ISSN: 0973-7510
E-ISSN: 2581-690X
Sexually transmitted infections (STIs) are a major public health problem worldwide with significant social and economic implications. Effective control and prevention strategies necessitate a thorough understanding of the prevalence, isolation, and identification of STI pathogens. The present study aims to provide a comprehensive analysis of the isolation, identification, prevalence, and antimicrobial susceptibility pattern of STI pathogens based on culture method analysis. Endocervical /vaginal swab samples from female patients symptomatic for STI were cultured on different selective and differential media and pathogens were identified by colony morphology and biochemical tests. Antimicrobial Susceptibility Test (AST) of isolated and identified culture pathogen was performed by using Kirby-Bauer disc diffusion method. Among 209 endocervical/vaginal swab samples from symptomatic patients, 126 (60.28%) tested positive and 83 (39.71%) negative. Ureaplasma spp. (n = 100) was the most prevalent isolate, constituting 79.36% of culture-positive samples, followed by N. gonorrhoea (n = 99) at 78.57%, and Mycoplasma spp. (n = 41) at 32.54% individually and in combination. AST analysis revealed erythromycin (74%), ofloxacin (69%), and roxithromycin (64%) as the most resistant antibiotics for Ureaplasma spp. N. gonorrhoea showed the highest resistance to cefixime (78.79%), followed by ofloxacin (75.76%) and erythromycin (69.7%). Azithromycin and erythromycin exhibited 100% resistance against Mycoplasma spp. The study provides information on the prevalent bacterial pathogens involved in STIs among women in Anuppur and Shahdol districts, Madhya Pradesh. Understanding the diversity, distribution patterns and antibiotic sensitivity of these pathogens is crucial for developing targeted interventions and effective prevention strategies in such resource-limited areas.
Sexually Transmitted Infections, Neisseria gonorrhoeae, Ureaplasma spp., Mycoplasma spp., Antimicrobial Susceptibility Test, Multidrug Resistance
Sexually Transmitted Infections (STIs) are clinical syndromes caused by micro-organisms that can be acquired and spread through sexual activity.1 It has a tremendous impact on national health. Failure to recognize and treat STIs early can lead to major complications and long-term consequences. They are responsible for a substantial proportion of maternal health issues, ectopic pregnancy, termination of pregnancies, fertility problems, infant mortality, and the birth of underweight babies.2 The World Health Organization reports that there are over 1 million daily cases of STIs worldwide, resulting in an estimated 374 million new infections annually.3
Neisseria gonorrhoeae (gonococcus) is the second most prevalent causative agent known to cause STI with a significant global public health impact.4 Non-gonococci STI pathogens like genital Mycoplasma spp. (Mycoplasma hominis & Mycoplasma genitalium) and Ureaplasma spp., which include Ureaplasma parvum and Ureaplasma urealyticum. These facultative anaerobic microorganisms in the lower urogenital tract are linked to urogenital issues in women, such as cervicitis, cystitis, bacterial vaginosis, pelvic inflammatory disease, chorioamnionitis, postpartum fever, infertility, prematurity, intrauterine growth retardation, and systemic neonatal infections.5
In India, it is estimated that approximately 6% of adults are affected by STIs.6,7 There are differences in STI rates among ethnic and racial groups that are important to consider. The epidemiology of STIs within rural communities seldom has been studied.8 Limited reporting and diagnostic tools in rural and tribal areas lead to underestimation of STI cases. Despite global advancements in diagnosis and treatment, isolated tribal communities, constituting around 110 million individuals in India, face significant socio-economic and health challenges.9
Tribal society adheres to traditional customs, mandating members to follow them. Marriage is generally permitted in Indian society to involve only one man and one woman at a time (monogamy). However, many tribal societies have allowed a man or woman to marry multiple women or men, which is known as polygyny and polyandry, respectively. Many researchers have found that tribal societies have a system of endogamy, multiple sex partners, and unprotected sex.10,11 As a result, the circumstances are favourable for the transmission of STIs among tribals.
Antibiotics are potent medications used in the treatment of bacterial infections. The inappropriate use of these drugs has led to the proliferation of antibiotic resistance across a wide spectrum of bacteria.12 STI pathogens are becoming increasingly resistant to antimicrobial agents, a global issue. New medications replace antibiotics that have lost their potency, but new strains emerge with new resistance determinants; this problem affects all antibiotic classes.13
As there is no data available on prevalence and antibiotic resistance patterns of causative organisms of STIs among tribal women in the Anuppur and Shahdol districts, Madhya Pradesh, the present study was carried out to address this issue.
Study area
The study was conducted among women in the Anuppur and Shahdol districts of Madhya Pradesh.
