ISSN: 0973-7510
E-ISSN: 2581-690X
Food is the primary cause for diseases in humans and carries high risk pathogens. Assessment of the safety in foods is needed to validate the presence of pathogenic bacteria. We used colony PCR for this approach to detect foodborne pathogens such as Escherichia coli, Lactobacillus and Bacillus cereus. Suitable primers were selected based on specific gene 1040 for Escherichia coli, gene S2 for Lactobacillus, and gene NVF for Bacillus cereus. Agarose gel electrophoresis is used for the detection of amplified products against a suitable marker. ImageJ is used for DNA band analysis, enabling precise quantification, normalization, and statistical comparisons. These studies have established a promising role in the detection of pathogens in various environmental samples. The insights gained from this study may serve as the foundation for rapid detection of foodborne diseases in the food industry.
Colony PCR, Primers, Escherichia coli, Lactobacillus, Bacillus cereus
Food technologists are currently showing their interest in molecular methods as a powerful tool for identifying and detecting contaminants in food specimens.1 Colony PCR Analysis is one of the widely used techniques that utilizes the principles of polymerase chain reaction (PCR) to observe contaminants at the molecular level.2,3 Powerful screening techniques are necessary as demand for the safety and integrity of food products has shot up due to the expansion of the food business globally.4 Colony PCR is cost-effective for detecting bacterial contamination because it directly amplifies DNA from colonies, avoiding expensive steps like DNA extraction and purification required in traditional methods. This reduces overall expenses while maintaining accuracy in identifying contaminants.5 One of the main important concerns for food manufacturers, agencies, and customers is quality and safety.6 Bacteria, fungi, viruses, toxins, and chemical residues pose hazardous health risks and economic implications.7 Conventional detection methods consume a lot of time and as well as money with hard labor work, which may lack sensitivity, specificity, or the capacity to effectively detect emerging pathogens.8,9 Furthermore, these techniques may not support rapid detection, identification, and delay in responses which can increase risk factors in public health.10 In response to these challenges, molecular techniques have transformed food safety by offering sensitive, specific, and rapid means of detecting and identifying contaminants.11 Analysis by colony PCR in food safety involves a broad spectrum of contaminants and food pathogens such as Salmonella, Escherichia coli, Listeria, Lactobacillus, Bacillus cereus and Staphylococcus due to their prevalence in foodborne infections.12,13 Fungal contaminants also exhibit health effects such as Aspergillus, Penicillium, and Fusarium species, which produce mycotoxins, are also investigated.14,15 Colony PCR offers rapid, sensitive and diverse solutions for detecting and identifying various contaminants in foods.16,17 With the advent increase in use of molecular approach methods in the field of molecular biology and recombinant technology, screening of contaminants in colony is possible with the use of primers in bacterial culture.18 This method offers quick identification of pathogens and the results are promising among the other methods in sensitivity and specificity.19
Preparation of food samples
Food samples were collected from the canteen and supermarkets of the KL University campus, Andhra Pradesh, India. The food specimens were collected in 5 ml screw cap tubes. Sample is collected carefully and sealed with parafilm until it is spread over agar plates. The list of food samples is shown in Table 1.
Table (1):
List of food samples collected from various sources
| 1-Chicken | 2-Biryani | 3-Noodles | 4-Pongal | 5-Tamrid Rice |
| 6-Jaggery | 7-Corn Flakes | 8- White Bread | 9-Palak Curry | 10-Corn |
| 11-Biscuit Cream | 12-Kaju Katle | 13-Canta Loupe | 14-Pineapple | 15-Mousambi |
| 16-Raw Milk | 17- Muskmelon | 18-Sapodilla | 19-Butter | 20-Brinjal Curry |
| 21-Potato Curry | 22-Capsicum Curry | 23-Tomato | 24-Ivy Gourd | 25-Sweet Neem Leaf |
| 26-Carrot Curry | ||||
Identification of food contaminants
Samples were collected in vials, and double-distilled water was added to dilute them. Following dilution, the samples were streaked onto LB agar plates, which were then incubated for 24 hours. After incubation, bacterial growth was observed, and colonies were collected into PCR tubes, to which master mix was added.20 Bacterial identification is accomplished using primers designed for specific genes at specific temperatures 56°C for Escherichia coli, 55.4°C for Lactobacillus and 54°C for Bacillus cereus which facilitate DNA amplification.
