ISSN: 0973-7510
E-ISSN: 2581-690X
Various microorganisms can grow in egg-based food products, and yeast is among the microorganisms that thrive in egg products. Yeasts naturally produce large enzymes by breaking down organic materials during growth. Yeast has a great biotechnological potential owing to its extracellular enzymatic activity. Therefore, it is important to study yeast species, especially those from food sources and the environment. Protease production and activity are affected by various factors such as temperature, time, and pH. In this study, 47 yeast isolates were identified and purified from traditionally processed Indonesian duck eggs via repeated sub-culturing and quadrant streaking. Screening of the yeast isolates on skim milk agar medium showed that 23 isolates exerted proteolytic activity, whereas the remaining 25 did not. Yeast isolates with proteolytic capabilities belonged to the Saccharomyces, Pichia, and Candida genera. The highest protease activities were observed in isolates TA-17 and TPi-08, with activity values of 0.618 and 0.098 U/mL, respectively.
Identification, Isolation, Characteristics, Duck Eggs, Yeast Isolates
Duck eggs, a readily available and inexpensive source of protein, are widely consumed by Indonesians. According to the data from the Central Agency of Statistics (2023), the average duck egg consumption per capita per week in 2022 was 3.66 eggs. Duck egg production increased to 355,187.10 tons in 2022 from 344,470.12 tons in 2021. Duck eggs are a renowned source of animal protein because they are delicious, easy to digest, and highly nutritious. Various microbes can develop in egg products and are particularly useful in the fermentation process. Therefore, certain microbes are widely used in the egg-processing industry.
Yeast is a microscopic unicellular fungus with sizes ranging from 5-20 µm in length and 1-10 µm in width.1 Yeast cells have various shapes, including cocci, cylindrical cells, bacilli, and apiculates.1 Yeast is widely used in several industrial products such as food, beverages, waste processing, and chemical manufacturing industries.2,3 These microorganisms play a significant role in the quality control and safety of processed food products. Yeast can naturally produce considerable amounts of enzymes by digesting organic materials during its growth.4 Therefore, yeast is classified according to the catalytic reactions, including oxidoreductase (aids in oxide-reduction), transferase (aids in molecular group transfer), hydrolase (aids in hydrolysis), lysis (bond-breaking), isomerase (aids in intramolecular change), and ligase (aids in covalent bond formation between two molecules with high energy consumption).5
Yeasts have great biotechnological potential for extracellular enzymatic activities, making it essential to study yeast species mainly from environmental sources.6 In addition, enzymes derived from yeast are more ecologically friendly and cost-effective than those derived from animals or plants. Yeasts are easy to obtain, have minimal nutritional requirements, short fermentation cycles, and a wide range of catalytic activities. Furthermore, yeast can easily survive in the environment and can be produced at large concentrations by genetic modification.7
Protease enzymes are a class of enzymes used in various industrial sectors. The main purpose of protease enzymes is to hydrolyze or catalyze peptide bonds in proteins. Protease production and activity are also affected by several factors, such as temperature, time, and pH. The optimal factor results in maximum enzyme activity.8 Enzymes are widely used in the food industry owing to their advantages. Enzymes have fewer negative side effects and waste byproducts than synthetic chemical products.2 Microbes are living organisms that produce enzymes for metabolic processes. The enzymes produced by microbes can be both extracellular and intracellular. Microbes are frequently used in the food industry owing to their numerous benefits.3
Several studies have identified the proteolytic activity of various yeast isolates. However, only a few studies have investigated yeast isolates obtained from raw and processed duck eggs. In this study, we screened and identified yeast from duck eggs and processed them to obtain the correct strain that could hydrolyze proteins to break down peptides in eggs.