Recruitment of subjects
Target Patients were females (age <55) visiting the OPD or admitted in the wards of Obstetrics & Gynaecology of the District Hospital, Annupur and Birsamunda Government Medical College, Shahdol, from May 2022 to September 2023, with a known history of reproductive tract infections were enrolled in the study. 209 endocervical/vaginal swab samples were collected and subjected to laboratory analysis. Patients experiencing symptoms included cloudy/bloody discharge, burning micturition, yellow/green discharge, painful intercourse, strong vaginal odour, vaginal itching/irritation, weight loss, lower abdominal pain, and abnormal menstruation was major focus for the collection of samples. A bilingual (Hindi and English) questionnaire/consent form was filled by patients or staff with patient consent before specimen collection.
Sample collection & transportation
Two endocervical/vaginal swabs were collected from each patient, i.e. sterile cotton swab (PW1280) and HiCulture™ Transport Swabs w/ Stuart Transport Medium (MS306S). To protect the viability of pathogens for isolation, the specimens were put onto a culture medium as soon as possible after collection.
Bacterial culture
The necessary differential and selective media along with supplements were purchased from HiMedia and prepared aseptically as per the instructions given by HiMedia. 0.01 ml of a collected sample is aseptically streaked over various selective and differential medium Thayer Martin agar base (M413), A-7 (Shepard’s Differential Agar Base: M1739), Chocolate agar base (M1548), and Urogenital mycoplasma broth base (M1374). The plates were incubated aerobically at 37°C for 24 hours.
Quantitative analysis
The quantitative analysis of pathogens in the swab sample was determined by measuring CFU/ml (colony-forming units per milliliter) on a culture plate using the plate count method, specifically the spread plate technique. Swab samples displaying bacterial growth equal to or exceeding 105 CFU/ml were classified as culture-positive and subjected to subsequent identification and confirmatory analysis. Samples that showed either no growth or growth less than or equal to 105 CFU/ml were considered as culture-negative.
Isolation, identification, and confirmatory analysis of pathogens
The pathogens were tentatively identified based on the pathogen’s growth on various culture media, colour production, and colony morphology. The method for the isolation and identification of N. gonorrhoeae infections was based on the laboratory analysis of N. gonorrhoeae.14 For N. gonorrhoeae, a rapid biochemical identification test kit (KB008 HiNesseriaTM identification kit, HiMedia) with seven conventional biochemical tests and five carbohydrate utilization tests was used. Ureaplasma spp. were isolated on A-7 or Shepard’s Differential Agar,15 while a specialized medium, Urogenital Mycoplasma Broth, was used for the isolation and identification of genital Mycoplasma spp. The culture response, observed after incubation at 37°C for 48 hours to 8 weeks, indicated Mycoplasma spp. growth through a pH shift, identified by a color change in the phenol red indicator. Mycoplasma spp. isolation through culture methods may take 2 to 5 days for M. hominis and up to 8 weeks for M. genitalium.16
After the identification, the pathogens were sub-cultured on species-specific media to get pure culture. Antimicrobial Susceptibility Test (AST) of isolated and identified pathogen was performed.
Antimicrobial Susceptibility Test (AST)
The assessment of antimicrobial susceptibility to the isolated and identified bacteria against the commonly prescribed antibiotics, as indicated in Table 1, was evaluated using the disc diffusion technique, specifically the Kirby-Bauer (KB) method. Sterile Mueller-Hinton agar (MHA) plates were prepared and AST was performed by the method described by Sharma et al.17
Table (1):
Interpretation of zones of inhibition of antibiotics by Kirby-Baurer method
| Antibiotics | Abbreviation | Class of antibiotics | Disc concentration | Diameter of growth inhibition zone (mm) | ||
|---|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||||
| Cefixime | CFM | β-Lactam | 15 µg | 20 or more | 11-19 | Below 11 |
| Ceftriaxone | CTR | β-Lactam | 30 µg | 20 or more | 14-19 | Below 14 |
| Ciprofloxacin | CIP | Fluoroquinolone | 5 µg | 25 or more | 15-24 | Below 15 |
| Levofloxacin | LE | Fluoroquinolone | 5 µg | 20 or more | 12-19 | Below 12 |
| Ofloxacin | OF | Fluoroquinolone | 5 µg | 20 or more | 13-19 | Below 13 |
| Norfloxacin | NX | Fluoroquinolone | 10 µg | 18 or more | 13-17 | Below 12 |
| Tetracycline | TE | Tetracycline | 30 µg | 22 or more | 15-20 | Below 15 |
| Doxycycline | DO | Tetracycline | 30 µg | 26 or more | 16-25 | Below 16 |
| Azithromycin | AZM | Macrolides | 15 µg | 19 or more | 13-18 | Below 13 |
| Erythromycin | E | Macrolides | 15 µg | 21 or more | 12-20 | Below 12 |
| Roxithromycin | RO | Macrolides | 30 µg | 18 or more | 13-17 | Below 13 |
Statistical analysis
Chi-square test, Pearson and Fisher’s exact test was used to analyse the age-wise prevalence and associated symptoms of pathogens. An intra-comparison of antimicrobial susceptibility was statistically analysed with one-way ANOVA using Tukey’s HSD post-hoc test. The level of significance was expressed as the p-value (p < 0.001, p < 0.005, p < 0.01 and p < 0.05). All statistical analyses were performed using IBM SPSS Statistics 20 (Version 20.0, Armonk, NY: IBM Corporation).
