ISSN: 0973-7510
E-ISSN: 2581-690X
Nine actinobacterial isolates were purified from the sediment sample of Kogilvai village, Warangal, Telangana, based on their capability to grow on the minimal medium with Ferulic acid (FA) as only Carbon (C) source. FA to Vanillin conversion capacity of these isolates was identified by Thin-layer chromatography (TLC). Biotransformation of FA to Vanillin was high by four isolates, S1, S3, O3 and O4 when compared to other five isolates (O1, O2, S2, S4 and S5) with initial pH 7 in basal medium. Among these four isolates, optimal and rapid FA to Vanillin bioconversion of 140 mg/L was shown by isolate S1 with UV-spectrophotometry. Its conversion was confirmed by High Performance Liquid Chromatography (HPLC) analysis with retention time of 2.9 min after 28hrs of incubation at 37°C with 1g/L ferulic acid in the medium with 150 rpm. Isolate S1 could utilize Lactose, Maltose, Glycerol, Fructose, Galactose, Sucrose, Dextrose, L-Arabinose, ONPG, Esculin and not other carbohydrates present in the Himedia Hicarbo kit. Molecular characterization showed that 16S rDNA gene sequence of isolate S1 was 98.27% similar to Limosilactobacillus fermentum CECT 562 with completeness of 96.7% and was identified as Limosilactobacillus sp. 16S rDNA gene sequence of isolate S1 was submitted to NCBI GenBank and its accession number was OR136396. As this isolate has high potential of FA to Vanillin biotransformation capacity, it can be further explored to be used for industrial setups for commercial exploitation.
Actinobacteria, Ferulic Acid, Molecular Characterization, High Performance Liquid Chromatography
Vanillin (4-hydroxy-3-methoxy benzaldehyde) & its related compounds have significant economic significance due to its rising demand and cost. It has numerous applications in the pharmaceutical, fragrance and food industries.
Vanillin is frequently made using environmentally harmful methods and also involves no substrate selectivity. Even though direct extraction was used for natural vanillin production from vanilla costs more, it is nevertheless sought after on the global market.1 There are numerous issues with vanilla extraction, including low concentration, which raises the cost of the extraction method.2,3
Microbial manufacturing is a potential and developing field for the creation of natural flavors. Recent advances have been explored in microbial Vanillin production through genetic and metabolic engineering methods.4
Consequently, using microbes which can bioconvert “cheap” substrate such as FA (trans-4-hydroxy-3-methoxycinnamic acid) to economically important Vanillin is an alternative cost-effective method.5
Studies on the microbial bioproduction of Vanillin have shown that several species of, Streptomyces and Amycolatopsis sp. ATCC 39116 are particularly effective at converting hydroxycinnamic acids specially FA into Vanillin.6
One of the most prevalent phenolic compounds is FA, which is found in the lignocellulose, plant cell walls, and agricultural plants’ cell walls like grain, corn and sugar crops.7 According to some reports, a feruloyl-CoA synthetase first converts FA to feruloyl-CoA, and after that the CoA thioester is hydrated then cleaves into Vanillin & acetyl-CoA by an aldolase/enoyl-CoA hydratase.8,9
Different microorganisms have varied capacity of biotransformation of FA to vanillic acid (VA) /Vanillin or both.10-15 Halomonas elognata strains quickly transformed FA to VA at 80% yield. However, in these studies, Vanillin was not formed which has considerably a greater value than VA.16,17
Recombinant Pseudomonas fluorescens BF13 yielded of Vanillin and VA, 81% and 16%, respectively, in 3 L stirred tank reactor with incubation of 24 h.18 Whereas, recombinant Pseudomonas putida strain KT2440 yield of Vanillin was 86% in 3 h.19 Halomonas sp. B15 yielded FA biotransformation of 36.5% with production of VA, 365 mg/L after 31 h at 100 g NaCl/L.1
Only a small number of microorganisms have so far been discovered which can bioconvert FA to Vanillin with increased yield. The main objective of this study was to isolate and identify the bacterial strain which can efficiently bioconvert FA to Vanillin.