Isolation of Yeast
Yeast isolates were collected from fresh and processed duck eggs (salted eggs, boiled eggs, pasteurized eggs, and egg flour). All products were serially diluted (101-105) using sodium chloride. The samples (20 µL) from each dilution factor were inoculated on malt extract agar medium. The medium consisted of malt extract 20 g, peptone 1 g, glucose 20 g, agar 20 g, and 1 L sterile distilled water, 50 µg chloramphenicol/mL, which were mixed and autoclaved. The inoculated agar plates were incubated at 30°C for 48 h. Each colony that grew and exerted different characteristics was sub-cultured on a different MEA plate and incubated at 30°C for 48 h.9
Screening for proteolytic activity
A proteolytic activity assay was carried out on 3% skim milk agar (SMA) medium supplemented with chloramphenicol as an antibiotic (100 mg/L). Isolates were incubated for 48 h at 30°C. The proteolytic activity was determined based on the size of the clear zone generated by casein degradation in skim milk. A clear zone (halo zone) around the growth of the yeast colony indicated a positive result for protease production and subsequent proteolytic activity. Proteolytic index was calculated using the following formulation10:
Proteolytic Index = (clear zone diameter – colony diameter)/(colony diameter )
Identification of Yeast
Macroscopic characteristics
Identifying yeast colony morphology involves directly observing the visible characteristics of yeast colonies such as their color, shape, texture, elevation, and surface.11
Microscopic characteristics
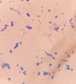
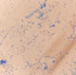
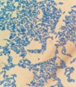
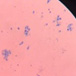
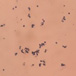
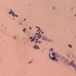
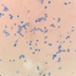
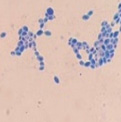
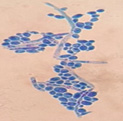
This yeast identification stage was conducted by observing the cell shape, budding, and the presence or absence of hyphae or pseudohyphae using methylene blue staining. The yeast was collected using an inoculation loop and streaked onto a glass slide. The aqueous solution was dropped onto a slide and homogenized using an inoculation loop to spread the yeast cells. The slide was fixed over a Bunsen burner flame and dripped with methylene blue solution. Observations were performed under an electron microscope at 1000׳ magnification using immersion oil. The parameters observed were the shape, budding, and hyphae or pseudohyphae of the yeast cells. 9,12
Sugar Fermentation Test
Sugar fermentation test medium (containing 1% glucose, 1% sucrose, 1% galactose, 1% maltose, and 1% lactose) was added to 1 g of peptone, and bromothymol blue was used as a fermentation indicator. A loop of yeast isolates obtained from the 24 h old solid medium was inoculated into a test tube using a Durham tube. The isolates were incubated for 72 h at 37°C. A positive result was indicated by the color change of the medium from green to yellow and the formation of gas bubbles in the Durham tube.13
Urease test
The urease test was carried out by inoculating the yeast onto the Urea Agar Base (Christensen) slant medium in a test tube, followed by incubation at 30°C for 7 days. Periodic checks were performed to determine the test results. Yeast that positively produced urease was indicated by a color change of the medium from yellow to purplish.13
Capsule staining test
The capsule staining test of yeast cells was performed using the smear method on glass slides. A loop of the isolate was placed on the slide, dripped with crystal violet for ±5 min and then rinsed with 1% CuSO4 solution. Observations were conducted under a microscope at 1000׳ magnification using immersion oil. The capsule appeared as a faint blue circle encircling the purple yeast vegetative cells.13
Yeast growth curve
Cell growth curves were obtained by measuring optical density at 600 nm (OD600 nm) using a UV-visible spectrophotometer.14,15
Characteristics of Yeast’s Protease (optimization of time, temperature, and pH)
Protease characterization includes the activity tests at optimal temperatures (such as 27, 30, 33, 37, and 40°C). The effect of incubation time was examined in the range of 0-48 h. The effect of pH was tested in the pH range of 4-8 using 1N HCl buffer and 1N NaOH buffer.16
SDS-PAGE
Purified proteins were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A discontinuous gel system consisting of stacking and separating gels was used for SDS-PAGE. A high molecular weight protein was used as a marker. The sample (15 µL) was mixed with 10 µL of sample buffer and heated for 1 min before being injected into the well. Electrophoresis was conducted at a current of 20 mA and voltage of 50 volts. Electrophoresis was terminated when the sample dye reached 0.3-0.5 cm from the bottom of the gel. The gel was soaked in Coomassie brilliant blue dye and slowly agitated for 2 h. The gel was then washed by soaking in the de-staining solution until it was clear and the ribbon was visible. Measurement of the molecular weight of the enzymes was based on a standard curve obtained from the linear equation between the molecular weight of the standard protein and its relative mobility.