Quantitative analysis of STI pathogens
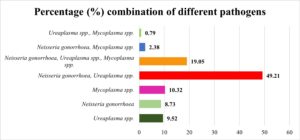
Among the 209 endocervical/vaginal swab samples collected from symptomatic patients, 126 (60.28%) showed bacteriuria ≤105cfu/ml, while 83 (39.71%) had no growth or growth ≤105cfu/ml. Polymicrobial culture growth was significant, revealing various pathogen combinations. Figure 1 illustrates prevalent combinations, aiding understanding of their distribution in culture-positive samples (n=126). N. gonorrhoea + Ureaplasma spp. was most common (49.21%), followed by N. gonorrhoea, Ureaplasma spp., and Mycoplasma spp. (19.05%). Ureaplasma spp. alone (9.52%), N. gonorrhoea alone (8.73%), and Mycoplasma spp. alone (10.32%) were detected. Ureaplasma spp. (79.36%) was the most prevalent isolate, followed by N. gonorrhoea (78.57%) and Mycoplasma spp. (32.54%) based on colony morphology and biochemical diagnosis in both single and combined growth.
Pathogen identification and confirmatory analysis
Figure 2 provides an overview of the identification and confirmation of N. gonorrhoea, Ureaplasma spp., and Mycoplasma spp. It includes information on the colony morphology/colour changes of pathogens and the specific biochemical tests used for their identification. N. gonorrhoea on chocolate media exhibits small, smooth, translucent, raised convex colonies measuring 0.5-1.0 mm. On modified Thayer Martin agar, colonies are small, greyish-white to colourless, mucoid, and well-defined. After 48 hours, colonies grow to 3mm with a less smooth texture. Biochemically, N. gonorrhoea is non-urease-producing, ONPG and VP tests are negative, but oxidase and catalase tests are positive. Nitrate reduction is negative, indicating no nitrate reduction, while the glucose test is positive. Lactose, maltose, sucrose, fructose, and mannose fermentation tests are negative.
Ureaplasma spp. is confirmed by identifying typical colonies on A-7 agar, a differential agar media that distinguishes it from other urease-producing bacteria. As conventional culture techniques cannot differentiate the two Ureaplasma spp. i.e., U. urealyticum & U. parvum and are sometimes invalidated by the overgrowth of other bacteria.15 Mycoplasma spp. utilizes arginine to produce ammonia through a three-enzyme process, leading to an elevation in the medium’s pH. This pH shift is detected by a colour change to red, with phenol red serving as the pH indicator.
Age-wise distribution and prevalence of pathogens
Table 2 provides a comprehensive overview of the age-wise distribution and prevalence of STI pathogens. The sample was collected from women of different age groups and was categorized into six different groups: ≤20, 21-27, 28-34, 35-41, 42-48, and 49-55.
Table (2):
Age-specific distribution and prevalence of STI pathogens
| Age Groups (Years) | |||||||
|---|---|---|---|---|---|---|---|
| Samples | ≤20 | 21-27 | 28-34 | 35-41 | 42-48 | 49-55 | p-value |
| Total Sample Collected (n=209) | 5 (2.39%) | 34 (16.27%) | 59 (28.23%) | 69 (33.01%) | 28 (13.40%) | 14 (6.70%) | – |
| Culture Positive (n=126) | 4 (80%) | 21 (61.76%) | 33 (55.93%) | 45 (65.22%) | 16 (57.14%) | 7 (50%) | – |
| Culture Negative (n=83) | 1 (20%) | 13 (38.23%) | 26 (44.07%) | 24 (34.78%) | 12 (42.86%) | 7 (50%) | – |
| Ureaplasma spp. (n=100) | 4(100%) | 17 (80.95%) | 29 (87.88%) | 34 (75.56%) | 11 (68.75%) | 5 (71.43%) | 0.498 |
| Neisseria gonorrhoea (n=99) | 4 (100%) | 16 (76.19%) | 26 (78.79%) | 34 (75.76%) | 13 (81.25%) | 6 (85.71%) | 0.740 |
| Mycoplasma spp. (n=41) | 1 (25%) | 6 (28.57%) | 11 (33.33%) | 15 (33.33%) | 5 (31.25%) | 3 (42.86%) | 0.846 |
The prevalence of Ureaplasma spp. infection was highest in the 28-34-year age group, with 87.88% of culture-positive cases. The lowest occurrence of infections was observed in individuals aged ≤20-year, with all collected samples testing positive for the presence of the bacteria. The prevalence of N. gonorrhoea infection remained relatively consistent across different age groups, ranging from 75.76% to 85.71% of culture-positive cases. The age groups with the lowest and highest prevalence were those aged 20 or younger and those aged 42 to 48, respectively. In the case of Mycoplasma spp. infection, the prevalence varied across age groups. The highest prevalence was found in the age group of 35 to 41 years, with 33.33% of culture-positive cases. The ≤20-year age group has the lowest prevalence of Mycoplasma spp. infection.
