ISSN: 0973-7510
E-ISSN: 2581-690X
The use of antibiotics against a range of pathogenic bacteria has increased in recent years, leading to the development of drug resistance, which makes disease control challenging. Thus, the need for the development of new antibacterial medications is critical. Natural resources, such as entomopathogenic bacteria (EPB), provide a rich source of metabolites with well-known antibacterial properties. The present study aimed to investigate the antibacterial activity of symbiotic (n = 1) and non-symbiotic (n = 8) entomopathogenic bacterial species associated with the entomopathogenic nematode (EPN) Steinernema feltiae against four multidrug-resistant bacterial species. Bacterial cells and filtrates from Xenorhabdus bovienii strongly inhibited the growth of Staphylococcus aureus (33.3 and 28.9 mm) and Escherichia coli (24.6 and 21.6 mm) in disk diffusion, minimum inhibitory concentration (MIC) (2 and 8 µl/ml) and minimal bactericidal concentration (MBC) (4 and 12.5 µl/ml) assays. In conclusion, the direct application of endogenous S. feltiae-associated EPB, especially X. bovienii, appears promising as an antibacterial agent against multidrug-resistant bacteria (MRBs).
Symbiotic and Non-symbiotic Entomopathogenic Bacteria, Multidrug-resistant Bacteria, Antibacterial Activity
Antibiotic multidrug resistance has adverse implications for public health worldwide. In terms of public health, Staphylococcus aureus, Escherichia coli, Enterococcus faecalis, Bacillus cereus, Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa are important multidrug-resistant bacteria.1 Antibiotics are commonly used due to their effectiveness against a wide range of pathogenic bacteria. However, such use has led to an increase in antibiotic drug resistance, which raises the risk of a lack of effective antibiotics in the future, with implications for public health.2-4 Multidrug-resistant bacterial infections, although difficult to measure accurately, are expected to result in nearly 10 million deaths annually by 2050.5 Therefore, treatments other than antibiotics are required to target bacteria that are resistant to multiple drugs.
One treatment strategy is the use of biological compounds derived from bacterial or natural sources. These compounds (secondary metabolites or natural products) are a key starting point when developing new pharmaceuticals. Novel compounds with unique structures, high activity, and high selectivity have been discovered via screening natural products.6 Fungi,7,8 plants,9 and bacteria6,10,11 are the three major producers of natural products in nature. An alternative that seems preferable is using antimicrobial peptide (AMP) molecules, which are synthesized by soil-dwelling organisms. Many bacterial species produce antimicrobial toxins in the form of secondary metabolites. In entomopathogenic nematode/bacterium symbiotic relationships, the prokaryotic symbiont’s antimicrobials have preserve the monoxenic environment for EPB in the intestine of infective dauer EPN juveniles.12 Xenorhabdus and Photorhabdus, members of the Enterobacteriaceae family of gram-negative bacteria, which form symbiotic associations with infected EPN juveniles of the Steinernema and Heterorhabditis genera, respectively, provide new bioactive pharmaceuticals for treating microorganisms.13 EPNs deliver cooperative bacteria into the insect hemolymph when they infect a target insect. These bacteria release toxins and enzymes, which kill the insect host within 48 hours.14,15 The nematode-infected carcass is kept from opportunistic pathogens, such as protozoa, fungi, bacteria, and viruses, by a variety of natural compounds produced by Xenorhabdus and Photorhabdus bacteria.16-18 These bacteria are motile rods, oxidase-negative, non-spore forming, chemoorganotrophic heterotrophs, with respiratory and fermentative metabolism and facultative anaerobes.19
In the past, the association between EPNs and their mutualistic bacteria was thought to be monoxenic. However, several recent studies have revealed that what exhibited decreased virulence when injected into insects.20-22 Xenorhabdus, the primary symbiont, and a dozen other regularly occurring microbiota (e.g. Deftia, Stenotrophomonas, Pseudomonas, Achromobacter, Alcaligenes, Pseudochrobactrum, Brevundimonas, and Ochrobactrum) constituted the bacterial group associated with laboratory-reared dauers from Steinernema carpocapsae, Steinernema feltiae, Steinernema glaseri and Steinernema weiseri.23 The general hypothesis is that non-symbiotic bacteria ‘hitchhike’ in infected juvenile vectors at random through the cuticle or inter-cuticular region and enter the insect haemocoel during infected juvenile penetration.24
Xenorhabdus strains are thought to have commercial potential in the production of novel antibiotics, which could be used to manage therapeutically significant pathogens, as well as oral insecticidal toxins for use in biocontrol.25 Previous research demonstrated that bacterial antimicrobial resources, including symbiotic or non-symbiotic bacterial cells and filtrates, successfully prevented the growth of K. pneumonia, E. coli, and Enterobacter cloacae,26 Bacillus subtilis and Botrytis cinerea,27 Streptococcus pyogenes and S. aureus,28,29 Rhizoctonia solani, Phytophthora capsici, and Bacillus anthracis30 and Fusicladium effusum.31 These bacteria produce numerous secondary metabolites with antibiotic activity.29 These metabolites include chaiyaphumine,32 1-carbapen- 2-em-3-carboxylic acid,26 3-hydroxy-2-isopropyl-5-phenethylphenyl carbamate,33 3,5-dihydroxy-4-isopropystilbene,27 2-isopropyl-5-(3-phenyl-2-oxiranyl0-benzene-1,3diol,34 and benzaldehyde.30 Data on the antibacterial potential of EPB linked to EPNs in South western part of Saudi Arabia are lacking. Thus, this study aimed to assess the antibacterial effectiveness of EPB associated with S. feltiae against four multidrug-resistant bacterial species (B. cereus, E. cloacae, E. coli, and S. aureus) using disk diffusion, minimum inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) assays.