Protease enzyme activity test
Protease activity was measured following a previously described method17 using Hammerstein casein as the substrate (2% casein in 0.01 M phosphate buffer solution at pH 7.0). The supernatant (0.5 mL) containing protease enzyme solution was added to 0.1 mL of 0.01 M phosphate buffer at pH 7. The reaction mixture was incubated at 37°C for 10 min. The reaction was terminated by the addition of 0.1 M trichloroacetic acid (0.5 mL). Subsequently, the supernatant was separated from the precipitate via centrifugation at 10,000 rpm for 10 minutes at 4°C. The filtrate (0.38 mL) was mixed with 1.25 mL of 0.4 M sodium carbonate and 0.25 mL of Folin-Ciocalteau reagent. The mixture was incubated for 20 min. The optical density (OD) was measured at 578 nm. The blank and standard values were determined similarly, in which the protease sample (0.5 mL) was replaced with 0.5 mL of distilled water (0.5 mL) and tyrosine (5 mM). One unit (U) of protease enzyme activity is the enzyme required to produce 1 µmol of tyrosine per minute under the test conditions. Protease activity was calculated as follows:
PU= (Asp-Abl)/(Ast-Abl) x 1/Q
PU: Protease activity units (units/mL)
Asp: Sample absorbance value
Ast: Standard absorbance value
Abl: Blank absorbance value
Q: Time
Measurement of protein concentration
Protein concentration was measured using a previously described microassay.18 The enzyme sample (0.4 mL) was mixed with 4 mL of Bradford solution, homogenized, and incubated at room temperature (26±0.5°C) for 10 min before measuring the absorbance at 595 nm. A standard curve was constructed using bovine serum albumin (BSA) with a protein concentration range of 0.01-0.1 mL. The specific protease activity (U/mg) was calculated as the ratio of protease activity (U/mL) to protein concentration (mg/mL).
Isolation of yeast from fresh and processed eggs
In this study, 47 yeast isolates were obtained from fresh eggs, boiled eggs, pasteurized eggs, egg flour, salted eggs, and Century eggs. Two eggs were used for each sample. Yeasts can grow in various nutrient-rich foods.19 Eggs have complete nutritional content that can be exploited for energy consumption. Therefore, this study identified several different types of yeasts. Furthermore, we conducted a protease screening of all isolates with proteolytic ability.
Isolation of microorganisms producing protease
In this study, 47 yeast isolates were identified and purified by repeated subculturing and quadrant streaking. The screening of yeast isolates on SMA medium revealed that 23 isolates had proteolytic activity, whereas the remaining 25 isolates did not. The use of skimmed milk has also been reported by other researchers.8,10 The proteolytic activity was characterized by the formation of a clear zone around the colony by the casein substrate, which appeared white in the colloidal suspension of SMA medium after digestion into peptides and amino acids. This hydrolysate is a source of proteins required for cell metabolism, growth, and development.1
Proteolytic yeast isolates show that the yeast can produce extracellular proteases. Proteolytic yeasts are microorganisms capable of producing extracellular proteases. The extracellular protease enzyme is a protein-degrading enzyme produced within cells and released outside the cell1.
This research presented 23 isolates with each proteolytic enzyme activity index shown by the protein hydrolysis index (IHP) (Table 1), namely yeast isolates from pindang eggs (TPi-01, TPi-03, TPi-08), fresh eggs (TS-02, TS-05, TS-07, TS-08, TS-11, TS-12, TS-15, TS-31, TS-32, TS-33), and salted eggs (TA-03, TA-06, TA-07, TA-09, TA-12, TA-16, TA-17, TA-18, TA-20, TA-22). Proteolytic activity is characterized by the presence of a clear zone in the medium, indicating the breakdown of protein molecules in the skim milk added to the medium. The proteolytic index was linear with the diffusion rate of the extracellular protease released by each isolate on the MEA solid medium. The highest proteolytic index was exhibited by the protease from TA-17 isolate (2.03) after 48 h of incubation at 30°C, whereas the lowest index was exhibited by the protease from TA-06 and TA-16 isolates (1.03).