Association between Age-specific distribution and prevalence of STI pathogens
The chi-square test was used to assess the link between various pathogens and age groups among tribal women in Madhya Pradesh. Results revealed no statistically significant associations. For Ureaplasma spp., the Pearson chi-square test yielded a non-significant value (c² = 4.368, df = 5, p = .498). Similarly, N. gonorrhoea (c² = 2.024, df = 5, p = 0.740) and Mycoplasma spp. (c² = 2.024, df = 5, p = 0.846) showed no significant association with age groups. In summary, the p-values for all three pathogen tests exceeded the commonly used threshold for statistical significance (typically set at 0.05 or 0.01), indicating that there was no significant association between pathogen infections (Ureaplasma spp., N. gonorrhoea, and Mycoplasma spp.) and the age group of tribal women in Madhya Pradesh. Consequently, the data did not provide enough evidence to suggest that age group had a significant impact on pathogen infections among tribal women in this study.
Symptoms associated with STI pathogens
Table 3 summarizes correlations between symptoms in individuals diagnosed with Ureaplasma spp., N. gonorrhoea, and Mycoplasma spp. It represents the total reported cases for each symptom and specifies the number associated with each STI pathogen. For example, cloudy/bloody discharge was reported by 71 individuals, with 38 cases linked to Ureaplasma spp., 38 to N. gonorrhoea, and 17 to Mycoplasma spp. Predominantly, lower abdominal pain followed by cloudy/bloody discharge was observed in patients with these STIs. Significance values denote the strength of the symptom-pathogen association, with a p-value less than 0.05 (*) indicating a strong link. This data aids in comprehending symptom patterns for each pathogen, facilitating diagnosis and management.
Table (3):
Comparative Analysis of Symptoms in STI Pathogens
Symptoms |
Total (n=209) |
Culture Positive (n=126) |
Ureaplasma spp. |
Neisseria gonorrhoea |
Mycoplasma spp. |
|---|---|---|---|---|---|
Cloudy/ bloody discharge |
71 (33.97%) |
47 (37.30%) |
38 (80.85%) |
38 (80.85%) |
17 (36.17%) |
Burning Micturition |
38 (18.18%) |
22 (17.46%) |
12* (54.54%) |
13 (59.09%) |
10 (45.45%) |
Yellow/Green discharge |
11 (5.26%) |
7 (5.56%) |
6 (85.71%) |
7 (100%) |
1 (14.28%) |
Painful intercourse |
6 (2.87%) |
4 (3.17%) |
3 (75%) |
3 (75%) |
3 (75%) |
Strong vaginal odour |
37 (17.70%) |
24 (19.05%) |
17 (70.83%) |
19 (79.17%) |
8 (33.33%) |
Vaginal Itching/Irritation |
57 (27.27%) |
36 (28.57%) |
28 (77.78%) |
27 (75%) |
16* (44.44%) |
Weight loss |
1 (0.48%) |
0 |
0 |
0 |
0 |
Lower abdominal pain |
108 (51.67%) |
62 (49.21%) |
42* (67.74%) |
44* (70.97%) |
18 (29.03%) |
Abnormal Menstruation |
30 (14.35%) |
23 (18.25%) |
18 (78.26%) |
17 (73.91%) |
12* (52.17%) |
*= p <0.05 represents significant association between symptoms and pathogens.
Antimicrobial susceptibility test
Tables 4, 5 and 6 represent a comparison of the significant levels of various antibiotics categorized in three categories namely sensitive, moderate, and resistant, with corresponding significance values denoted by unique codes. The top three antibiotics from each group were analysed in terms of their sensitivity, along with their respective significance values. Significance values (p-values) as mentioned in Table no. 4, 5 and 6 were used to compare their effectiveness and are shown in the Figure 3, 4 and 5 respectively.