Chemicals
FA (99%) & Vanillin (99%) used in this research were bought from the Sigma-Aldrich company. Other chemicals purchased were Analytical Grade.
Enrichment culture & media
The sediment sample from Kogilvai Village, Warangal, Telangana was collected from the initial stages of paddy growing field in sterile polythene bags from around 5.0 cm below the soil surface and were stored in the laboratory until use. This sediment sample was then mixed with CaSO4 and kept aside for about 20 days.20 Isolation of microorganisms was performed using the serial dilution technique. Stock sediment solution was made by suspending 1 g of sediment sample in 100 ml sterile distilled water, then 1 ml of this suspension was inoculated in 9 mL sterile distilled water and mixed well. Then, 1 mL of this first dilution was used to prepare different dilutions using serial dilution method. Different dilutions (0.1 mL) of these were spread evenly on the sterilized Starch Casein Agar (SCA) and Oatmeal agar (OMA) plates. The SCA medium consists of (g/L): soluble starch (10), casein (0.3), K2HPO4 (2), NaCl (2), KNO3 (2), CaCO3 (0.02), MgSO4.7H2O (0.05), FeSO4.7H2O (0.01), agar (18) pH at 7.2.21 Oatmeal medium preparation: Oatmeal 20 g was cooked in 1000 ml distilled water for 20 min, filtered using cheese cloth and prepared the volume of filtrate to 1000 ml with distilled water. 20 g of agar was added, pH 7.2. Inoculated plates were incubated for 7 to 14 days at 30°C. The petri plates were noticed often for the growth of actinobacterial isolates. The isolates were randomly picked, purified & maintained on SCA and OMA plates at 4°C until further studies.
Screening of isolates converting ferulic acid to vanillin
The purified Actinobacterial isolates were plated on minimal medium supplemented with 0.2 μm nylon filter sterilized Ferulic acid (500 mg/l) as only C source. After incubation of 48 h at 37°C, the colonies formed were later purified on the same media plates. Single bacterial colonies obtained were later maintained on the same media at 4°C until further research work.22 Minimal medium consists of (g/L): NH4NO3 (3.0) as a nitrogen source, KH2PO4 (1.0), Na2PO4 (1.0), NaCl (0.2), MgSO4.7H2O (0.2) CaCl2 (0.05) and agar (18) pH was maintained at 7.
Effect of Ferulic acid on the growth of actinobacterial isolates
Purified isolates were grown in nutrient broth and incubated at 37°C for 24 h to prepare seed culture. Growth was measured at 600 nm in terms of OD. To determine the Ferulic acid biotransformation capacity of each isolate, seed culture (4% v/v) of each and every purified isolate was transferred to 30 ml of sterile basal media supplemented with 1g/L FA as C & energy source after 18 hrs of culture growth. The basal medium comprised of (g/L): Na2HPO4 (4), KH2PO4 (1), glucose (1), yeast extract (1), MgSO4.7H2O (0.2), NaCl (0.2), CaCl2.2H2O (0.05), pH at 7.2.23 A control experiment was simultaneously performed by adding 1 g/L FA in the same medium without isolates. Ferulic acid was pre-dissolved in 37 g/L NaOH solution and sterilized with 0.2 µm syringe filters before use. Samples were withdrawn at random intervals (6, 24, 28, 30 and 48 hrs) to analyze the growth of culture. Cultures’ growth was determined by taking spectrophotometric readings at 600 nm and were examined for FA bioconversion to Vanillin by TLC. The isolate with the quick and highest capacity of bioconversion of FA to Vanillin was later selected for further work.
FA to Vanillin bioconversion analysis
FA to Vanillin bioconversion by nine actinobacterial isolates was checked initially by TLC. Methylene chloride, methanol, and anhydrous acetic acid (98.5:1:0.5, v/v/v) were used as the mobile phase to create the chromatograms in a vertical glass chamber.24 Ideal chamber saturation time of 30 minutes at room temperature (25 + 2°C) was used for the mobile phase.