S. feltiae-associated bacteria
EPB associated with the EPN S. feltiae were originally isolated and characterized by Prof. Dr. Ahmed Noureldeen, Department of Biology, Faculty of Sciences, Taif University, Taif, Saudi Arabia.35-37 Nine isolates of EPB (Xenorhabdus bovienii, Stenotrophomonas maltophilia, Stenotrophomonas tumulicola, Pseudomonas mosselii, Pseudochrobactrum saccharolyticum, Serratia liquefaciens, Lysinibacillus xylanilyticus, Advenella kashmirensis and Aeromonas hydrophila) were maintained on NBTA medium (nutrient agar with 0.025% bromothymol blue and 0.004% triphenyl tetrazolium chloride) and incubated 2 days at 28°C. To produce cell suspensions or cell-free conditioned filtrates, Five millilitres of Luria-Bertani (LB) broth were inoculated with one colony of each bacterial isolate, and they were stirred constantly (220 rpm) overnight at 28°C. Then, 400 ml of LB medium with 100 ml aliquots of the culture that had been shaken for 24 hours at room temperature were transferred to flasks and agitated at 200 rpm for five days. The multiplied bacterial culture was centrifuged (13,000 rpm for 30 minutes) at 4°C to obtain the supernatant and bacterial pellets. After that, a 0.22 m Millipore filter was used to separate a filtrate free of cells, and the pellet was resuspended in sterile distilled water. Further dilution of the filtrate with sterilized- distilled water was done to obtain concentrations of 300, 150, 75, 50, 25, 12.5, 8, 4, 2 and 1 μl/ml and then stored at 4°C. A spectrophotometer was used to adjust the bacterial cell culture from OD600 to 1.0. A spread plate with a 10-fold serial dilution generated a bacterial culture containing 1 × 106 (CFU/ml).
Multidrug-resistant bacteria preparation
Four isolates of MRBs (E. coli, S. aureus, B. cereus and E. cloacae) were employed. The bacteria were plated onto Mueller–Hinton agar (MHA), which was then incubated at 37°C for 24 hours. In order to adjust the 0.5 McFarland standard for turbidity, one colony was dissolved in 0.85% sodium chloride. A sample of the bacterial culture (100 μl) was then spread on the MHA for a disk diffusion test.38
Antibacterial activity
An MHA plate containing the MRBs was coated with 20 μl of each isolate’s cell suspension (106 CFU/ml) or filtrate (150 μl/ml) to test the antibacterial potential of the nine entomopathogenic bacterial species. Following that, the plates were left to incubate for 24 hours at 37°C. A clean area from the edge of a bacterial colony that was expanding read positive results. A disc sensitivity test was conducted on EPB isolates that suppressed at least one multidrug-resistant bacterium.
Disk diffusion method
Twenty microliters of each filtrate (150 μl/ml) or cell suspension (106 CFU/ml) from the nine bacterial isolates were loaded onto sterile 6 mm paper discs, which were then placed on MHA plates with the isolates that were resistant to multiple drugs. Positive controls included antibiotic discs with ampicillin and penicillin, and negative controls included discs with distilled water. Eight replicates of each entomopathogenic bacterial isolate were tested against various MRBs. The plates were then stored at 37°C for 24 hours. Using a ruler, the clear zone’s diameter (mm, representing the zone of inhibition) was estimated.
MIC and MBC assays
To determine the MIC, the broth microdilution technique was employed, utilizing the bacterial filtrates that give the best results for disk diffusion. Ten serial dilutions of the bacterial filtrate (300, 150, 75, 50, 25, 12.5, 8, 4, 2 and 1 μl/ml) were undertaken in a 96-well micro-titre plate. In a control treatment, multidrug-resistant bacteria were incubated in sterile Mueller–Hinton broth. After that, the plates were kept for 24 hours at 37°C. The MIC was the minimal filtrate amount that caused the well-located bacteria were not proliferating. To determine the MBCs, a subculture of 10 µl from each well of the 96-well MIC micro-titre plate was placed onto MHA plates. The plates were then incubated at 37°C for 24 hours. The lowest concentration of each filtrate at which bacteria did not multiply was the MBC. All MIC and MBC assays were performed in triplicate.
Statistical analysis
Results are displayed as mean±standard error (M±SE). A one-way analysis of variance was conducted to statistically analyse the data, using the CoStat program, followed by multiple comparisons. P-values that were 0.05 or less were regarded as significant.