Table (4):
Antibiotic Susceptibility of Ureaplasma spp. at different significance levels
| Sig. | CIP | DO | TE | DO | AZM | TE & OF | E | OF | AZM & NX |
|---|---|---|---|---|---|---|---|---|---|
| P<0.05 | a | α | w | a | α | w | a | α | w |
| P<0.01 | b | β | x | b | β | x | b | β | x |
| P<0.005 | c | γ | y | c | γ | y | c | γ | y |
| P<0.001 | d | δ | z | d | δ | z | d | δ | z |
| Sensitive | Moderate | Resistant | |||||||
Table (5):
Antibiotic Susceptibility of Neisseria gonorrhoea at different significance levels
| Sig. | CIP | TE | DO | DO | LE | CIP | CFM | OF | E |
|---|---|---|---|---|---|---|---|---|---|
| P<0.05 | a | α | w | a | α | w | a | α | w |
| P<0.01 | b | β | x | b | β | x | b | β | x |
| P<0.005 | c | γ | y | c | γ | y | c | γ | y |
| P<0.001 | d | δ | z | d | δ | z | d | δ | z |
| Sensitive | Moderate | Resistant | |||||||
Table (6):
Antibiotic Susceptibility of Mycoplasma spp. at different significance levels
| Sig. | CIP & DO | RO | TE & OF | DO & NX | OF & CIP | RO | AZM & E | TE & LE | OF |
|---|---|---|---|---|---|---|---|---|---|
| P<0.05 | a | α | w | a | α | w | a | α | w |
| P<0.01 | b | β | x | b | β | x | b | β | x |
| P<0.005 | c | γ | y | c | γ | y | c | γ | y |
| P<0.001 | d | δ | z | d | δ | z | d | δ | z |
| Sensitive | Moderate | Resistant | |||||||
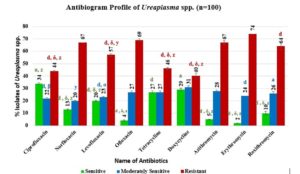
Antibiotic sensitivity profiling of Ureaplasma spp.
Figure 3 shows results from an antibiogram analysis on Ureaplasma spp. isolates, revealing their antibiotic susceptibility. Antibiotics (ciprofloxacin, norfloxacin, levofloxacin, ofloxacin, tetracycline, doxycycline, azithromycin, erythromycin, roxithromycin) were used for AST, and sensitivity was assessed through the zone of inhibition measurements (Table 1). The figure illustrates varying sensitivity, with ciprofloxacin (34%), doxycycline (29%), and tetracycline (27%) being the most effective, and erythromycin (74%), ofloxacin (69%), roxithromycin (64%) being most resistant. Valuable insights from Figure 3 aid healthcare professionals in prescribing antibiotics for Ureaplasma infections.
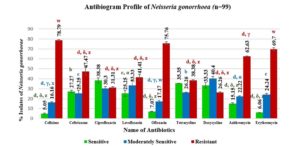
Antibiotic sensitivity profiling of Neisseria gonorrhoea
Figure 4 displays the antibiogram of N. gonorrhoea, showing the bacteria’s sensitivity, moderate sensitivity, and resistance to common antibiotics. Ciprofloxacin, a DNA gyrase inhibitor, exhibits the highest sensitivity at 38.38%, followed by tetracycline and doxycycline. Levofloxacin, ceftriaxone, and tetracycline also demonstrate relatively higher sensitivity (25.25% to 27.27%). The bacteria show significant resistance to cefixime (78.79%), ofloxacin (75.76%), and erythromycin (69.7%), with azithromycin also showing notable resistance (62.63%). These findings offer insights into antibiotic effectiveness against N. gonorrhoea.
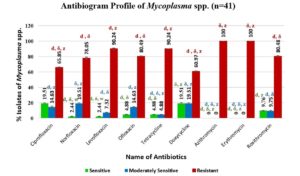
Antibiotic sensitivity profiling of Mycoplasma spp.
Figure 5 summarizes the AST profile of Mycoplasma spp. Notably, azithromycin and erythromycin exhibit 100% resistance, while levofloxacin, tetracycline, and ofloxacin show high resistance (80.49% to 90.24%). Conversely, doxycycline and ciprofloxacin are the most effective antibiotics, with a sensitivity rate of 19.51%. Continuous research and surveillance are essential to monitor evolving resistance profiles and guide appropriate antibiotic therapy for Mycoplasma spp.
Resistance to multi-drug
AST results reveal MDR in all three STI pathogens across 9 antibiotics. N. gonorrhoea exhibits a 56.57% MDR rate (56/99 cases), Ureaplasma spp. at 47% (47/100 cases), and Mycoplasma spp. with an 80.49% MDR rate (33/41 cases). These percentages signify the resistance of strains within each pathogen to multiple drugs. The analysis underscores a troubling trend of rising resistance in these pathogens, posing challenges for effective medical intervention.
