ISSN: 0973-7510
E-ISSN: 2581-690X
Antarctica is renowned as the most inhospitable environment where microorganisms are thriving in the frontiers of life. In the past few years, many novel bacterial species have been reported from the Antarctic environment. During taxonomic re-evaluation of novel bacterial species from Antarctica, it was noticed that Kocuria polaris shared high 16S rRNA gene sequence similarity with Kocuria rosea. In the present study, the taxonomic position, metabolic potentials, and stress survival strategy of K. polaris were evaluated through genome analysis. K. polaris encodes genes for glycolysis, citrate cycle, pentose phosphate pathway, dissimilatory nitrate reduction, assimilatory sulfate reduction, etc. In addition, K. polaris also encodes genes for cold and salt stress. The 16S rRNA gene sequence extracted from K. polaris and K. rosea genomes showed 99.7% similarity. In the phylogenomic tree, K. polaris and K. rosea clustered together. The average nucleotide identity and digital DNA–DNA hybridization values between K. polaris and K. rosea exceeded the threshold (95-96% for ANI and 70% for dDDH) value for distinguishing species, showing that they are similar species. The present study shed light on K. polaris survival strategy in extreme conditions. We further propose to reclassify Kocuria polaris as a later heterotypic synonym of Kocuria rosea.
Kocuria polaris, Kocuria rosea, Reclassification, Heterotypic Synonym, Metabolic Potentials
Antarctica is renowned as one of the planet’s most inhospitable environments, defined by its extreme cold, arid conditions, relentless winds, intense UV radiation, and extremes of light and darkness.1 Despite being on the frontiers of life´s limits, the frozen realm likely harbors an extensive and undiscovered array of microorganisms.2 The distinctive and inhospitable environment leads to the preferential selection of microbial species exhibiting atypical metabolic capabilities and/or the synthesis of uncommon metabolites and substances.3,4 In the past few years, many novel microbial species from the Antarctic environment have been reported.5-7 In this regard, a Gram-positive, orange-pigmented novel psychrophilic bacterium Kocuria polaris (K. polaris) was reported from an Antarctic cyanobacterial mat.8 During the taxonomic assessment of Antarctic bacteria, an intriguing observation emerged, K. polaris exhibited a high degree of 16S rRNA gene sequence similarity to Kocuria rosea (K. rosea). As a result, this study aims to provide a comprehensive clarification of the taxonomic classification of K. polaris employing genome analysis. Further, its metabolic potential and survival strategy in the cold environment were evaluated through genome analysis.
Genome attributes
To evaluate the taxonomic position of K. polaris, all the type species genomes of the genus Kocuria were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). Nesterenkonia flava CCTCC AB 207010T (GCF_031432335.1) genome was also downloaded to use as an outgroup for the construction of a phylogenomic tree. The quality of the genomes was evaluated using CheckM.9 Since K. polaris exhibited a high degree of 16S rRNA gene sequence similarity to K. rosea their genomes were visualized and compared using Proksee.10,11 The tRNAs were predicted using tRNAscan-SE.12 Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values were determined to evaluate the genomic relatedness between K. polaris and K. rosea. Pyani with ANIm parameter.13 and the Genome-to-Genome Distance Calculator (http://ggdc.dsmz.de/ggdc.php version 3.0; local alignment tool BLAST+ using formula 2).14,15 were used to estimate the ANI and dDDH values, respectively.
Phylogenomic tree construction
Phylogenomic tree was constructed using the Anvi’o tool (version 7.1).16,17 The process of converting FASTA files into contigs-db and identifying open reading frames and matching genes in the contigs to single-copy core genes was carried out using the program anvi-gen-contigs-database and anvi-run-hmms.18,19 The genes present in HMM source ‘Bacteria 71’ 20 were taken and aligned using MUSCLE.21 The generated tree was displayed using MEGA version 7.0.22
16S rRNA gene comparison and functional annotation
To compare K. polaris and K. rosea the 16S rRNA gene from the genomes was extracted using Tthe script “anvi-get-sequences-for-hmm-hits” (hmm-source Ribosomal RNA 16S) (https://github.com/tseemann/barrnap). The EzBioCloud server’s pairwise alignment function was used to assess the 16S rRNA gene (extracted from the genome) sequence similarity between K. polaris CMS 76orT and K. rosea ATCC 186T (www.ezbiocloud.net/tools/pairAlign). To evaluate K. polaris metabolic potentials and survival strategy in the cold environment functional annotation was performed by KofamKOALA23 using the anvi-run-kegg-kofams program.