Screening of EPB isolates against multidrug-resistant bacteria
The filtrates and the cell suspension of bacteria associated with S. feltiae inhibited most of the tested multidrug-resistant bacteria, with the level of inhibition varying from weak to strong (Table 1). E. coli was sensitive to eight of the EPB isolates, while seven of them were effective against S. aureus. B. cereus and E. cloacae were susceptible to six of the bacterial isolates. Among all the EPBs examined, X. bovienii filtrates and cells showed the strongest inhibition activity (+++) against all the tested multidrug-resistant bacteria. Cells of S. maltophilia strongly inhabited E. coli, E. cloacae, and S. aureus. Filtrates of this bacterium showed strong inhibition potential against E. cloacae and S. aureus and moderate inhibition activity (++) on B. cereus and E. coli. Although S. tumulicola and P. mosselii cells exhibited strong inhibition activity against E. coli and S. aureus, their filtrates exhibited moderate or weak (+) inhibition activity against these bacteria. No inhibition activity was detected when B. cereus, E. coli and E. cloacae bacteria were treated with P. saccharolyticum cells or filtrates. Except for E. coli, which was susceptible only to L. xylanilyticus cells, all the MRBs were resistant to its cells or filtrates (Table 1).
Table (1):
Antibacterial activity of filtrates or cells of nine EPB species against multidrug-resistant bacteria
| EPBs | Bacterial suspension form | The growth inhibition a | |||
|---|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | ||
| Xenorhabdus bovienii | Filtrates | +++ | +++ | +++ | +++ |
| Cells | +++ | +++ | +++ | +++ | |
| Stenotrophomonas maltophilia | Filtrates | +++ | +++ | ++ | ++ |
| Cells | +++ | +++ | +++ | ++ | |
| Stenotrophomonas tumulicola | Filtrates | ++ | + | ++ | + |
| Cells | +++ | ++ | +++ | ++ | |
| Pseudomonas mosselii | Filtrates | + | + | ++ | + |
| Cells | +++ | + | +++ | + | |
| Pseudochrobactrum saccharolyticum | Filtrates | + | – | – | – |
| Cells | + | – | – | – | |
| Serratia liquefaciens | Filtrates | + | + | + | + |
| Cells | ++ | ++ | +++ | ++ | |
| Lysinibacillus xylanilyticus | Filtrates | – | – | – | – |
| Cells | – | – | + | – | |
| Advenella kashmirensis | Filtrates | – | + | + | – |
| Cells | – | + | + | – | |
| Aeromonas hydrophila | Filtrates | – | – | + | – |
| Cells | + | – | + | + | |
| Penicillin | – | – | – | – | |
| Ampicillin | – | – | – | – | |
aNo inhibition (-): 0–5 mm; weak inhibition (+): 6–10 mm; moderate inhibition (++): 11–15 mm; strong inhibition (+++): > 15 mm.
Disk diffusion method
The method of disk diffusion was used to verify the antimicrobial effect of the nine symbiotic and non-symbiotic bacteria associated with S. feltiae (Table 2, Figure). At a concentration of 150 μl/ml filtrates and 106 CFU/ml cells for all nine EPBs, inhibitory zones (means±SE) ranging from 0.0 to 33.3 mm (P < 0.05) were observed. The nine isolates’ bacterial cells were more efficient than their bacterial filtrates against all of the studied MRBs, causing a mean inhibition zone of 8.86 mm for the cells and 6.53 mm for the filtrates. The largest clear zones (33.3 and 28.9 mm) were obtained when S. aureus bacterium was exposed to X. bovienii cells and filtrates, respectively. Moreover, E. coli, B. cereus, and E. cloacae growth was inhibited, with average zones of (24.6 and 21.6 mm), (21.6 and 17.0 mm), and (19.5 and 17.9 mm), respectively. S. maltophilia cells and filtrates also inhibited the growth of S. aureus, E. cloacae, E. coli and B. cereus, with clear zones of 20.4 and 19.3 mm, 19.4 and 18.0 mm, 17.3 and 14.5 mm, and 14.0 and 12.0 mm, respectively. With inhibition zones of 17.9 and 13.6 mm, 12.9 and 9.4 mm, 17.1 and 13.3 mm, and 11.8 and 7.8 mm for S. aureus, E. cloacae, E. coli, and B. cereus, respectively, the cells and filtrates of S. tumulicola were the third most lethal to the examined bacteria. S. liquefaciens ranked fourth in terms of inhibition zone activity against S. aureus (11.8 and 7.6 mm, 11.5 and 7.4 mm, 18.9 and 8.8 mm and 13.1 and 7.9 mm, respectively). Contrarily, S. aureus treated with filtrates of A. kashmirensis and A. hydrophila exhibited the smallest inhibitory zone (0.38 mm). L. xylanilyticus cells and filtrates showed no inhibition activity on B. cereus and E. cloacae (Table 2).
