- Fahmi A, Syukur S, Chaidir Z, Melia SRI. Isolation and molecular characterization of probiotic from Sidamanik green tea (Camellia sinensis) fermentation. Biodiversitas J Biol Divers. 2023;24(8).
Crossref - Odafe OJ, Olorode OA. The Assessment of Palm Wine for its Composition of Probiotic Lactic Acid Bacteria and their Therapeutic Implication on Diabetic Management. Asian J Med Princ Clin Pract. 2023;6(2):366-375.
- Orlando L, Allaby R, Skoglund P, et al. Ancient DNA analysis. Nat Rev methods Prim. 2021;1(1):14.
Crossref - Masago K, Fujita S, Oya Y, et al. Comparison between fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of DNA and RNA extracted from frozen and FFPE tissues from lung cancer patients: A real-world use of genomic tests. Medicina (B Aires). 2021;57(12):1375.
Crossref - Omar SBA, Hidayan AA, Yanuarita D, Umar MT, Andriyono S. Phylogenetic Analysis of Endemic Fish from the Maros Karst Region, South Sulawesi, Indonesia. Int J Agric Biol. 2021;26(6):661-666.
Crossref - Sharga BM, Chromiak UI, Pylypiv DB, Feketa VP. Molecular Biology Practicals. 2022.
- Widiastuti A, Hartono DM, Moersidik SS, Gusniani I. Characteristics of leachate and their effect on shallow groundwater quality (case study: TPA Cipayung, Depok). IOP Conf Ser: Earth Environ Sci. 2018;120:12003.
Crossref - Li W, Feng Z, Zhu X, Gong W. Efficient removal of Cr (VI) from coal gangue by indigenous bacteria-YZ1 bacteria: Adsorption mechanism and reduction characteristics of extracellular polymer. Ecotoxicol Environ Saf. 2024;272:116047.
Crossref - Zhou B, Zhang T, Wang F. Microbial-based heavy metal bioremediation: Toxicity and eco-friendly approaches to heavy metal decontamination. Appl Sci. 2023;13(14):8439.
Crossref - Ahmed F, Fakhruddin ANM, Fardous Z, Chowdhury MAZ, Rahman MM, Kabir MM. Accumulation and Translocation of Chromium (Cr) and Lead (Pb) in Chilli Plants (Capsicum annuum L.) Grown on Artificially Contaminated Soil. Nat Environ Pollut Technol. 2021;20(1):63-70.
Crossref - Susanto A, Hasari AA, Putro EK, Yochu WE, Mahlisa R, Manuel AA. Identification and management of Toxic & Hazardous Wastes (THW) based on the Indonesian Government Regulation Number 22 of 2021. IOP Conf Ser: Earth Environ Sci. 2023;1180:12023.
Crossref - Larbi-Koranteng S, Awuah RT, Kankam F, et al. Morphological, biochemical and molecular identification of rhizobacteria isolates with potential for biocontrol of fungal plant pathogens. Arch Phytopathol Plant Prot. 2021;54(17-18):1346-1359.
Crossref - Bekele GK, Gebrie SA, Mekonen E, et al. Morphological, biochemical and molecular identification of rhizobacteria isolates with potential for biocontrol of fungal plant pathogens. Int J Microbiol. 2022;2022:5655767.
Crossref - Fibriarti BL, Feliatra F, Amin B, Darwis D. Biodegradation of LDPE plastic by local strain of Bacillus sp. isolated from dump soil of Pekanbaru, Indonesia. Biodiversitas J Biol Divers. 2021;22(12).
Crossref - Bergey’s Manual of Determinative Bacteriology and Bergey’s International Society for Microbial Systematics (BISMiS): past, present and future. Acta Microbiol Sin. 2023;63(5):1714–23.
- Hadi SN, Dewi PS, Widiyawati I, Ahadiyat YR. The Characteristics of Buprofezin Resistant Bacteria From Cassava Rhizosphere. In: 3rd International Conference on Sustainable Agriculture for Rural Development (ICSARD 2022). Atlantis Press. 2023:373-82.
Crossref - Miglani R, Parveen N, Kumar A, et al. Degradation of xenobiotic pollutants: an environmentally sustainable approach. Metabolites. 2022;12(9):818.
Crossref - Ogodo AC, Agwaranze DI, Daji M, Aso RE. Microbial techniques and methods: Basic techniques and microscopy. Analytical Techniques in Biosciences. 2022:201-220.
Crossref - Bruns MA. Bacteria and archaea. Principles and Applications of Soil Microbiology. 2021:111-148.
Crossref - Yaqoob AA, Bakar MABA, Kim H-C, Ahmad A, Alshammari MB, Yaakop AS. Oxidation of food waste as an organic substrate in a single chamber microbial fuel cell to remove the pollutant with energy generation. Sustain Energy Technol Assessments. 2022;52(Part C):102282.
Crossref - Legg M. Advancing understanding of secondary cell wall polymer binding and synthesis in S-layers of Gram-Positive bacteria. 2022.
- Cote CK, Heffron JD, Bailey SO, Welkos SL, Bozue JA. Bacillus anthracis and other Bacillus species. Mol Med Microbiol. 2024:1681-1742.
Crossref - Hederstedt L. Molecular biology of Bacillus subtilis cytochromes anno 2020. Biochem. 2021;86(1):8-21.
Crossref - Ghosh S, Chatterjee S. Isolation and characterization of Bacillus tequilensis from gut content of Perionyx excavatus and evaluation of its starch hydrolyzing property. Biosc Biotech Res Comm. 2020;13(2):670-675.
Crossref - Sunkad G, Khadarbi, Patil MS, Joshi R. Exploration of the Potential of Bacillus spp. as an Antagonist and PGPR against Stem and Pod Rot of Groundnut. Legum Res. 2023;46(11):1501-1509.
Crossref - Sayaniya AV, Patel P. Isolation and Characterization of Detergent Compatible Alkaline Protease Producing Bacillus Subtilis APO-1. Biomed J Sci Tech Res. 2021;35:27949-55.
Crossref - Fusco V, Chieffi D, Fanelli F, Montemurro M, Rizzello CG, Franz CMAP. The Weissella and Periweissella genera: up-to-date taxonomy, ecology, safety, biotechnological, and probiotic potential. Front Microbiol. 2023;14:1289937.
Crossref - Kabir MS, Tasmim T. Isolation of pectinase producing bacteria from the rhizosphere of Andrographis paniculata nees and 16S rRNA gene sequence comparison of some potential strains. Adv Microbiol. 2019;9(1):1-13.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.