Concerns regarding the high prevalence of STIs remain crucial, as these infections can lead to various individual and community health complications. Furthermore, the emergence of drug-resistant organisms and the substantial economic burden associated with STIs underscore the need for ongoing efforts to enhance disease management.18 The result of this study showed that about 79.36% of symptomatic women harboured Ureaplasma spp. This rate is slightly higher to the prevalence of Ureaplasma spp. in China was 54.5%,19 to that in an Italian study (23%) and the rate obtained in Croatian study (34%) as mentioned by Karim et al.20 Highest prevalence had seen between the age group 35-41 years. Based on the bacterial culture result, the prevalence of N. gonorrhoea in the Anuppur district is 78.57% while Mycoplasma spp. possessed a 32.54% prevalent rate. The overall highest prevalence for all three pathogens occurred in the 35-41 age group, while the second highest prevalence for all three pathogens was observed in the 28-34 age group. A similar prevalence age group (30-34 years followed by the 35-39 years) pattern in women was observed in a study conducted in the tribal population of central India.1
This suggests that individuals in their mid to late thirties and early forties may be particularly susceptible to these infections, possibly due to certain behavioural or biological factors prevalent in this age range. The finding might reflect a lack of awareness about safe sexual practices and the importance of regular screening for sexually transmitted infections in their mid to late thirties and early forties.
Antibiotics are generally biologically active, inhibiting protein, nucleic acid, and cell wall synthesis, as well as DNA replication and cell division.22 Misuse of these drugs has led to the proliferation of antibiotic resistance in many bacterial strains.12 The pathogen N. gonorrhoeae has a remarkable potential to acquire resistance to clinically utilized antimicrobial medicines within 10-20 years.23 New medications replace antibiotics that have lost their potency, but new strains emerge with new resistance determinants; this problem affects all antibiotic classes.13 Single-dose antibiotics effectively treat gonococcal and non-gonococcal sexually transmitted infections, but rising resistance, as reported,24 poses a growing threat. Antibiotic resistance has surged over the past few decades, fuelled by overuse and abuse, exacerbated by factors like international travel and commerce. This crisis stems from a long history of neglect despite repeated warnings from researchers and clinicians since the early 1960s.25
N. gonorrhoea rapidly develops resistance to newly introduced antimicrobials for gonorrhoea treatment within 1-2 decades, showcasing its persistent adaptability since the antimicrobial era’s inception in the 1930s.26 This study found high resistance of N. gonorrhoeae to commonly prescribed drugs, notably 78.79% for cefixime, 75.76% for ofloxacin, 69.7% for erythromycin, and 62.63% for azithromycin.
The absence of a cell wall makes Ureaplasma spp. and Mycoplasma spp. inherently resistant to all medications that target the cell wall, including β-Lactam.27 Ureaplasma spp. exhibit antibiotic resistance via distinct mechanisms like Macrolide resistance entails 23S rRNA gene mutations, fluoroquinolone resistance stems from parC gene mutations in quinolone resistance-determining regions, and tetracycline resistance is linked to acquiring the TetM gene on Tn916-like mobile elements.28 In a Tunisian study, Ureaplasma spp. isolates showed resistance to ciprofloxacin and erythromycin, with intermediate resistance to azithromycin. Ofloxacin resistance was noted in 73.27% of isolates, and levofloxacin resistance in 17.82%. Additionally, 37.62% of isolates exhibited resistance to tetracycline.29 AST result of Ureaplasma spp. of this study showed a resemblance to the work done by Boujemaa and co-workers.
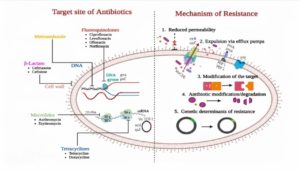
M. hominis exhibits intrinsic resistance to C14 and C15-membered macrolides, including erythromycin, and azithromycin. However, promising antibiotics that display excellent efficacy against M. hominis are doxycycline and tetracycline.30 Among the antibiotics tested in this study, azithromycin and erythromycin show the highest resistance, with 100% of Mycoplasma spp. being resistant to it. Levofloxacin, tetracycline, and ofloxacin also exhibit high resistance rates, ranging from 80.49% to 90.24%. Some previous studies also documentation of 100% erythromycin resistance.31,32 The resistance mechanism probably involves a target alteration in the DNA gyrase and topoisomerase IV subunits.33 Doxycycline and ciprofloxacin with a sensitivity rate of 19.51% can be considered as a drug of choice for treating genital Mycoplasma infection. Tetracyclines showed only moderate sensitivity. Tetracycline resistance due to the acquisition of tetM gene is widely reported.33 Knowledge of local patterns of antimicrobial resistance facilitate clinicians to choose best treatment options for the patients. Most of the known resistance mechanisms, including antibiotic inactivation, drug binding site modification, membrane permeability decrease, and enhanced drug efflux.34 The site of action and mechanism of resistance of antibiotics is depicted in Figure 6.
As suggested by Gonzalez and his colleagues in 2009 that the high rates of poverty, income inequality, unemployment and low educational attainment make it more difficult for individuals to protect their sexual health.36 Tribal females need special attention for the prevention of STIs as in tribal areas, there is little or no access to the health delivery system. also, many researchers have found that tribal societies have a system of multiple sex partners and unprotected sex.