Table (2):
Antibacterial potential of filtrates or cells of nine EPBs against multidrug-resistant bacteria according to disk diffusion analysis
| EPBs | Bacterial suspension form | Inhibition Zone (mm) a | |||
|---|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | ||
| Xenorhabdus bovienii | Filtrates | 28.9±0.77 | 17.9±0.52 | 21.6±0.38 | 17.0±0.46 |
| Cells | 33.3±0.62 | 19.5±0.33 | 24.6±0.49 | 21.6±0.49 | |
| Stenotrophomonas maltophilia | Filtrates | 19.3±0.37 | 18.0±0.46 | 14.5±0.33 | 12.0±0.27 |
| Cells | 20.4±0.32 | 19.4±0.42 | 17.3±0.37 | 14.0±0.27 | |
| Stenotrophomonas tumulicola | Filtrates | 13.6±0.53 | 9.4±0.26 | 13.3±0.56 | 7.8±0.37 |
| Cells | 17.9±0.39 | 12.9±0.39 | 17.1±0.39 | 11.8±0.25 | |
| Pseudomonas mosselii | Filtrates | 7.0±0.33 | 6.4±0.18 | 12.9±0.48 | 6.4±0.18 |
| Cells | 16.9±0.29 | 6.8±0.25 | 16.5±0.27 | 6.8±0.25 | |
| Pseudochrobactrum saccharolyticum | Filtrates | 6.0±0.0 | 0.75±0.41 | 1.0±0.33 | 0.0±0.0 |
| Cells | 6.4±0.18 | 2.8±0.49 | 1.9±0.47 | 0.75±0.41 | |
| Serratia liquefaciens | Filtrates | 7.6±0.38 | 7.4±0.26 | 8.8±0.37 | 7.9±0.39 |
| Cells | 11.8±0.31 | 11.5±0.19 | 18.9±0.29 | 13.1±0.39 | |
| Lysinibacillus xylanilyticus | Filtrates | 0.0±0.0 | 0.0±0.0 | 0.50±0.19 | 0.0±0.0 |
| Cells | 0.88±0.29 | 0.0±0.0 | 6.3±0.16 | 0.0±0.0 | |
| Advenella kashmirensis | Filtrates | 0.38±0.18 | 6.0±0.0 | 6.4±0.18 | 0.50±0.19 |
| Cells | 1.3±0.41 | 6.4±0.18 | 7.1±0.13 | 0.88±0.29 | |
| Aeromonas hydrophila | Filtrates | 0.38±0.18 | 0.0±0.0 | 6.0±0.0 | 0.63±0.18 |
| Cells | 6.8±0.16 | 0.50±0.19 | 7.0±0.0 | 6.0±0.0 | |
| Penicillin | 0.38±0.18 | 0.0±0.0 | 0.50±0.27 | 0.0±0.0 | |
| Ampicillin | 0.63±0.32 | 0.25±.16 | 0.63±0.26 | 0.0±0.0 | |
aIn this experiment, each treatment was represented by four repeats with two plates. The inhibition zone diameters ± standard errors are depicted by numbers in each column.
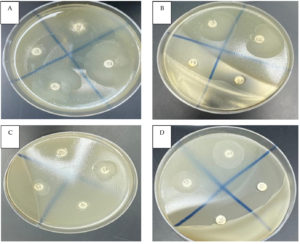
Figure. Disk diffusion test of the symbiotic entomopathogenic bacterium X. bovienii’s filtrate against multidrug-resistant bacteria. Clear zone of S. aureus (A13), E. coli (B18), B. cereus (C17) and E. cloacae (D14). Penicillin (B17), ampicillin (B20) and negative control (D15)
MICs
The values of MIC for the nine symbiotic and non-symbiotic bacteria linked to the EPN S. feltiae against four bacterial species were also assessed, as indicated in Table 3. EPB cell-free conditioned media with MICs between 2 and 300 µl/ml prevented the multidrug-resistant bacteria from growing. Among the bacteria tested, S. aureus (2 µL/ml) exhibited the highest estimated susceptibility to the X. bovienii filtrate, the next is E. coli (8 µl/ml), E. cloacae and B. cereus (12.5 µl/ml). All the tested bacteria were also sensitive to the S. maltophilia filtrate, which showed the strongest inhibitory activity (12.5 µl/ml) on S. aureus and E. cloacae, followed by E. coli (25 µL/ml) and B. cereus (50 µl/ml). S. tumulicola exhibited strong activity against S. aureus and E. coli (50 µl/ml) and moderate activity against E. cloacae and B. cereus (75 µl/ml). The MIC of S. liquefaciens was 75 µl/ml for all the tested bacteria. At a high MIC value (300 µl/ml), the L. xylanilyticus filtrate was only effective against E. coli. A. kashmirensis, A. hydrophila, penicillin and ampicillin also exhibited low activity against S. aureus, with an average MIC value (300 µl/ml).
Table (3):
MICs of EPB filtrates (µl/ml) against multidrug-resistant bacteria
| EPBs | MIC (µL/mL) a | |||
|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | |
| Xenorhabdus bovienii | 2 | 12.5 | 8 | 12.5 |
| Stenotrophomonas maltophilia | 12.5 | 12.5 | 25 | 50 |
| Stenotrophomonas tumulicola | 50 | 75 | 50 | 75 |
| Pseudomonas mosselii | 75 | 75 | 50 | 75 |
| Pseudochrobactrum saccharolyticum | 75 | 150 | 150 | ND |
| Serratia liquefaciens | 75 | 75 | 75 | 75 |
| Lysinibacillus xylanilyticus | ND | ND | 300 | ND |
| Advenella kashmirensis | 300 | 75 | 75 | 300 |
| Aeromonas hydrophila | 300 | ND | 75 | 150 |
| Penicillin | 300 | ND | 300 | ND |
| Ampicillin | 300 | 300 | 300 | ND |
a There were three repetitions of each treatment in this test. The numbers in each column indicate the MIC of each symbiotic or non-symbiotic bacterium. ND = not detected.