After chromatograms were developed, the TLC plates were dried & the components were observed under UV irradiation at 254 nm. Every analysis was performed in duplicate. After that, Vanillin concentration in the sample converted by isolate, S1 (with quick and high ferulic acid to vanillin transformation capacity) was analyzed by UV-spectrophotometry at 363 nm.25 The Vanillin concentration present in the sample (isolate, S1 filtered culture broth) after 28 hrs of incubation period was extrapolated from Vanillin standard graph. Control experiment was performed by adding FA into the same media without isolate.
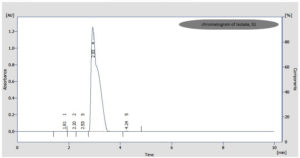
HPLC (Ultimate 3000, Thermo Scientific® with C18 column) analysis was also used to confirm the biocoversion of FA to Vanillin by isolate, S1. The analysis was done at 24 + 2°C or room temperature. The mobile phase was made up of 60% of Acetonitrile, 0.2% of Acetic acid and 38% water. Injection volume, 20 μl with flow rate of 1 mL/min. & detection wavelength, 280 nm.
Biochemical and Molecular characterization
The isolate with the quick and highest potential of bioconversion of FA to Vanillin was selected (isolate, S1) and characterized biochemically by testing different carbohydrates utilization using Hicarbo kit, Himedia.
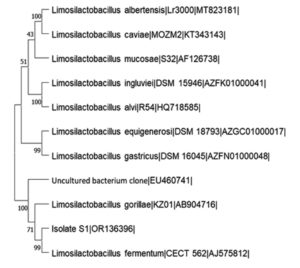
The DNA of the isolate, S1 was extracted subjecting bacterial colony to incubation at high temperature (96°C). After DNA extraction, a PCR reaction was performed by employing two primers: 8F: 5’-AGA GTT TGA TCC TGG CTC AG 3’ and 1525R: 5’-AAG GAG GTG ATC CAG CCG CA 3′. The PCR reaction was performed to amplify its 16S rDNA gene26 according to the following protocol: Initial denaturation for 5 min at 95°C, then denaturation for 30 sec. at 95°C, primer annealing at 46.2°C for 1 min, and extension at 72°C for 1 min for 30 cycles. The last extension was performed at 72°C for 5 min and Amplicons were stored by keeping at 4°C. A PCR product with 1.5 kb size was eluted from agarose gel after PCR products underwent agarose gel electrophoresis, was purified and sequenced. 16S rDNA gene sequencing was outsourced. The EZ Biocloud public portal for data and analytics (https://www.ezbiocloud.net/apps) was utilized to identify the isolate using its 16S rDNA gene sequence. The 16S rDNA gene sequence of the isolate, S1 was later submitted to NCBI GenBank to get its accession number. Phylogenetic analysis was done using MEGA 11 software.
Isolates growing on FA as sole C source
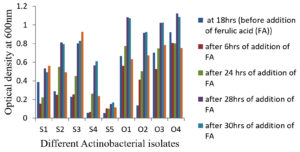
Nine actinobacterial isolates (four on OMA (O1, O2, O3, O4) and five on SCA – (S1, S2, S3, S4, S5) were isolated and purified from the diluted sediment sample of Kogilvai Village, Warangal, Telangana on the minimal media with FA as sole C source. Growth of these nine actinobacterial isolates (optical density at 600 nm) before and after addition of FA in the media was shown in Figure 1.
