Forensic Microbiology is noted as an emerging sector in forensic science research, and the demand for its application is increasing. Its use plays a pivotal role in refining the evidence used in criminal investigations; for example, in estimating post-mortem intervals, the cause of death, and characterizing clandestine burials. The use of such advanced strategies in recent years has been made possible through the integration of two sectors, microbiology and forensic science research. In this respect, this article reviews the forensic utility of microbial populations supported by advances in sequencing and bioinformatics in the context of its various applications, such as microbial profiling for identification, estimating the postmortem interval, analyzing the cause of death, characterizing trace evidence, detecting clandestine burials. The limitations and future prospects of Forensic Microbiology are also investigated. The various inferences reviewed in this article can be used to develop specific guidelines for the ongoing development of efficient strategies employed in criminal investigations.
Forensic Microbiology, Bacteria, Fungi, DNA, Criminal Investigation
Refining trace evidence in criminal investigations is a central focus of research specialists in the forensic science sector. One of the conceivable approaches that has emerged is the applicability of microorganism-based research in forensic science, which has resulted in the development of a sector referred to as “Forensic Microbiology”. This sector depends on the identification of distinct microbiomes linked to a crime scene, and its prominence increased enormously following the 2001 terrorist attacks in the United States, driven by concerns regarding potential biological threats.
Early forensic investigations involved establishing pathogenicity and the cause of death in both people and animals. Recent advancements in the use of microbiome techniques for forensic research, particularly in the fields of estimating the post-mortem interval (PMI), identifying clandestine burials, and gathering soil and skin trace evidence, have been made possible by advances in DNA sequencing and computational methods. For example, metagenomic analysis, along with sequencing the 16S rRNA gene for bacteria and the ITS rRNA gene for fungi, have been utilized to identify and characterize the microorganisms that inhabit the body or surroundings.1 Prominent researchers around the globe, including Robert Koch, Joseph Lister, and Louis Pasteur, have also fueled interest in postmortem microbiology.2 The Human Microbiome Project (HMP) found that a typical healthy human body houses ten times more microorganisms than human cells, and these microbes exert significant influences on both human health and diseases. However, knowledge about how these microbes change after death was initially limited. This led to research on the “Thanatomicrobiome” and its potential uses in forensic science research.3 The thanatomicrobiome (from Thanatos, Greek for “death”) is a relatively new concept referring to the study of microbial communities that colonize and proliferate within internal organs and body orifices after death.4
Forensic Microbiology is also used to investigate unexplained infectious disease-associated deaths and unveil the significant influence of gut microbiota on behaviors and responses to specific addictive substances. In this respect, an individual’s microbiota and its metabolites can offer various information regarding a plethora of conditions such as alcoholism, drug addiction, and mental disorders, emphasizing the gut-brain link in the context of substance addiction.5
The current review focuses primarily on emphasizing and examining the forensic use of human microbiome analysis, with the aim of offering guidance for employing this emerging field while also recognizing and addressing its limitations.