MBCs
Except for EPB L. xylanilyticus, the multidrug-resistant bacterium S. aureus was most vulnerable to all filtrates, with MBCs ranging from 4 to 300 µl/ml (Table 4). With an MBC of 4 µl/ml, the X. bovienii filtrate had higher activity against S. aureus than any of the other tested bacteria, but a MIC value of 12.5 µl/ml was reported for E. coli, E. cloacae, and B. cereus. S. aureus was the least susceptible bacterium that was suppressed by A. kashmirensis and A. hydrophila. E. coli had less sensitivity to L. xylanilyticus and B. cereus to A. kashmirensis, with these having the highest MBC values (300 µl/ml).
Table (4):
MBCs of EPB filtrates (µl/ml) against multidrug-resistant bacteria
| EPBs | MBC (µL/mL) a | |||
|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | |
| Xenorhabdus bovienii | 4 | 12.5 | 12.5 | 12.5 |
| Stenotrophomonas maltophilia | 12.5 | 25 | 25 | 50 |
| Stenotrophomonas tumulicola | 75 | 75 | 75 | 75 |
| Pseudomonas mosselii | 150 | 75 | 75 | 150 |
| Pseudochrobactrum saccharolyticum | 150 | 150 | 150 | ND |
| Serratia liquefaciens | 150 | 75 | 75 | 75 |
| Lysinibacillus xylanilyticus | ND | ND | 300 | ND |
| Advenella kashmirensis | 300 | 150 | 75 | 300 |
| Aeromonas hydrophila | 300 | ND | 150 | 150 |
| Penicillin | 300 | ND | 300 | ND |
| Ampicillin | 300 | 300 | 300 | ND |
a All experiments were conducted in triplicate. The numbers in each column indicate the MBC of each symbiotic or non-symbiotic bacterium. ND = not detected.
The present study investigated nine symbiotic and non-symbiotic entomopathogenic bacterial species previously isolated from the EPN S. feltiae against four multidrug-resistant bacteria. The findings support those reported in a previous study, in which the five EPBs associated with S. feltiae were identified, and their efficiency against the two aphid species Aphis illinoisensis and A. punicae, was evaluated.37 The results of the current study also confirm those of earlier work, in which the authors isolated Photorhabdus and Xenorhabdus spp. from Heterorhabditis and Steinernema spp., respectively, in the same province and revealed their complex behaviours against Meloidogyne incognita infecting pomegranate under greenhouse conditions.35 The current research clearly demonstrates that entomopathogenic bacterial isolates provide a novel approach for preventing the spread of several multidrug-resistant bacteria.
Based on their antibacterial activity, the majority of the nine species of S. feltiae-associated bacteria have been shown to prevent the proliferation of pathogenic bacteria, with variation in their antibacterial activity range. This might be as a result of each bacterium’s ability to synthesize therapeutic molecules or the multidrug-resistant bacteria’s sensitivity. In addition, it was clear that the bacterial cells of the nine species were more potent than were the filtrates at eliminating multidrug-resistant bacteria, despite having low toxicity. Among the evaluated bacterial isolates, the strongest inhibitory impact was observed in cells and filtrates from the symbiont X. bovienii, and L. xylanilyticus displayed the weakest inhibition activity against all the tested multidrug-resistant bacteria. These results are consistent with those of previous studies on E. cloacae, K. pneumoniae, E. coli, B. subtilis, B. cinerea, S. pyogenes, S. aureus, B. anthracis, P. capsici, R. solani and F. effusum.25-31 Similarly, these results agree with those of other research on the pathogenic soybean fungus Sclerotinia sclerotiorum and the larvae of the fall webworm (Hyphantria cunea).39,40
Thus far, 29 species of Xenorhabdus and 20 species of Photorhabdus that produce different types of natural products have been isolated worldwide, including Europe, Australia, America, Africa and Asia.19,41 Xenorhabdus strains of EPB have strong commercial potential in the area of new antibiotic production, with such antibiotics targeting clinically relevant bacteria.25 With the exception of P. saccharolyticum, the data obtained showed that E. coli was susceptible to all of the tested bacterial filtrates and cells. S. aureus was susceptible to all isolates, except L. xylanilyticus and A. kashmirensis. E. cloacae showed resistance to P. saccharolyticum, L. xylanilyticus and A. hydrophila. Furthermore, B. cereus appeared resistant to P. saccharolyticum, L. xylanilyticus and A. kashmirensis. Accordingly, based on values of the inhibition zone, MIC and MBC, the toxicity of EPBs toward the tested MRBs could be arranged as follows, in descending order: X. bovienii > S. maltophilia > S. tumulicola > S. liquefaciens > P. mosselii > A. kashmirensis > A. hydrophila > P. saccharolyticum > L. xylanilyticus. In descending order, the susceptibility of the MRBs was as follows: E. coli > S. aureus > E. cloacae > B. cereus. These findings are in accordance with those of earlier studies, which showed that X. budapestensis, X. szentirmaii, X. innexi, X. ehlersii, X. nematophila, X. bovienii and X. cabanillassii, as well as P. luminescens, inhibited the growth of clinical and multidrug-resistant isolates of S. aureus, E. coli, K. pneumoniae, and B. subtilis.26,42 It is evident that Xenorhabdus strains possess a wide variety of antibacterial components, making them potential sources of novel antibiotics against S. aureus, E. coli, E. cloacae and B. cereus that resist conventional antibiotics.43 These results were in