The genus Kocuria was proposed by Stackebrandt et al.24 and at the time of writing the genus includes 26 validly published species names.25 Among Kocuria species, K. polaris was isolated from the Antarctic cyanobacterial mat, and as mentioned above it was reported to share high 16S rRNA gene sequence similarity with the type strain of K. rosea,8 hence the present study evaluates its taxonomic position. We further evaluated the metabolic potentials and survival strategy of K. polaris in a cold environment.
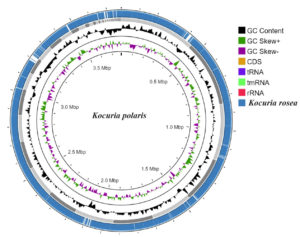
The genome size of K. polaris CMS 76orT was 3779800 (bp) with 72.8% G+C content while the genome size of K. rosea ATCC 186T was 3946651 (bp) with 72.7% G+C content. The genome completeness of K. polaris CMS 76orT and K. rosea ATCC 186T was 99.1 and 98.6%, respectively with zero contamination. A total of 48 tRNAs were predicted in both K. polaris CMS 76orT and K. rosea ATCC 186T. The graphical representation of the genome’s comparison is mentioned in Figure 1.
Metabolic potential and survival strategy of K. polaris
K. polaris CMS 76orT encodes genes for glycolysis, citrate cycle, and pentose phosphate pathway. Nitrate stands as the most highly oxidized variant among fixed nitrogen compounds, constituting a vital nutrient crucial for the sustenance of microbial and plant life.26 In prokaryotes, dissimilatory nitrate reduction mechanisms have been extensively explored.26-28 Reduction of nitrate to nitrite by respiratory membrane-bound NarG or periplasmic nitrate reductase NapA is the first step in dissimilatory nitrate reduction. Nitrite is next reduced to ammonia by cytoplasmic nitrite reductase NirB or periplasmic nitrite reductase NrfA.27 In the present study, the genes encoding dissimilatory nitrate reduction (NarGHI and NirBD) were noticed in K. polaris CMS 76orT. In addition, genes related to nitrate assimilation were also noticed in K. polaris CMS 76orT.
Microorganisms use assimilatory sulfate reduction to convert inorganic sulfate to sulfide.29 In the present study, it was noticed that K. polaris CMS 76orT encodes genes (CysND, CysH and Sir) for assimilatory sulfate reduction. K. polaris CMS 76orT encodes genes for various amino acid metabolism (like betaine, methionine, lysine, ornithine, arginine biosynthesis, etc). A detailed list of metabolic potentials of K. polaris CMS 76orT is mentioned in Table S1.
Low temperatures significantly limit cellular function by impacting cell structure, water thickness, solute movement, membrane flexibility, enzyme activity, and large molecule interactions.30 Microorganisms that survive in cold environments rely on adaptive strategies to keep their fundamental cellular processes intact.30 In reaction to a quick temperature drop, many bacteria produce small cold shock proteins.31 The cold-shock protein, CspA, was found to be significantly upregulated during the cold-shock response.32 In the present study, CspA was also noticed in K. polaris CMS 76orT. Universal stress proteins (USP) are key regulatory stress proteins that help bacteria survive in stressful environments.33 In the present study, USP (ABCDEFG) was noticed in K. polaris CMS 76orT. Trehalose production was reported to play a significant role in resistance to freezing in cold environments.34 In the present study, The genes encoding trehalose biosynthesis were noticed in K. polaris CMS 76orT.
A sudden drop in temperature can cause phase separation of cell membrane phospholipids, resulting in decreased membrane fluidity and increased permeability.35 Palmitoleate has been shown to increase the flexibility of cell membranes while decreasing the temperature at which phase transition occurs. This helps to mitigate the adverse effects of cold temperatures. When the temperature drops, in certain bacteria, the cold-induced acyltransferase LpxP induces the attachment of palmitoleate to lipid A.36 The adaptation of membrane fluidity also involves the fast desaturation of fatty acids in pre-existing phospholipids. This is accomplished by the activation of fatty acid desaturase (Des), which is regulated by the sensor kinase DesK and the response regulator DesR.37 Genome analysis of K. polaris CMS 76orT revealed the presence of LpxP, DesK and DesR.