Microbial dynamics and post-mortem interval
Microbial activity plays a prominent role in decomposing vertebrate remains, and a plethora of constituents in human cells, including the proteins, carbohydrates, and lipids, offer the microorganisms a nutrient-rich environment in which to flourish.6 When oxygen levels fall after death, the body’s natural microorganisms (particularly those in the gut) metabolize the cellular products (including the various macromolecules released after the breakdown of cells), leading to tissue liquefaction and gas accumulation, causing bloating in corpse.7 Due to this breakdown of cellular contents during post-mortem, the compositions of the human microbial communities are significantly altered. Taxonomic investigations of microbial populations have revealed that each specific body part harbors a prevalent taxonomic group unique to it; for example, Proteobacteria on the skin, Lactobacillus in the vagina, Bacteroidetes and Fusobacteria in the mouth, and Firmicutes in the intestines.8
In the examination of postmortem bacterial migration in various body samples using culturing and real-time quantitative PCR techniques, an increase in the proportion of DNA from intestinal bacteria such as Bacteroides, Enterobacter, Clostridia and Bifidobacteria has been reported.9 Certain studies have reported that various foreign microorganisms begin to colonize body surfaces and orifices after death.10 Bacteria prevalent in the bloating and purge stages, such as Wohlfahrtimonas and Ignatzschineria, and linked to flies and are commonly present until tissues are dried or eliminated. After drying and skeletonization, soil-related bacteria, including Acinetobacter, are widespread in most body locations.11 Haem-based analyses have revealed that within the first week following death (1-7 days), blood sterility rapidly decreases, and within the initial five days after death, the liver and pericardial fluids are the most suitable locations for postmortem microbiological sampling.9
Research conducted under field conditions examined the changes in soil microbes around pig carcasses during summer and winter. Contrary to prevailing beliefs in forensic science, the results revealed the substantial presence of soil microorganisms within the post-mortem microbial community. Additionally, a discernible and predictable evolutionary pattern in these bacterial communities was identified.12
The necrobiome therefore specifically relates to the surface microbial communities associated with a human cadaver, while the thanatomicrobiome characterizes the microbial dynamics within the internal human organs.4 The acknowledgement of these differences directed the research community to search for efficient microbiome-based strategies for forensic applications.
Microbial succession in decomposition
A human decomposition experiment conducted at the University of Tennessee’s Forensic Anthropology Centre revealed substantial alterations in the fungal and bacterial populations within the soil during decomposition, and a unique pattern was identified as the process
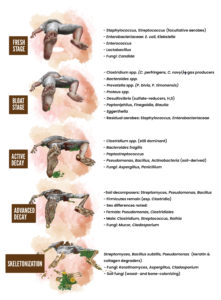
progressed.13 These observed changes in the soil’s microbial community suggest that specific microbes could be potential time-of-death indicators. However, the varying decomposition rates among different cadavers in the study highlighted the need to consider both bacterial and fungal communities to ensure the reliability of such a forensic approach. Another investigation proved that during the decomposition of a solitary adult pig carcass placed directly on a grass patch in Hammond, Louisiana, lipid biodegradation by soil bacteria was more prevalent than protein biodegradation during degradation of the carcass. This study also suggested the potential of predicting specific microorganisms linked to distinct stages of decay by identifying particular microbial assemblages (such as Group I lipase genes, Acinetobacter spp., etc.) over time. The findings and perspective reported by this study validated that the insects on carrion may alter the microbial community structure and its functioning.14 Their principal findings also showed that at different stages of decay, four dominant phyla showed distinct changes. Among the various clusters discussed, Proteobacteria, the most prevalent phylum, decreased over time, while Firmicutes became more abundant.15 Bacteroidetes were present only in the fresh and bloated stages, while the Actinobacteria communities found on the skin vanished during the dry stage. Another study suggested that the bacterial community composition varied significantly at different body locations throughout the decomposition process.11,16 In this respect, distinct shifts were noticed during the stages of bloat and purge, where fly-associated bacteria such as Wohlfahrtimonas and Ignatzschineria were prevalent until tissue desiccation or removal. Subsequently, soil-related bacteria, particularly Acinetobacter, became dominant during the later stages of dehydration and skeletonization in most examined body sites. The bacterial communities in the caecum of decaying corpses highlighted the potential of studying specific microbial taxa, such as Bacteroides and Parabacteroides, which showed a decline over time and were inversely correlated with the PMI.17 In addition, Clostridium was a strong positive predictor of the PMI. The influence of larval mass microbiology and chemistry in indicating time since death was also noted.18 According to these findings, three pig carcasses were left to decompose over three days at various intervals, and subsequent microbial