Table (1):
Proteolytic index
| Isolate code | Proteolytic index | |
|---|---|---|
| TPi | 01 | 1.24 |
| 03 | 1.21 | |
| 08 | 1.23 | |
| TS | 02 | 1.17 |
| 05 | 1,11 | |
| 07 | 1.17 | |
| 08 | 1.33 | |
| 11 | 1.67 | |
| 12 | 1.18 | |
| 15 | 1.74 | |
| 31 | 1.82 | |
| 32 | 1.87 | |
| 33 | 1.95 | |
| TA | 03 | 1.62 |
| 06 | 1.03 | |
| 07 | 1.37 | |
| 09 | 1.24 | |
| 12 | 1.18 | |
| 16 | 1.03 | |
| 17 | 2.03 | |
| 18 | 1.28 | |
| 20 | 1.67 | |
| 22 | 1.06 | |
The proteolytic capability of yeast was determined using SMA medium containing casein.10 Skim milk contains casein, which is an enzyme substrate. Casein hydrolysis demonstrates the hydrolytic activities of proteases and peptidases. Proteases and peptidases are involved in the hydrolysis of casein into soluble peptides and amino acids. Casein in the hydrolyzed SMA medium was marked by the presence of a clear zone around the colony. Proteolytic activity was indicated by widening of the clear zone. The results of protein polymer degradation are only shown by the presence of a clear zone, indicating that the protein was broken down into peptides and amino acid compounds soluble in the medium.20
The protein hydrolysis index was calculated for each yeast isolate obtained from the fresh and processed duck eggs. The index value, defined as the ratio of the diameter of the clear zone to the diameter of the yeast isolate colony on the SMA plate, describes the proteolytic capacity of the yeast to hydrolyze proteins. The ratio represents relative protease activity. The protein is degraded by proteolytic yeast into amino acids and further broken down into CO2, H2O, and Ammonia (NH3), which are released into the environment.1
Macroscopic and microscopic identification
Based on the results of macroscopic and microscopic identification (Table 2), three genera were identified: Pichia (TPi-01,03, TS-12,15,31,32, TA-17), Saccharomyces (TPi-08, TS-02, 05, 07, TA-03, 09, 12, 16, 18, 20, 22), and Candida (TS-08, 11, 33, TA-06, 07).12, 21-23 The Pichia genus has a cocci shape, mucoid texture, white color, convex surface, and an entire margin.24 The Saccharomyces genus exhibits a cocci shape/oval, white, and smooth surface colony,25 whereas the Candida genus has a cocci shape, membrane texture, and cream-smooth surface.21
Table (2):
Macroscopic and Microscopic Characterization of Yeast Isolates
The macroscopic characteristics of yeast isolates from the Pichia genus are round shape, viscous, white, cream color, dull surface, slightly prominent elevation, and 2-7 mm colony diameter. These yeast cell characteristics were observed in isolates TA-17 and TS-12. The isolates from Pichia exhibited a round, elliptical, elongated ovoid shape, reproduced by multipolar budding and forming 1-4 ascospores per ascus. The isolates formed single cells, pairs, chains, and groups. The isolates could also ferment sucrose and glucose, but could not ferment lactose or produce gas. Pichia yeast had pseudohyphae and white residues in the test medium (MEA). According to the results, the TA-17 and TS-12 isolates shared characteristics with Pichia. Pichia cells are round, elongated, ovoid, and elliptical. These cells also possess pseudohyphae and exhibit single cell pairs, chains, and group formations. Additionally, cells can reproduce vegetatively through multipolar budding and generatively use ascospores to ferment glucose and sucrose.21