K. polaris CMS 76orT was also reported to tolerate NaCl up to 2.9%.8 Salt-in and salt-out mechanisms help microorganisms regulate osmoregulation.38,39 Microorganisms employ the salt-in strategy to maintain osmotic equilibrium through the accumulation of large amounts of inorganic salts or ions within their cytosol.38,39 The salt-out strategy entails the removal of salt ions from the cytoplasm while concurrently accumulating large amounts of compatible solutes.38-40 Genome analysis of K. polaris CMS 76orT revealed the presence of genes related to potassium uptake protein (Trk system), and multicomponent Na+:H+ antiporter. In addition, it was also noticed that K. polaris CMS 76orT encodes genes for compatible solutes like betaine, proline, and trehalose.
Taxonomic position re-evaluation
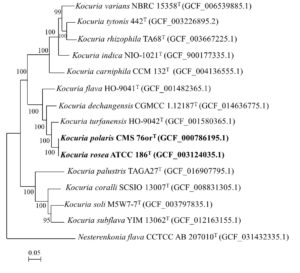
In the present study, the 16S rRNA gene sequence extracted from K. polaris CMS 76orT and K. rosea ATCC 186T genome showed 99.7% similarity to each other. Even in the original article K. polaris CMS 76orT was reported to share 99.8 and 71% 16S rRNA gene sequence and DNA–DNA hybridization (DDH) similarity with K. rosea ATCC 186T, respectively.8 The 16S rRNA gene sequence similarity was above the threshold value (98.7%), while the DDH value was close to the threshold value (70%) for species delineation.41 In the phylogenomic tree (Figure 2), K. polaris CMS 76orT and K. rosea ATCC 186T clustered together.
Figure 2. Phylogenomic tree (based on 71 bacterial single-copy genes) showing the relationships of Kocuria polaris. Bootstrap values greater than 50% are shown at branch points. Bar, 0.05 substitutions per nucleotide position
The proposed cut-off values for ANI and dDDH values for species delineation were 95-96% and 70%, respectively.14,42,43 The ANI value and dDDH value between K. polaris CMS 76orT and K. rosea ATCC 186T were 98.7 and 87.6%, respectively which were above the cut-off value indicating they are the same species. Based on the above results we propose to reclassify Kocuria polaris as a later heterotypic synonym of Kocuria rosea.
In the present study, the survival strategy under cold stress, metabolic potential, and taxonomic position of K. polaris was evaluated through genome analysis. K. polaris encodes genes for glycolysis, citrate cycle, pentose phosphate pathway, dissimilatory nitrate reduction, assimilatory sulfate reduction, etc. To overcome cold stress, K. polaris encodes genes for cold shock proteins, universal stress proteins, and mechanisms to enhance membrane fluidity. In addition, it also encodes genes related to potassium uptake protein, multicomponent Na+:H+ antiporter, and genes related to the synthesis of compatible solutes like betaine, proline, and trehalose involved in overcoming salt stress. The ANI, dDDH, and phylogenomic analysis suggest that K. polaris and K. rosea are similar species.
SUPPLEMENTARY INFORMATION
Additional file: Additional Table S1.
ACKNOWLEDGMENTS
None.
FUNDING
This study was funded by the Deanship of Scientific Research at Northern Border University, Arar, Kingdom of Saudi Arabia, through project number NBU-FFR-2024-2046-04.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by the author.
- Severin I, Stal LJ. Diazotrophic Microbial Mats. In Seckbach J, Oren A (eds.), Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Springer Netherlands, Dordrecht. 2010:321-339.
Crossref - Antony R, Sanyal A, Kapse N, Dhakephalkar PK, Thamban M, Nair S. Microbial communities associated with Antarctic snow pack and their biogeochemical implications. Microbiol Res. 2016;192:192-202.
Crossref - Doytchinov VV, Dimov SG. Microbial Community Composition of the Antarctic Ecosystems: Review of the Bacteria, Fungi, and Archaea Identified through an NGS-Based Metagenomics Approach. Life. 2022;12(6):916.
Crossref - Silva TRe, Silva LCF, de Queiroz AC, et al. Pigments from Antarctic bacteria and their biotechnological applications. Crit Rev Biotechnol. 2021;41(6):809-826.
Crossref - Noh HJ, Park Y, Yang J, Jang S, Lee H, Lee YM. Polymorphobacter megasporae sp. nov., isolated from an Antarctic lichen. Int J Syst Evol Microbiol. 2022;72(9).
Crossref - Guo XH, Wang N, Yuan XX, et al. Poseidonibacter antarcticus sp. nov., isolated from Antarctic intertidal sediment. Int J Syst Evol Microbiol. 2019;69(9):2717-2722.