The research provides valuable insights regarding the prevalence and antibiotic resistance pattern of STI pathogens in District Anuppur. The increased frequency of STI pathogens may be due to polygamy, endogamy and little or no access to the health delivery system. STI pathogens also acquire resistance to the currently recommended antibiotics either due to the exhibition of an extraordinary ability of phenotypic and genotypic variation or due to misuse and overuse of drugs. In view of the present findings, an urgent need for appropriate interventions is required to address the growing threat of STI pathogens and antimicrobial resistance in this region.
ACKNOWLEDGMENTS
The authors are thankful to Indira Gandhi National Tribal University, Amarkantak, Madhya Pradesh, India, for providing lab facilities for this work. The DST-SEED, New Delhi, is acknowledged for providing the needed fund for this study. The Council of Scientific & Industrial Research (CSIR), India, is acknowledged for providing financial assistance in the form of Senior Research Fellowship (SRF) to Ms Juhi. The authors also thank to the Birsamunda Government Medical College Shahdol, and the District Hospital Anuppur (Madhya Pradesh), India, for providing needed samples for this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PS conceptualized and designed the study. VH and SX collected the samples from district hospital Anuppur. SS and NJ helped to facilitate the swab sample collection from Birsamunda Government Medical College, Shahdol. VH perform bacterial culture of pathogens, analysed the sample data on the Age-wise and symptoms-wise prevalence of STI patients. J isolated and identified the pathogens, performed antimicrobial susceptibility test experiments. RS analysed the results. J wrote the manuscript. PS and RS edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by the Department of Science and Technology (DST) under the Science for Equity and Empowerment and Development (SEED) division, project Number SEED/WS/2019/351.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This work was approved by the Institutional Ethical Committee, Indira Gandhi National Tribal University, Amarkantak, wide Ref. No., IGNTU/IEC/01/2019, dated 11/05/2019 and Govt. Medical College Shahdol wide Ref. no. IERC/22/06/001, Dated 16/06/2022.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Apers L, Crucitti T, Verbrugge R, Vandenbruaene M. Sexually transmitted infections: what’s new? Acta Clin Belg. 2012;67(3):154-159.
Crossref - Sarkar S, Patra AC, Srinivas P, Ghosh A, Kushbaha G, Saha S. Pattern of sexually transmitted infections: A profile from a rural- and tribal-based sexually transmitted infections clinic of a tertiary care hospital of Eastern India. J Family Med Prim Care. 2018;7(5):1042-1046.
Crossref - Bui HTV, Bui HT, Chu SV, et al. Simultaneous real-time PCR detection of nine prevalent sexually transmitted infections using a predesigned double-quenched TaqMan probe panel. PLoS One. 2023;18(3):e0282439.
Crossref - Fentaw S, Abubeker R, Asamene N, Assefa M, Bekele Y, Tigabu E. Antimicrobial susceptibility profile of Gonococcal isolates obtained from men presenting with urethral discharge in Addis Ababa, Ethiopia: Implications for national syndromic treatment guideline. PLoS One. 2020;15(6):e0233753.
Crossref - Lee JY, Yang JS. Prevalence and Antimicrobial Susceptibility of Mycoplasma hominis and Ureaplasma Species in Nonpregnant Female Patients in South Korea Indicate an Increasing Trend of Pristinamycin-Resistant Isolates. Antimicrob Agents Chemother. 2020;64(10):e01065-20.
Crossref - Desai VK, Kosambiya JK, Thakor HG, Umrigar DD, Khandwala BR, Bhuyan KK. Prevalence of sexually transmitted infections and performance of STI syndromes against aetiological diagnosis, in female sex workers of red light area in Surat, India. Sex Transm Infect. 2003;79(2):111-115.
Crossref - Dhabhai N, Chaudhary R, Wi T, et al. Prevalence of reproductive tract infections including sexually transmitted infections among married women in urban and peri-urban mid to low socioeconomic neighbourhoods of Delhi, North India: an observational study protocol. BMJ Open. 2022;12(3):e059583.
Crossref - Thomas JC, Schoenbach VJ, Weiner DH, Parker EA, Earp JA. Rural gonorrhea in the southeastern United States: a neglected epidemic? Am J Epidemiol. 1996;143(3):269-277.
Crossref - Bang A. The why & the how of research for the tribal people’s health. Indian J Med Res. 2022;156(2):171-173.
Crossref - Anvikar AR, Rao VG, Savargaonkar DD, et al. Seroprevalence of sexually transmitted viruses in the tribal population of Central India. Int J Infect Dis. 2009;13(1):37-39.
Crossref - Saha KB, Saha UC, Sharma RK, Pandey A. Reaching tribal men to improve awareness to sexual morbidities: experience from Baiga tribe of Central India. Indian J Med Res. 2013;137(5):928-934.
- Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa – Mechanisms, epidemiology and evolution. Drug Resist Updat. 2019;44:100640.