The Saccharomyces isolate has a round colony shape, viscous texture, white color, dull surface, convex elevation, flat edges, and 0.5-5 mm colony diameter. Yeast have round, oval, and elongated cylindrical shapes with single cells, pairs, and group formations. Yeasts reproduce vegetatively using multipolar budding and generatively by forming 1-10 ascospores per ascus. This yeast can rapidly ferment sucrose, glucose, and galactose but cannot ferment lactose and maltose. Based on the above results, isolates A-18 and TPi-08 were comparable to Saccharomyces.21
The morphology of the Saccharomyces colony exhibited a round shape, white and cream color, cream texture, smooth and dull surface, and flat, slightly protruding, folded, and convex elevation. This yeast displays elongated round, oval, and cylindrical cell shapes; forms pseudohyphae; and exhibits single-cell, pair, and group formations. In addition, this yeast reproduces vegetatively by multipolar shoot multiplication and generatively by 1-4 or more ascospores per ascus. This yeast could also ferment sugars. This yeast grows on MEA and forms white or cream residues at the bottom of the test medium without pellicle.26
Candida yeasts show many cell shapes ranging from round, oval, and cylindrical to elongated, rare-apiculate, ogival, triangular, or bottle-shaped, with or without pseudohyphae. This yeast species reproduces asexually through multilateral budding. Candida yeast do not produce ascospores, arthrospores, teliospores, or ballistospores, but chlamydospores may occur in some species. Its color ranges from white to cream because it lacks carotenoid pigments. Some Candida species can ferment, whereas others cannot.21 These characteristics were observed in the TS-33 isolate, which exhibited white colonies, an elongated oval cell shape, multilateral reproduction, and glucose fermentation capability. The absence of capsules supported the hypothesis that this isolate was Candida yeast.
Proteolytic enzyme activity
In the salted egg samples, isolates TA-17 and TA-18 showed the most significant enzyme activity (0.618 U/mL). The TS-12 isolate obtained from fresh egg samples exhibited the highest activity. The TPi-08 isolate displayed the highest activity (0.0976 U/mL) among the yeast isolates obtained from pindang egg samples. Macroscopic and microscopic identification revealed that the TA-17 isolate was comparable to the TS-12 isolate, whereas the TA-18 isolate was similar to the TPi-08 isolate. Therefore, the TA-17 and TPi-08 isolates were subjected to characterization tests that involved the optimization of time, temperature, and pH (Table 3).
Table (3):
Three genera identified in this study
No |
Parameter |
Isolate Ta-17 |
Isolate TPi-08 |
|---|---|---|---|
1 |
Macroscopic |
||
Shape |
Cocci |
Cocci |
|
Texture |
Viscous |
Butyrous |
|
Colors |
White |
White |
|
Surface |
Dull |
Glistening and smooth |
|
Elevation |
Raised |
Convex |
|
Margin |
Entire |
Entire |
|
2 |
Microscopic |
||
Shape |
Cocci |
Cocci |
|
Budding |
Monopolar |
Multilateral |
|
Hifa |
– |
Pseudohifa |
|
Gram strain |
+ |
+ |
|
4 |
Capsule Staining Test |
+ |
+ |
5 |
Sugar Fermentation Test |
||
Glukosa |
+ |
+ |
|
Sukrosa |
+ |
+ |
|
Maltose |
– |
– |
|
Laktosa |
– |
– |
|
Galaktosa |
+ |
– |
|
6 |
Urease test |
– |
– |
7 |
pH |
4.69 |
5.16 |
8 |
Identification proximity |
Saccharomyces |
Pichia |
Description: (+) positive; (-) negative
The potential proteolytic yeast candidates test was carried out based on the highest enzyme activity value in the casein protein substrate (Table 4).