Ferulic acid to vanillin bioconversion analysis
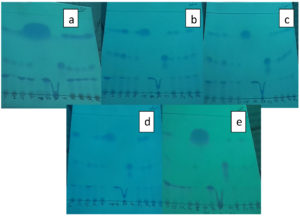
Ferulic acid biotransformation capacity of these nine isolates was determined using sterile basal medium supplemented with of 1 g/L Ferulic acid as substrate by drawing samples and analyzing for bioconversion capacity at different, random incubation times (6, 24, 28, 30 and 48 hrs) by TLC (Figure 2). Biotransformation of FA by some Bacillus sp. (B. subtilis, B. coagulans and B. megaterium) accumulated only minute concentrations of Vanillin and VA.27-29
Figure 2. TLC of nine Actinobacteria with Bioconversion capacity of FA to Vanillin at different incubation periods
Bioconversion process was shown by all the nine purified isolates with just incubation of 6 hrs after the addition of FA in the medium and continued till 28 hrs of incubation. After that, a substantial reduction in the quantity of Vanillin was seen as reaction time increased to 30 and 48 hrs, which was observed through analysis of TLC (Figure 2) and UV-spectrophotometry. HPLC confirmed the FA to Vanillin bioconversion by isolate, S1 (Figure 3). The current study has offered the first proof of Limosilactobacillus sp. strain, S1 with efficient FA biotransformation to Vanillin.
Research has been done on numerous bacteria, yeasts and filamentous fungi for the biotransformation of FA to Vanillin or VA. Low product concentrations were produced by the majority of the examined bioconversion processes. The yield of Vanillin formed by the biotransformation of FA has been extensively studied utilizing optimization techniques.
Among the nine isolates tested in this report, four isolates, S1, S3, O3 and O4 showed increased FA bioconversion to Vanillin when compared to other five isolates with initial medium’s pH 7. In a report, Amycolatopsis sp. began bioconverting FA present in broth after culturing the bacterial cells for 18 hours at various initial pH values between 7.0 and 8.5. It displayed an increased OD 600 value at pH 8.0 during the transformation than that at increased or decreased pH values.6
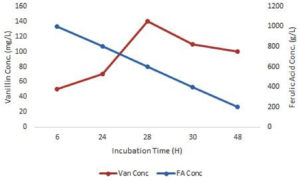
Among these four isolates (S1, S3, O3 and O4), optimal bioconversion was shown by isolate, S1 after the addition of FA in the medium. It was converting 1 g/L FA in the medium to 140 mg/L Vanillin with just 28 hrs of incubation (Figure 4) with 150 rpm as analyzed by UV-spectrophometry and confirmed by HPLC (Figure 3).
Biochemical and molecular characterization
Biochemical characterization (particularly carbohydrates utilization) of isolate S1 showed, it could utilize Lactose, Maltose, Glycerol, Fructose, Galactose, Sucrose, Dextrose, L-Arabinose, ONPG and Esculin. Whereas, other carbohydrates mentioned in Table were not utilized.
Table:
Carbohydrates utilization test by Isolate, S1
No. |
Carbohydrate utilization |
Result |
|---|---|---|
1 |
Fructose |
+ |
2 |
Xylose |
– |
3 |
Maltose |
+ |
4 |
Lactose |
+ |
5 |
Galactose |
+ |
6 |
Dextrose |
+ |
7 |
Trehalose |
– |
8 |
Raffinose |
– |
9 |
Melibiose |
– |
10 |
Glycerol |
+ |
11 |
L-Arabinose |
+ |
12 |
Mannose |
– |
13 |
Salicin |
– |
14 |
Sodium gluconate |
– |
15 |
Sucrose |
+ |
16 |
Inulin |
– |
17 |
Inositol |
– |
18 |
Dulcitol |
– |
19 |
Sorbitol |
– |
20 |
Arabitol |
– |
21 |
Adonitol |
– |
22 |
Mannitol |
– |
23 |
Erythritol |
– |
24 |
α-Methyl-D-glucoside |
– |
25 |
Rhamnose |
– |
26 |
Xylitol |
– |
27 |
Melizitose |
– |
28 |
α-Methyl-D-Mannoside |
– |
29 |
Cellobiose |
– |
30 |
Esculin |
+ |
31 |
ONPG |
+ |
32 |
D-Arabinose |
– |
33 |
Malonate |
– |
34 |
Citrate |
– |
35 |
Sorbose |
– |
Note: – = negative test ; + = positive test
16S rDNA gene sequence of the isolate, S1 was amplified with PCR and the PCR product was subjected to sequencing. The 16S rDNA gene sequence of isolate, S1 was compared with other 16S rDNA gene sequences in EZ Biocloud public data and analytics portal. 16S rDNA sequence of isolate, S1 showed 98.27% similarity to Limosilactobacillus fermentum CECT 562 with completeness of 96.7% and was identified as Limosilactobacillus sp. 16S rDNA gene sequence of this isolate, S1 was submitted to NCBI GenBank and its accession number was OR136396. The Neighbor-Joining tree was constructed to study phylogenetic analysis using MEGA 11 software (Figure 5).