agreement with those of previous research, which reported that Xenorhabdus produced derivatives of xenocoumacin and amicoumacin,44,45 each of which has been demonstrated to be an effective antibiotic against S. aureus.29 All Photorhabdus spp. produce isopropylstilbene,46,47 which has a wide range of biological properties, including antibiotic activity against E. coli and S. aureus.33 It has been proven that the S. aureus strains ATCC20475, PB36, and PB57 are sensitive to the inhibitory action of Photorhabdus extracts.48 Interestingly, the antibacterial activity of complete cell-free media was significantly higher than that of any isolated, recognized or patented molecules (e.g. nematophin).49 Xenorhabdus strains’ antibiotically active, unpurified, cell-free liquid cultures are effective against a wide range of pathogens, including bacteria, fungus and protozoa. In earlier research, the protease inhibitor protein-encoding gene from the symbiotic bacterium X. bovienii strains BJFS526 and Xbpi-1 was discovered, produced, and evaluated for its impact on the pea aphid Acyrthosiphon pisum.50,51
According to Fuchs et al.,52 X. szentirmaii is a uniquely important source of peptides with excellent antibacterial properties that inhibit almost all known phytopathogens. In the present study, S. maltophilia and S. tumulicola isolates, either cells or filtrates, ranked second to X. bovienii in inhibiting the growth of the tested multidrug-resistant bacteria. The results agree with those of a previous study, which revealed the termiticidal activity associated with S. maltophilia‘s synthesis of bacterial chitinases.53 In addition, they are in accordance with prior research that discovered S. maltophilia might have antagonistic effect against a spectrum of fungi and bacteria that are multidrug resistant.54 Berg55 stated that S. maltophilia suppressed the phytopathogen Rhizoctonia solani growth, probably because of antibiosis and the production of certain lytic enzymes that destroy pathogenic fungi. Further studies showed that the metabolic complexity of S. maltophilia is responsible for the creation of novel bioactive compounds, including metabolites that could be used in the biocontrol of bacteria, in addition to the existence of enzymes that can be employed in therapeutic applications.56 In the present study, S. liquefaciens and P. mosselii exhibited moderate inhibition activity against the tested bacteria. The other EPB isolates, L. xylanilyticus, P. saccharolyticum and A. hydrophila, exhibited less bactericidal activity. In contrast, a previous study reported that crude chitinase from A. hydrophila may act as a potent biocontrol agent against insects and that it may be a good alternative to chemical pesticides.57 The present study confirmed the potential of one symbiotic and eight non-symbiotic entomopathogenic bacterial species associated with the EPN S. feltiae in Saudi Arabia as antibacterial agents. The present findings are in line with previous studies on nematicidal and insecticidal activities conducted in the Taif distinct of Saudi Arabia.35-37
In the current study, the antibacterial efficacy of one symbiotic and eight non-symbiotic entomopathogenic bacterial species associated with S. feltiae against four important multidrug-resistant bacterial species was investigated. Overall, the results indicated that the EPB isolates had varying effects on several multidrug-resistant bacterial species. This might be due to the capacity of each bacterial species to produce beneficial compounds or the sensitivity of the MRBs to the specific metabolites produced by each symbiont or non-symbiont. Among the species tested, the symbiont X. bovienii exhibited excellent antibacterial activity against the four tested pathogenic bacterial species. The findings might serve as a starting point for the discovery of new bioactive substances.
ACKNOWLEDGMENTS
None.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed
by the author.
- Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2018;197(8):1079-1081.
Crossref - Siedlecka A. The occurrence of antibiotic resistance genes in tap water-a review. E3S Web of Conferences. 2018;30:9.
Crossref - Vicar EK, Acquah SEK, Williams W, Kuugbee ED, Saba CKS, Mensah GI. Antibiotic resistant bacteria infecting wounds of rural community dwellers in northern Ghana. Eur J Med Health Sci. 2021;3(1):112-117.
Crossref - Roe VA. Antibiotic resistance;a guide for effective prescribing in women’s health. J Midwifery Women’s Health. 2018;53(3);216-226.
Crossref - Chokshi A, Sifri Z, Cennimo D, Horng H. Global Contributors to Antibiotic Resistance. J Glob Infect Dis. 2019;11(1):36-42.
Crossref - Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311-335.
Crossref - Brian PW, Hemming HG. Production of antifungal and antibacterial substances by fungi;preliminary examination of 166 strains of fungi imperfecti. Microbiol. 1947;1(2):158-167.
Crossref - Zhang XY, Bao J, Wang GH, He F, Xu XY, Qi SH. Diversity and antimicrobial activity of culturable fungi isolated from six species of the South China Sea gorgonians. Microb Ecol. 2012;64(3):617-627.
Crossref - Cowan MM. Plant products as antimicrobial agents. Clin Microbiol Rev. 1999;12(4):564-582.
Crossref - Kirkup BC. Bacteriocins as oral and gastrointestinal antibiotics;theoretical considerations, applied research, and practical applications. Curr Med Chem. 2006;13(27):3335-3350.