Table (4):
Proteolytic enzyme activity
| Isolate Code | Enzyme Activity | Protein content | Specific enzyme activity |
|---|---|---|---|
| U/mL | mg/mL | U/mg | |
| ta 03 | 0.0383 | 0.0084 | 4.5578 |
| ta 06 | 0.0441 | 0.0402 | 1.0974 |
| ta 07 | 0.1378 | 0.0235 | 5.8661 |
| ta 09 | 0.1203 | 0.0181 | 6.6493 |
| ta 12 | 0.0819 | 0.0244 | 3.3552 |
| ta 16 | 0.0249 | 0.0166 | 1.5026 |
| ta 17 | 0.6180 | 0.0099 | 62.4253 |
| ta 18 | 0.6180 | 0.0080 | 77.2513 |
| ta 20 | 0.0817 | 0.0191 | 4.2775 |
| ta 22 | 0.0678 | 0.0460 | 1.4740 |
| ts 02 | 0.0374 | 0.0069 | 5.4187 |
| ts 07 | 0.0408 | 0.0200 | 4.3350 |
| ts 08 | 0.0951 | 0.0288 | 3.3040 |
| ts 11 | 0.1323 | 0.0167 | 7.9248 |
| ts 12 | 1.3934 | 0.0188 | 74.1193 |
| ts 15 | 0.0296 | 0.0271 | 1.0924 |
| ts 31 | 0.0170 | 0.0515 | 0.3296 |
| ts 32 | 0.0892 | 0.1341 | 0.6656 |
| ts 33 | 0.0817 | 0.4730 | 0.1727 |
| tpi 01 | 0.0442 | 0.0073 | 6.0578 |
| tpi 03 | 0.0488 | 0.0054 | 9.0394 |
| tpi 08 | 0.0976 | 0.0161 | 6.0638 |
Production time optimization
In this study, the OD was used to indicate yeast growth performance. According to the changes in the OD value during the growth of the TA-17 isolate (Figure 1), the OD value increased until 42 h, before slightly decreasing at 48 h. TPi-08 isolate showed an elevated OD value after 48 h. According to a previous study,27 the growth of Pichia continued to increase until 48 h and then declined as it was incubated until 72 h. Similarly, a previous study28 revealed that the highest OD value for Saccharomyces isolates was observed at 48 h. In the balanced state, OD values did not show any significant changes.
Enzyme production can be determined by measuring the enzyme activity. The enzyme concentration is linearly proportional to the enzyme activity. The enzymatic activities of the two isolates exhibited different optimization times. The highest enzymatic activity of TA-17 isolate was shown at 18 h, whereas that of TPi-08 isolate was shown at 48 h. An increased number of yeast cells increased their ability to break down proteins in the substrate, as shown by the activity of the TPi-08 isolate. Enzymes and other acid products can inhibit the formation of the enzyme-substrate complexes. Therefore, structural changes can occur, causing the active site of the enzyme to change, preventing it from properly binding to the substrate.
Effect of temperature
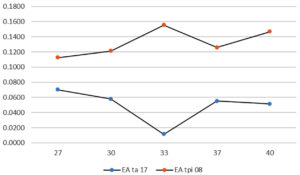
As the temperature increased, the enzyme activity increased until it reached its maximum. However, constantly raising the temperature after the enzyme reaches its maximum phase can cause proteins to denature, decreasing the enzyme activity until it returns to the minimum phase.29 Each enzyme exhibited optimal activity. The optimum activity of the protease enzyme from the TA-17 isolate was exhibited at 27°C, whereas that from the TPi-08 isolate was exhibited at 33°C (Figure 2).
The enzyme activity of the TPi-08 isolates elevated as the incubation temperature increased, although its highest activity was found at 33°C (0.14 U/mL). In contrast, the enzyme activity of the TA-17 isolate decreased as the temperature increased, although its highest activity was observed at 27°C (0.07 U/mL). According to several previous studies, Saccharomyces exhibited optimum activity at 25-35°C and ideal growth at 28-33°C.25,30
Effect of pH
In addition to temperature, pH also affects enzyme activity. pH plays an essential role in enzyme activity. The environmental conditions of the enzyme must be compatible with its characteristics so that it can function optimally and produce maximum outcomes.29 When pH is altered, the ionization process of the enzyme and substrate can change, thereby affecting the rate of substrate binding. The ideal pH value for various yeast species ranged from 4.2–6.0.31 Isolates TA-17 and TPi-08 showed the highest enzyme activities at pH 5 (0.134 U/mL and 0.11 U/mL, respectively). Saccharomyces and Pichia genera grew ideally in the pH range of 5-6.27 Furthermore, as the pH increased above 6, protease activity declined in the range of 0.03–0.07 U/mL. The decline in protease activity may have been caused by the pH not being within the ideal range of 6–8. Therefore, the TA-17 and TPi-08 isolates from salted and boiled eggs exhibited the highest protease activity at pH 5.