All nine Actinobacteria isolated from the sediment sample of Kogilvai Village, Warangal, Telangana, have the capacity of FA biotransformation to Vanillin. But, isolate S1 has the quick and potential conversion capacity of producing 140 mg/L (analyzed by HPLC) Vanillin with just 28 hrs of incubation after supplementation of 1 g/L FA in the medium at
37°C with 150 rpm. This study proved that the Limosilactobacillus sp. strain, S1 has efficient FA biotransformation to Vanillin. Further research is required in this regard to determine the isolate’s optimal conditions for the FA to Vanillin bioconversion process so that it can be applied in industrial setups for the commercial bioconversion process.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Ioannis V, Maria A, Rodothea D, et al. Novel Halomonas sp. B15 isolated from Larnaca Salt Lake in Cyprus that generates vanillin and vanillic acid from ferulic acid. W J Microbiol Biotechnol. 2015;31(8):1291-1296.
Crossref - Zamzuri NA, Abd-Aziz S, Rahim RA, Phang LY, Alitheen NB, Maeda T. A rapid colorimetric screening method for vanillic acid and vanillin-producing bacterial strains. J Appl Microbiol. 2014;116(4):903-910.
Crossref - Serra S, Fuganti C, Brenna E. Biocatalytic preparation of natural flavours and fragrances. Trends in Biotechnol. 2005;23(4):193-198.
Crossref - Xu L, Liaqat F, Sun J, Khazi MI, Xie R, Zhu D. Advances in the vanillin synthesis and biotransformation: A review. Renew Sustain Energy Rev. 2024;189(Part A):113905.
Crossref - Xu P, Hua D, Ma C. Microbial transformation of propenylbenzenes for natural flavor production. Trends Biotechnol. 2007;25(12):571-576.
Crossref - Xiao-kui M, Andrew JD. Transformation of Ferulic Acid to Vanillin Using a Fed-Batch Solid-Liquid Two-Phase Partitioning Bioreactor. ABiotechnol Prog. 2013;30(1):207-214.
Crossref - Ashengroph M, Nahvi I, Zarkesh-Esfahani H, Momenbeik F. Novel strain of Bacillus licheniformis SHL1 with potential converting ferulic acid into vanillic acid. Ann Microbiol. 2012;62:553-558.
Crossref - Overhage J, Priefert H, Rabenhorst J, Steinbuchel A. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl Microbiol and Biotechnol.1999;52(6):820-828.
Crossref - Calisti C, Ficca AG, Barghini P, Ruzzi M. Regulation of ferulic catabolic genes in Pseudomonas fluorescens BF13: involvement of a MarR family regulator. Appl Microbiol and Biotechnol. 2008; 80(3):475-483.
Crossref - Motedayen N, Ismail MB, Nazarpour F. Bioconversion of ferulic acid to vanillin by combined action of Aspergillus niger K8 and Phanerochaete crysosporium ATCC 24725. Afr J of Biotechnol. 2013;12(47):6618-6624.
Crossref - Li X, Yang J, Li X, Gu W, Huang J, Zhang KQ. The metabolism of ferulic acid via 4-vinylguaiacol to vanillin by Enterobacter sp. P6-4 isolated from Vanilla root. Process Biochem. 2008;43(10):1132-1137.