Crossref - Gillor O, Ghazaryan L. Recent advances in bacteriocin application as antimicrobials. Recent Pat. Antiinfect Drug Discov. 2007;2(2):115-122.
Crossref - Fodor A, Gualtieri M, Zeller M, et al. Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC). Pathogens. 2022;11(3):342.
Crossref - Boemare NE, Akhurst RJ, Mourant RG. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43(2):249-255.
Crossref - Gulcu B, Cimen H, Raja RK, Hazir S. Entomopathogenic nematodes and their mutualistic bacteria;Their ecology and application as microbial control agents. Biopestic Int. 2017;13(2):79-112.
- Shapiro-Ilan DI, Hazir S, Glaser I. Advances in use of entomopathogenic nematodes in integrated pest management in Integrated management of insect pests;Current and future developments (Kogan, M.; Heinrichs E.A. eds), Burleigh Dodds Science Publishing, Cambridge, UK. 2020;91-105.
Crossref - Shi YM, Bode HB. Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat Prod Rep. 2018;35(4):309-335.
Crossref - Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus; two roads to the same destination. Mol Microbiol. 2007;64(2):260-268.
Crossref - Chaston JM, Sven G, Tucker SL, et al. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus; convergent lifestyles from divergent genomes. PLoS One. 2011;6(11);e27909.
Crossref - Yimthin T, Fukruksa C, Muangpat P, et al. A study on Xenorhabdus and Photorhabdus isolates from Northeastern Thailand; Identification, antibacterial activity, and association with entomopathogenic nematode hosts. PLoS ONE. 2021;16(8):e0255943.
Crossref - Bisch G, Pages S, McMullen JG, et al. Xenorhabdus bovienii CS03, the bacterial symbiont of the entomopathogenic nematode Steinernema weiseri, is a non-virulent strain against lepidopteran insects. J Invertebr Pathol. 2015;124:15-22.
Crossref - Kim IH, Aryal SK, Aghai DT, et al. The insect pathogenic bacterium Xenorhabdus innexi has attenuated virulence in multiple insect model hosts yet encodes a potent mosquitocidal toxin. BMC Genom. 2017;18(1):927.
Crossref - McMullen JG, McQuade R, Ogier JC, Pages S, Gaudriault S, Stock SP. Variable virulence phenotype of Xenorhabdus bovienii (g-Proteobacteria; Enterobacteriaceae) in the absence of their vector hosts. Microbiology. 2017;163(4):510-522.
Crossref - Ogier JC, Pages S, Frayssinet M, Gaudriault S. Entomopathogenic nematode-associated microbiota; From monoxenic paradigm to pathobiome. Microbiome. 2020;8(1):25.
Crossref - Singh S, Reese JM, Casanova-Torres AM, Goodrich-Blair H, Forst S. Microbial population dynamics in the hemolymph of Manduca sexta infected with Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl Environ Microbiol. 2014;80(14):4277-4285.
Crossref - Sergeant M, Baxter L, Jarrett P, Shaw E, Ousley M. Identification, Typing, and Insecticidal Activity of Xenorhabdus Isolates from Entomopathogenic Nematodes in United Kingdom Soil and Characterization of the xpt Toxin Loci. Appl Environ Microbiol. 2006;72(9):5895-5907.
Crossref - Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. Identification, Characterization, and Regulation of a Cluster of Genes Involved in Carbapenem Biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol. 2002;68(8):3780-3789.
Crossref - Chen G. Antimicrobial Activity of the Nematode Symbionts, Xenorhabdus and Photorhabdus (Enterobacteriaceae), and the Discovery of Two Groups of Antimicrobial Substances, Nematophin and Xenorxides. In;Canada (Doctoral dissertation). British Columbia;Simon Fraser University. 1996.
- McInerney BV, Taylor WC, Lacey MJ, Akhurst RJ, Gregson RP. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J Nat Prod. 1991;54(3):785-795.
Crossref - Bode HB. Bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13(2):224-230.
Crossref - Ullah I, Khan AL, Ali L, et al. Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. J Microbiol. 2015;53(2):127-133.
Crossref - Bock CH, Shapiro-Ilan DI, Wedge DE, Cantrell CL. Identification of the antifungal compound, trans-cinnamic acid, produced by Photorhabdus luminescens, potential biopesticide against pecan scab. J Pest Sci. 2014;87(1):155-162.
Crossref - Grundmann F, Kaiser M, Schiell M, et al. Antiparasitic Chaiyaphumines from entomopathogenic Xenorhabdus sp. PB61.4. J Nat Prod. 2014;77(4):779-783.
Crossref - Shi D, An R, Zhang W, Zhang G, Yu Z. Stilbene derivatives from Photorhabdus temperata SN259 and their antifungal activities against phytopathogenic fungi. J Agric Food Chem. 2017;65(1):60-65.
Crossref - Hu KJ, Li JX, Li B, Webster JM, Chen G. A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae). Bioorg Med Chem. 2006;14(13):4677-468.
Crossref - Nour El-Deen AH, Al-Barty AF, Darwesh HY, Al-Ghamdi AS. Eco-Friendly Management of Root-Knot Nematode, Meloidogyne incognita Infecting Pomegranate at Taif Governorate, KSA. Res J Pharm Biol Chem Sci. 2016;7(1);1070-1076.