A previous study32 reported that Saccharomyces isolates exhibit optimal activity at pH 4-6. Similarly, Pichia isolate had an optimal pH of 4-5.27 Extreme pH alterations may cause enzyme denaturation by disrupting various non-covalent interactions that maintain the stability of the enzyme’s three-dimensional structure.33
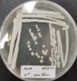
SDS-PAGE
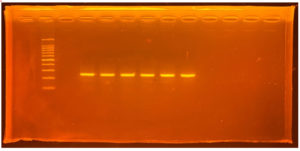
The molecular weight of the proteins from the TA-17 isolates was characterized using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Electropherograms of the isolates and reference standards are shown in Figure 3. The molecular weights of the isolates were in the range of 20-30 kDa.
In conclusion, the yeast isolates with proteolytic abilities discovered in this study were derived from the Saccharomyces, Pichia, and Candida genera. Isolates TA-17 and TPi-08 showed the highest protease activity, with activity values of 0.618 and 0.098 U/mL, respectively.
ACKNOWLEDGMENTS
The authors would like to thank the Unpad Doctoral Dissertation Research (RDDU) grant scheme provided by the Directorate of Research and Community Service (DRPM) at Padjadjaran University.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
The research was funded by the Unpad Doctoral Dissertation Research (RDDU), Indonesia, with grant number 1595/UN6.3.1/PT.00/2021.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Kaiser G. Microbiology. Baltimore: LibreTexts. 2019.
- Ma J, Sun Y, Meng D, Zhou Z, Zhang Y, Yang R. Yeast proteins: The novel and sustainable alternative protein in food applications. Trends Food Sci Technol. 2023;135(2022):190-201.
Crossref - Sadeghi A, Ebrahimi M, Shahryari S, Kharazmi MS, Jafari SM. Food applications of probiotic yeasts; focusing on their techno-functional, postbiotic and protective capabilities. Trends Food Sci Technol. 2022;128:278-295.
Crossref - Gopinath SCB, Anbu P, Hilda A. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience. 2005;46(2):119-126.
Crossref - Cai YD, Chou KC. Predicting enzyme subclass by functional domain composition and pseudo amino acid composition. J Proteome Res. 2005;4(3):967-971.
Crossref - Carvalho JK, Panatta AAS, Silveira MAD, et al. Yeasts isolated from a lotic continental environment in Brazil show potential to produce amylase, cellulase and protease. Biotechnol Rep. 2021;30.
Crossref - Vyas S, Chhabra M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour Technol. 2017;223:250-258.
Crossref - Suganthi C, Mageswari A, Karthikeyan S, Anbalagan M, Sivakumar A, Gothandam KM. Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J Genet Eng Biotechnol. 2013;11(1):47-52.
Crossref - Bitew D, Tesfaye A, Andualem B. Isolation, screening and identification of ethanol producing yeasts from Ethiopian fermented beverages. Biotechnol Rep. 2023;40:e00815.
Crossref - Shaikh IA, Turakani B, Malpani J, et al. Extracellular Protease Production, Optimization, and Partial Purification from Bacillus nakamurai PL4 and its Applications. J King Saud Univ – Sci. 2023;35(1):102429.
Crossref - Ali NM, Khan MM. Screening, identification and characterization of alcohol tolerant potential bioethanol producing yeasts. Curr Res Microbiol Biotechnol. 2014;2(1):316-324.
- Widiastutik N, Alami NH. Isolasi dan Identifikasi Yeast dari Rhizosfer Rhizophora mucronata Wonorejo. J Sains Dan Seni Pomits. 2014;3(1):11-16.
Crossref - john GH, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey’s Manual of Determinative Bacteriology. 9th. Baltimore, Maryland: Lippincott Williams & Wilkins; 1994.
- Nassar F, Abdelhafez A, El-Taye T, Abu-Hussein S. Proteases Production by a Bacterial Isolate Bacillus amyloliquefaciens 35s Obtained from Soil of the Nile Delta of Egypt. Br Microbiol Res J. 2015;6(6):315-330.