Forward Primer |
Backward Primer |
|
|---|---|---|
Escherichia coli |
AACTGCAGATGCCCAAATCCAACGATG |
GGTACCTCAATCGTGATTTACTGAGAGA |
Lactobacillus |
ATGATTGATCTAACGAGTCGATTG |
TTATTTGGCGGTGTAGGTGG |
Bacillus cereus |
CGGCGGCAACTACGAGAC |
CCGGTGATGCTGTCGCTCTCC |
Colony PCR process
PCR Cocktail- 2X Master mix- 5 µl Forward Primer- 0.5 µl Backward Primer- 0.5 Nuclease Free Water- 4. PCR Program- Initial Denaturation (94° for 5 min), Final Denaturation ( 94° for 1 min), Annealing Temperature (55.4°, 54°, 56° for), Extension ( 72° for 1 min), Final Extension ( 72° for 5 min). The PCR was performed for 35 cycles.
Characterization of contaminant by AGE
The samples after PCR were taken out and loaded into 1% agarose gel after the electrophoresis the gel was observed under gel doc.21
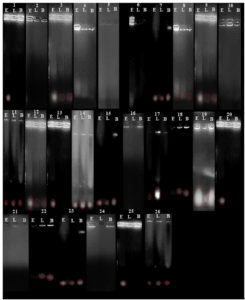
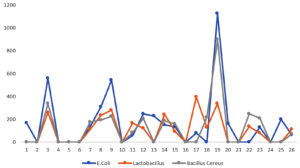
The assessment of specific bacterial strains in food samples was conducted by observing bands in gel electrophoresis. The presence of bands serves as an indicator of successful primer annealing to bacterial genomic DNA, resulting in amplification. This process allows for the identification of specific bacteria targeted by each primer set. Conversely, the absence of bands in certain gels suggests the non-existence of the targeted bacteria in those food samples. This absence might be attributed to the presence of other bacterial strains not covered by the three different primers designed for the specific bacteria. The gel electrophoresis results are visually represented in Figure 1. For quantitative analysis of the amplified bands, ImageJ software was utilized to measure the band areas, and the corresponding values are presented in Table 2. A graphical representation of the relationship between food sample and the area of the bands is shown in Figure 2. This graphical analysis offers a comprehensive insight into the bacterial composition across various food samples, emphasizing differences in band areas and facilitating the identification of specific bacterial strains. The graph indicates that E. coli exhibits higher intensity compared to Lactobacillus and Bacillus cereus. Bacillus cereus demonstrates the lowest intensity among the three bacterial strains.
Table (2):
The dataset comprises information regarding food samples and their corresponding band areas
Sample. No |
E. Coli |
Lacto-bacillus |
Bacillus cereus |
|---|---|---|---|
1 |
166 |
0 |
0 |
2 |
0 |
0 |
0 |
3 |
560 |
260 |
336 |
4 |
0 |
0 |
0 |
5 |
0 |
0 |
0 |
6 |
0 |
0 |
0 |
7 |
138 |
112 |
177 |
8 |
309 |
235 |
194 |
9 |
543 |
280 |
225 |
10 |
0 |
0 |
0 |
11 |
58 |
166 |
84 |
12 |
248 |
120 |
208 |
13 |
228 |
0 |
0 |
14 |
151 |
244 |
186 |
15 |
133 |
92 |
160 |
16 |
0 |
0 |
0 |
17 |
78 |
396 |
0 |
18 |
0 |
128 |
216 |
19 |
1123 |
336 |
898 |
20 |
164 |
0 |
0 |
21 |
0 |
0 |
0 |
22 |
0 |
136 |
247 |
23 |
130 |
79 |
210 |
24 |
0 |
0 |
0 |
25 |
199 |
0 |
0 |
26 |
64 |
112 |
70 |
Figure 1. Amplicon bands were observed for 26 contaminated food samples. (Chicken Curry, Biryani, Noodles, Pongal, Tamrid Rice, Jaggery, Corn Flakes, Bread, Palak Curry, Sweet Corn, Cream Biscuit, Kaju Katle, Canta Loupe, Pineapple, Mousambi, Raw Milk, Muskmelon, Sapodilla, Butter, Brinjal Curry, Potato Curry, Capsicum Curry, Tomato, Ivy Gourd, Sweet Neem Leaf, Carrot Curry). Here E- Escherichia coli, L- Lactobacillus, B- Bacillus cereus
In this research endeavour, colony PCR was conducted on a total of 26 food samples suspected of contamination. The DNA extraction and amplification process were conducted using PCR for 35 cycles. To verify the presence of PCR amplicons, 12 µl of the PCR product was loaded onto a 1% agarose gel. This investigation introduces a rapid method for detecting contaminants in food samples. However, for enhanced accuracy and reliability, it is advisable to optimize further and validate the colony PCR technique, particularly when dealing with larger sample sets. Additional efforts towards optimization and validation can contribute to the robustness and effectiveness of the colony PCR method in identifying contaminants, thereby strengthening its utility in food safety and quality assurance protocols.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not Applicable.