- Alotaibi SS, Darwish H, Zaynab M, et al. Isolation, Identification, and Biocontrol Potential of Entomopathogenic Nematodes and Associated Bacteria against Virachola livia (Lepidoptera; Lycaenidae) and Ectomyelois ceratoniae (Lepidoptera;Pyralidae). Biology. 2022;11(2):295.
Crossref - Baazeem A, Alotaibi SS, Khalaf LK, et al. Identification and environment-friendly biocontrol potential of five different bacteria against Aphis punicae and Aphis illinoisensis (Hemiptera; Aphididae). Front Microbiol. 2022;13:961349.
Crossref - Seier-Petersen MA, Jasni A, Aarestrup FM, et al. Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. J Antimicrob Chemother. 2014;69(2)343- 348.
Crossref - Chacon-Orozco JG, Bueno Jr C, Shapiro-Ilan DI, Hazir S, Leite LG, Harakava R. Antifungal activity of Xenorhabdus spp. and Photorhabdus spp. against the soybean pathogenic Sclerotinia sclerotiorum. Sci Rep. 2020;10(1):1-12.
Crossref - Yuksel E, Ozdemir E, Delialioglu RA, Canhilal R. Insecticidal activities of the local entomopathogenic nematodes and cell-free supernatants from their symbiotic bacteria against the larvae of fall webworm, Hyphantria cunea. Experimental Parasitology. 2022;242:108380.
Crossref - Muangpat P, Suwannaroj M, Yimthin T, et al. Antibacterial activity of Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes against antibiotic-resistant bacteria. PLoS ONE. 2020;15(6):e0234129.
Crossref - Fodor A, Fodor AM, Forst F, et al. Comparative analysis of antibacterial activities of Xenorhabdus species on related and non-related bacteria in vivo. J Microbiol Antimicrob. 2010;2(4):36-46.
- Fodor A, Clarke DJ, Dillman AR, Tarasco E, Hazir S. New Antimicrobial Peptides from Bacteria/Invertebrate Obligate Symbiotic Associations. Front Microbiol. 2022;13:862198.
Crossref - Reimer D, Luxenburger E, Brachmann AO, Bode HB. A new type of pyrrolidine biosynthesis is involved in the late steps of Xenocoumacin production in Xenorhabdus nematophila. Chem Bio Chem. 2009;10(12):1997-2001.
Crossref - Park H, Perez C, Perry E, Crawford JM. Activating and attenuating the amicoumacin antibiotics. Molecules. 2016;21(7):E824.
Crossref - Li J, Chen G, Wu H, Webster JM. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol. 1995;61(12):4329-4333.
Crossref - Buscato EL, Buttner D, Bruggerhoff KFM, et al. From a multipotent stilbene to soluble epoxide hydrolase inhibitors with antiproliferative properties. Chem Med Chem. 2013;8(6):919-923.
Crossref - Muangpat P, Yooyangket T, Fukruksa C, et al. Screening of the Antimicrobial Activity against Drug Resistant Bacteria of Photorhabdus and Xenorhabdus Associated with Entomopathogenic Nematodes from Mae Wong National Park, Thailand. Front Microbiol. 2017;8:1142.
Crossref - Brachmann AO, Forst S, Furgani GM, Fodor A, Bode HB. Xenofuranones A and B; phenylpyruvate dimers from Xenorhabdus szentirmaii. J Nat Prod. 2006;69(12):1830-1832.
Crossref - Zeng F, Xue R, Zhang H, Jiang T. A new gene from Xenorhabdus bovienii and its encoded protease inhibitor protein against Acyrthosiphon pisum. Pest Mang Sci. 2012;68(10):1345-1351.
Crossref - Jin D, Zeng F, Dong S, Zhang H. Effects of a protease inhibitor protein from Xenorhabdus bovienii on physiology of pea aphid, Acyrthosiphon pisum. Pestic Biochem Physiol. 2014;108:86-91.
Crossref - Fuchs SW, Grundmann F, Kurz M, Kaiser M, Bode HB. Fabclavines;Bioactive peptide-polyketide-polyamino hybrids from Xenorhabdus. Chembiochem. 2014;15(4):512-516.
Crossref - Jabeen F, Ali H, Manzoor M, Younis T, Rasul A, Qazi JI. Potential of bacterial chitinolytic, Stenotrophomonas maltophilia in biological control of termites. Egypt J Biol Pest Cont. 2018;28:86.
Crossref - Amer A, Hamdy B, Mahmoud D, et al. Antagonistic activity of bacteria isolated from the Periplaneta americana L. gut against some multidrug-resistant human pathogens. Antibiotics. 2021;10(3):294.
Crossref - Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4(11):430-435.
Crossref - Ribitsch D, Heumann S, Karl W, et al. Extracellular serine proteases from Stenotrophomonas maltophilia; Screening, isolation and heterologous expression in E. coli. J. Biotechnol. 2012;157(1):140-147.
Crossref - Aly HH, Abdel Naser AK, Korany SM, El-Hendawy HH, Mansour AN. Physiological effects of crude chitinase from Aeromonas hydrophila on the greater wax moth; Galleria mellonella L. (Lepidoptera;Pyralidae). Egypt J Desert Res. 2019;69(3):101-111.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.