Crossref - Bozal N, Tudela E, Rossello-Mora R, Lalucat J, Guinea J. Pseudoalteromonas antarctica sp. nov., isolated from an Antarctic coastal environment. Int J Syst Bacteriol. 1997;47(2):345-351.
Crossref - Reddy GSN, Prakash JSS, Prabahar V, et al. Kocuria polaris sp. nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int J Syst Evol Microbiol. 2003;53(1):183-187.
Crossref - Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043-1055.
Crossref - Grant JR, Enns E, Marinier E, et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023;51(W1):W484-W492.
Crossref - Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-410.
Crossref - Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955-964.
Crossref - Pritchard L, Glover RH, Humphris S, John G, Elphinstone, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8(1):12-24.
Crossref - Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60.
Crossref - Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Goker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022;50(D1):D801-d807.
Crossref - Eren AM, Esen OC, Quince C, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. Peer J. 2015;3:e1319.
Crossref - Eren AM, Kiefl E, Shaiber A, et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2021;6(1):3-6.
Crossref - Hyatt D, Chen GL, Locascio PF, Miriam Land L, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Crossref - Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7(10):e1002195.
Crossref - Lee MD. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. 2019;35(20):4162-4164.
Crossref - Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-1797.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Aramaki T, Blanc-Mathieu R, Endo H, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36(7):2251-2252.
Crossref - Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Bacteriol. 1995;45(4):682-692.
Crossref - Parte AC, Sarda Carbasse J, Meier-Kolthoff JP, Reimer LC, Goker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607-5612.
Crossref - Kamp A, Hogslund S, Risgaard-Petersen N, Stief P. Nitrate Storage and Dissimilatory Nitrate Reduction by Eukaryotic Microbes. Front Microbiol. 2015;6:1492.
Crossref - Sun Y, De Vos P, Willems A. Influence of nitrate and nitrite concentration on N2 O production via dissimilatory nitrate/nitrite reduction to ammonium in Bacillus paralicheniformis LMG 6934. Microbiologyopen. 2018;7(4):e00592.
Crossref - Keren R, Lawrence JE, Zhuang W, et al. Increased replication of dissimilatory nitrate-reducing bacteria leads to decreased anammox bioreactor performance. Microbiome. 2020;8(1):7.
Crossref - Koprivova A, Melzer M, von Ballmoos P, Mandel T, Brunold C, Kopriva S. Assimilatory Sulfate Reduction in C3, C3-C4, and C4 species of Flaveria. Plant Physiol. 2001;127(2):543-550.
Crossref - De Maayer P, Anderson D, Cary C, Cowan DA. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 2014;15(5):508-517.
Crossref - Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindstrom M, Korkeala H. Cold shock proteins: A minireview with special emphasis on csp-family of enteropathogenic Yersinia. Front Microbiol. 2016;7:1151.
Crossref - Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272(1):196-202.
Crossref - Nachin L, Nannmark U, Nystrom T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. 2005;187(18):6265-6272.
Crossref - Park C, Park W. Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential. Front Microbiol. 2018;9:1081.
Crossref - Barria C, Malecki M, Arraiano CM. Bacterial adaptation to cold. Microbiology. 2013;159(Pt 12):2437-2443.
Crossref - Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ, Raetz CRH. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12°C. J Biol Chem. 2002;277(16):14186-14193.
Crossref - Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, et al. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. Embo J. 2001;20(7):1681-1691.
Crossref - Saini A, Kumar A, Singh G, Giri S. Survival Strategies and Stress Adaptations in Halophilic Archaebacteria. In Dhiman SS, Gnimpieba EZ, Gadhamshetty V (eds.), Microbial Stress Response: Mechanisms and Data Science, American Chemical Society, Washington, D.C. 2023;1-21.
Crossref - Weinisch L, Kuhner S, Roth R, et al. Identification of osmoadaptive strategies in the halophile, heterotrophic ciliate Schmidingerothrix salinarum. PLoS Biol. 2018;16(1):e2003892.
Crossref - Liu KH, Ding XW, Narsing Rao MP, et al. Morphological and transcriptomic analysis reveals the osmoadaptive response of endophytic fungus Aspergillus montevidensis ZYD4 to high salt stress. Front Microbiol. 2017;8:1789.
Crossref - Chun J, Oren A, Ventosa A, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68(1):461-466.
Crossref - Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(1):81-91.
Crossref - Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126-19131.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.