Crossref - Achterholt S, Priefert H, Steinbuchel A. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl Microbiol and Biotechnol. 2000;54(6):799-807.
Crossref - Hua D, Ma C, Song L, et al. Enhanced vanillin production from ferulic acid using adsorbent resin. Appl Microbiol Biotechnol. 2007;74(4):783-790.
Crossref - Muheim A, Lerch K. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl Microbiol and Biotechnol. 1999;51(4):456-461.
Crossref - Brunati M, Marinelli F, Bertolini C, Gandolfi R, Daffonchio D, Molinari F. Biotransformations of cinnamic and ferulic acid with Actinomycetes. Enzyme Microbial Technol. 2004;34(1):3-9.
Crossref - Abdelkafi S, Sayadi S, Gam ZBA, Casalot L, Labat M. Bioconversion of ferulic acid to vanillic acid by Halomonas elongata isolated from table-olive fermentation. FEMS Microbiol Lett. 2006;262(1):115-120.
Crossref - Abdelkafi S, Labat M, Gam ZBA, Lorquin J, Casalot L, Sayadi S. Optimized conditions for the synthesis of vanillic acid under hypersaline conditions by Halomonas elongata DSM 2581T resting cells. W J Microbiol Biotechnol. 2008;24(5):675-680.
Crossref - Di Gioia D, Luziatelli F, Negroni A, Ficca AG, Fava F, Ruzzi M. Metabolic engineering of Pseudomonas fluorescens for the production of vanillin from ferulic acid. J Biotechnol. 2010;156(4):309-316.
Crossref - Graf N, Altenbuchner J. Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl Microbiol Biotechnol. 2014;98(1):137-149.
Crossref - Balwos A, Schaal K, Pulverer G, Starr MP, Truper HG, Schlegel HG. The prokaryotes – A handbook of habitats, isolation and identification of bacteria. Published by springer verlag. 1981:II.
- Shahidi Bonjar GH, Aghighi S. Chitinolytic and microsclerostatic activity of Iranian strains of Streptomyces plicatus and Frankia sp. on olive isolate of Verticillium dahliae. Biotechnol. 2005;4(2):108-113.
Crossref - Shashank M, Meenakshi K, Ashish S, Ambarish SV, Shashwati GS. Bioconversion of ferulic acid to vanillic acid by Paenibacillus lactis SAMS-2001. Annals of Microbiol. 2016;66:875-882.
Crossref - Xiao-kui M, Andrew JD. Effect of bioconversion conditions on vanillin production by Amycolatopsis sp. ATCC 39116 through an analysis of competing by-product formation. Bioprocess and Biosys Engg. 2014;37(5):891-899.
Crossref - Magda F, Melissa S, Gustavo C, Amelia H, Luiz Alberto LS. Analysis of vanillin by TLC and HPLC-PDA in herbal material and tincture from Vanilla planifolia Jacks ex. Andrews. Drug Analytical Research. 2018;02:1-7.
Crossref - Zhao J, Xia H, Yu T, et al. colorimetric assay for vanillin detection by determination of the luminescence of o-toluidine condensates. PLoS One. 2018;13(4):e0194010.
Crossref - Chen YL, Lee CC, Lin YL, Yin KM, Ho CL, Liu T. Obtaining long 16S rDNA sequences using multiple primers and its application on dioxin-containing samples. BMC Bioinfo. 2015;16(Suppl. 18):S13.
Crossref - Gurujeyalakshmi G, Mahadevan A. Dissimilation of ferulic acid by Bacillus subtilis. Curr Microbiol. 1987;16:69-73.
Crossref - Crawford RL, Olson PP. Microbial catabolism of vanillate: decarboxylation to guaiacol. Appl Env Microbiol. 1978;36:539-543.
Crossref - Karmakar B, Vohra RM, Nandanwar H, Sharma P, Gupta KG, Sobti RC. Rapid degradation of ferulic acid via 4-vinylguaiacol and vanillin by a newly isolated strain of Bacillus coagulans. J of Biotech. 2000;80(3):195-202.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.