Crossref - Shaskolskiy B, Dementieva E, Leinsoo A, et al. Drug Resistance Mechanisms in Bacteria Causing Sexually Transmitted Diseases and Associated with Vaginosis. Front Microbiol. 2016;7:747.
Crossref - Meyer T, Buder S. The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands. Pathogens. 2020;9(2):91.
Crossref - Frolund M, Bjornelius E, Lidbrink P, Ahrens P, Jensen JS. Comparison between culture and a multiplex quantitative real-time polymerase chain reaction assay detecting Ureaplasma urealyticum and U. parvum. PLoS One. 2014;9(7):e102743.
Crossref - Stellrecht KA, Woron AM, Mishrik NG, Venezia RA. Comparison of multiplex PCR assay with culture for detection of genital mycoplasmas. J Clin Microbiol. 2004;42(4):1528-1533.
Crossref - Sharma P, Netam AK, Singh R. Prevalence and in vitro antibiotic susceptibility pattern of bacterial strains isolated from tribal women suffering from urinary tract infections in District Anuppur, Madhya Pradesh, India. Biomed Res Ther. 2020;7(8):3944-3953.
Crossref - Mah ND, Birmingham AR, Treu CN, Bodkin RP, Awad NI, Acquisto NM. Sexually Transmitted Infection Review for the Acute Care Pharmacist. J Pharm Pract. 2020;33(1):63-73.
Crossref - Song T, Ye A, Xie X, et al. Epidemiological investigation and antimicrobial susceptibility analysis of ureaplasma species and Mycoplasma hominis in outpatients with genital manifestations. J Clin Pathol. 2014;67(9):817-820.
Crossref - Karim S, Bouchikhi C, Banani A, et al. Detection of Ureaplasma Biovars and Subtyping of Ureaplasma parvum among Women Referring to a University Hospital in Morocco. Infect Dis Obstet Gynecol. 2020;2020:7286820.
Crossref - Rao VG, Anvikar A, Savargaonkar D, et al. Prevalence of sexually transmitted disease syndromes in tribal population of central India. J Epidemiol Community Health. 2009;63(10):805-806.
Crossref - Yang C, Song G, Lim W. A review of the toxicity in fish exposed to antibiotics. Comp Biochem Physiol C Toxicol Pharmacol. 2020;237:108840.
Crossref - Unemo M, Nicholas RA. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 2012;7(12):1401-1422.
Crossref - Tien V, Punjabi C, Holubar MK. Antimicrobial resistance in sexually transmitted infections. J Travel Med. 2020;27(1):taz101.
Crossref - Krupp K, Madhivanan P. Antibiotic resistance in prevalent bacterial and protozoan sexually transmitted infections. Indian J Sex Transm Dis AIDS. 2015;36(1):3-8.
Crossref - Unemo M, Shafer WM. Antibiotic resistance in Neisseria gonorrhoeae: origin, evolution, and lessons learned for the future. Ann N Y Acad Sci. 2011;1230:E19-E28.
Crossref - Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482-501.
Crossref - Beeton ML, Spiller OB. Antibiotic resistance among Ureaplasma spp. isolates: cause for concern? J Antimicrob Chemother. 2017;72(2):330-337.
Crossref - Boujemaa S, Mlik B, Allaya AB, Mardassi H, Mardassi BBA. Spread of multidrug resistance among Ureaplasma serovars, Tunisia. Antimicrob Resist Infect Control. 2020;9(1):19.
Crossref - Song J, Wu X, Kong Y, et al. Prevalence and antibiotics resistance of Ureaplasma species and Mycoplasma hominis in Hangzhou, China, from 2013 to 2019. Front Microbiol. 2022;13:982429.
Crossref - Kechagia N, Bersimis S, Chatzipanagiotou S. Incidence and antimicrobial susceptibilities of genital mycoplasmas in outpatient women with clinical vaginitis in Athens, Greece. J Antimicrob Chemother. 2008;62(1):122-125.
Crossref - Redelinghuys MJ, Ehlers MM, Dreyer AW, Lombaard HA, Kock MM. Antimicrobial susceptibility patterns of Ureaplasma species and Mycoplasma hominis in pregnant women. BMC Infect Dis. 2014;14:171.
Crossref - Rajan S, Nair SR. Prevalence and Antibiotic Susceptibility Patterns of Mycoplasma hominis and Ureaplasma urealyticum in Females with Genital Infections from Central Kerala, India. J Clin Diagn Res. 2021;15(11):DC08-DC11.
Crossref - Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27(3):587-613.
Crossref - Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20(6):356-370.
Crossref - Gonzalez JS, Hendriksen ES, Collins EM, Duran RE, Safren SA. Latinos and HIV/AIDS: examining factors related to disparity and identifying opportunities for psychosocial intervention research. AIDS Behav. 2009;13(3):582-602.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.