- Zhang M, Wu J, Shi Z, et al. Molecular Methods for Identification and Quantification of Foodborne Pathogens. Molecules. 2022;27(23):8262.
Crossref - Sanz M, Lau L, Herrera D, Morillo JM, Silva A. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a review. J Clin Periodontol. 2004;31(12):1034-1047.
Crossref - Bergkessel M, Guthrie C. Colony PCR. Methods Enzymol. 2013;529:299-309.
Crossref - Rychlik M, Zappa G, Anorga L, et al. Ensuring Food Integrity byetrology and FAIR Data Principles. Front Chem. 2018;6:49.
Crossref - Nakano S, Kobayashi T, Funabiki K, Matsumura A, Nagao Y, Yamada T. Development of a PCR assay for detection of Enterobacteriaceae in foods. J Food Prot. 2003;66(10):1798-1804.
Crossref - Lin P, Tsai H, Ho T. Food Safety Gaps between Consumers’ Expectations and Perceptions: Development and Verification of a Gap-Assessment Tool. Int J Environ Res Public Health. 2020;17(17):6328.
Crossref - Gizaw Z. Public health risks related to food safety issues in the food market: a systematic literature review. Environ Health Prev Med. 2019;24(1):68.
Crossref - Kabiraz MP, Majumdar PR, Mahmud MMC, Bhowmik S, Ali A. Conventional and advanced detection techniques of foodborne pathogens: A comprehensive review. Heliyon. 2023;9(4):e15482.
Crossref - Law JW, Ab Mutalib NS, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2015;5:770.
Crossref - Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4(6):337-348.
Crossref - Engleberg NC, Eisenstein BI. Detection of microbial nucleic acids for diagnostic purposes. Annu Rev Med. 1992;43:147-55.
Crossref - Aladhadh M. A Review of Modern Methods for the Detection of Foodborne Pathogens. Microorganisms. 2023;11(5):1111.
Crossref - Moseley SL, Huq I, Alim ARMA, So M, Samadpour-Motalebi M, Falkow S. Detection of enterotoxigenic Escherichia coli by DNA colony hybridization. J Infect Dis. 1980;142(6):892-898.
Crossref - Paterson RRM, Lima N. Filamentous Fungal Human Pathogens from Food Emphasising Aspergillus, Fusarium and Mucor. Microorganisms. 2017;5(3):44.
Crossref - Yu J, Pedroso IR. Mycotoxins in Cereal-Based Products and Their Impacts on the Health of Humans, Livestock Animals and Pets. Toxins (Basel). 2023;15(8):480.
Crossref - Zhang D, Lu X, Liao Y, et al. Rapid and Simple Detection of Trichosporon asahii by Optimized Colony PCR. Biomed Res Int. 2019;2019:1803278.
Crossref - Woodman ME, Savage CR, Arnold WK, Stevenson B. Direct PCR of Intact Bacteria (Colony PCR). Curr Protoc Microbiol. 2016;42:A.3D.1-A.3D.7.
Crossref - Santis G, Evans TW. Molecular biology for the critical care physician. Part II: where are we now? Crit Care Med. 1999;27(5):997-1003.
Crossref - Nomura S, Sukowati EW, Shigeno Y, et al. Blautia coccoides JCM1395T Achieved Intratumoral Growth with Minimal Inflammation: Evidence for Live Bacterial Therapeutic Potential by an Optimized Sample Preparation and Colony PCR Method. Pharmaceutics. 2023;15(3):989. MC10058202.
Crossref - Patil-Joshi A, Rangaswamy BE, Apte-Deshpande A. Paper-based PCR method development, validation and application for microbial detection. J Genet Eng Biotechnol. 2021;19(1):37.
Crossref - Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp. 2012;(62):3923.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.