Crossref - Rachman SD, Maulida N, Safari A, Anggraeni NI, Fadhlillah M. Peranan Ion Logam Kobalt Terhadap Kinerja Fermentasi dan Toleransi Cekaman Lingkungan Sel Ragi Saccharomyces cerevisiae. Chimica et Natura Acta. 2019;7(3):114- 124.
Crossref - Sumardi S, Farisi S, Ekowati CN, Diana MS. Aktivitas Dan Karakterisasi Enzim Protease Isolat Bacillus Sp. (Uj132) Secara Kualitatif Dan Kuantitatif. J Ris Akuakultur. 2019;14(3):193.
Crossref - Walter H. Proteinase (Protein Substrates). Method with Haemoglobin, Casein and Azocoll as Substrate. In: Bergmeyer J, Grass M (Eds). Methods of Enzymatic Analysis. third edition. Weinheim Germany: Verlag Chemie; 1984.
- Bradford MM. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248-254.
Crossref - Riesute R, Salomskiene J, Moreno DS, Gustiene S. Effect of yeasts on food quality and safety and possibilities of their inhibition. Trends Food Sci Technol. 2021;108:1-10.
Crossref - Hastuti US, Nugraheni FSA, Asna PM Al. Identifikasi dan penentuan indeks hidrolisis protein pada bakteri proteolitik dari tanah mangrove di Margomulyo, Balikpapan. Proceeding Biol Educ Conf. 2017;14(1):265-270.
- Kurtzman CP, Fell JW, Boekhout T. The Yeasts, a Taxonomic Study Volume 1 Fifth Edition. Elsevier publications. 2011.
- Lina L, Rusmiyanto E, Kurniatuhadi R. Khamir Potensial Probiotik Hasil Isolasi dari Fermentasi Jus Jeruk Siam (Citrus nobilis var. microcarpa). Biol Samudra. 2022;3(2):115-132.
Crossref - Anggraini I, Ferniah RS, Kusdiyantini E. Isolasi Khamir Dari Batang Tanaman Tebu Dan Identifikasinya Berdasarkan Sekuens Internal Transcribed Spacer. J Bioteknol Biosains Indones. 2019;6(1):39.
Crossref - Gerard LM, Corrado MB, Davies CV, Soldב CA, Dalzotto MG, Esteche S. Isolation and identification of native yeasts from the spontaneous fermentation of grape musts. Arch Microbiol. 2023;205(9):1-14.
Crossref - Li Y, Liu T, He G. Isolation and identification of yeasts from Tibet Kefir. Adv J Food Sci Technol. 2015;7(3):199-203.
Crossref - Kurtzman CP, Robnett CJ, Yarrow D. Three new species of Candida from apple cider: C. anglica, C. cidri and C. pomicola. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol. 2001;80(3-4):237-244.
Crossref - Muche N, Geremew T, Jiru TM. Isolation and characterization of potential probiotic yeasts from Ethiopian injera sourdough. 3 Biotech. 2023;13(9):1-16.
Crossref - Olivares-Marin IK, Gonzalez-Hernandez JC, Regalado-Gonzalez C, Madrigal-Perez LA. Saccharomyces cerevisiae exponential growth kinetics in batch culture to analyze respiratory and fermentative metabolism. J Vis Exp. 2018;2018(139):1-10.
Crossref - Robinson PK. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1-41.
Crossref - Lip KYF, Garcia-Rios E, Costa CE, et al. Selection and subsequent physiological characterization of industrial Saccharomyces cerevisiae strains during continuous growth at sub- and- supra optimal temperatures. Biotechnol Rep. 2020;26.
Crossref - Blanco P, Sieiro C, Villa TG. Production of pectic enzymes in yeasts. FEMS Microbiol Lett. 1999;175(1):1-9.
Crossref - Graff S, Chaumeil JC, Boy P, Lai-Kuen R, Charrueau C. Influence of pH conditions on the viability of Saccharomyces boulardii yeast. J Gen Appl Microbiol. 2008;54(4):221-227.
Crossref - Hames BD. Biochemistry: The Instant Notes. Second Edition. Hongkong: Springer-Verlag. 2000.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.