analysis revealed distinct shifts in three different communities before, during, and after the 98-h mark. These variations appeared to be influenced by the oxidation-reduction potential of the larval masses. Notably, significant interactions were observed between time and microbial taxa, especially within the Firmicutes and Proteobacteria phyla. The findings suggested a promising potential for using these shifts as a tool to estimate PMI, and they offered new insights into the microbial and chemical attributes of larval masses.18 During human decomposition, microbial succession follows a predictable timeline, with shifts from facultative aerobes in the fresh stage to obligate anaerobes (such as Clostridium and Bacteroides) during bloat, followed by mixed anaerobes and soil-associated taxa in active and advanced decay, and keratin- and collagen-degrading microbes in the dry stage. Figure summarizes the predominant microorganisms associated with each stage of human body decomposition.4,19,20
Similar findings were obtained in a study that used two distinct DNA extraction techniques, classic microbiologic techniques, and 16S ribosomal sequencing at the beginning and end of decomposition, with the aim of evaluating the microbiome and likely bacterial succession.21 Over three days (Day 1, Day 2, and Day 6), bacterial phyla distributions in various anatomical samples differed significantly. There was a distinct phyla prevalence in cheek/ear (Firmicutes) and mouth/nose (Actinobacteria, Proteobacteria) samples on Day 1, and specific bacterial species typically found in human and animal microbiomes were present (Kurthia gibsonii, K. sibirica, Staphylococcus sciuri, S. lentus, and Serratia marcescens). On Day 2, the following new bacterial species were observed: Staphylococcus lentus, Micrococcus lylae, M. albus, Klebsiella oxytoca, and Marinococcus albus. By Day 6, a marked shift occurred; samples from mouth, nose, and genitalia were dominated by Actinobacteria and Proteobacteria instead of Firmicutes. In addition, two previously unseen bacterial species emerged, suggesting a shift in decomposition stages. The difference in bacterial diversity between the onset and end of decomposition suggested that a bacterial succession timeline could be developed to support postmortem interval estimation. Furthermore, the variation in bacterial composition among different anatomical regions indicates that microbial colonization during decomposition is site-specific, with distinct communities characterizing each sampling site.21
In agreement with these reported findings, another study revealed a strong linear relationship between microbial communities and post-mortem intervals, and this provided justification for consistent succession during the decomposition process.22 The microbial communities differed in relation to both the PMI and body sites samples, leading to increased similarities between sites over time due to body cavity ruptures. In particular, the rectum displayed a more stable microbial composition due to its enclosed nature compared to grave soil and skin.
Decaying carcasses in stagnant ponds were examined over two seasons and distinct variations in surface (epinecrotic) and rock-associated (epilithic) bacterial communities were identified.23 These findings suggested the potential use of epilithic biofilm, coexisting with epinecrotic biofilm in aquatic habitats, as a temporal control for estimating the PMI and considering the influences of seasonal and environmental factors.
Microbial profiling techniques
Various microbiological techniques, including both traditional and novel methods, are employed to track specific microbial sources or changes in microbial communities. The conventional methods used to track microbial sources in environmental forensic investigation encompass the following strategies.
- Identifying indicator bacteria and viruses;
- Analyzing microbial ratios;
- Using phenotypic techniques (such as MAR and immunological approaches);
- Assessing soil microbial biomass growth indicators;
- Employing community-level physiological profiling (CLPP); and
- Conducting phospholipid fatty acid analysis (PLFA).
The above-mentioned methods principally rely on cultivating diverse microbial groups on artificial media, limiting their forensic application to microbial communities inhabited in humans and warm-blooded animals that thrive under these conditions.24 Among the various strategies implemented in this regard, the analysis of Variable Number Tandem Repeats (VNTRs) is becoming increasingly crucial in bacterial genotyping for identifying pathogens. In this respect, Multi Locus VNTR analysis (MLVA) is noted as the foundation for genetic fingerprinting in forensic investigations. Choosing suitable VNTRs for Multi Locus analysis is pivotal; while 6-8 markers are generally adequate, 20-40 might be needed for less diverse species or specific evolutionary studies. However, certain circumstances produce complexities associated with the homoplasmy, and it has been necessary to examine multiple markers to rectify the concerns allied with the homoplasmy, where different alleles share the same repeat number despite different histories. The data can also be checked against a central database to find similarities with known individuals (microbial or others) or matched against other samples discovered in an investigation.25 Recent research trends have also validated the integration of MLVA with the detection of Single Nucleotide Polymorphism (SNP) markers to assist in augmenting specificity. Furthermore, various computational algorithms have been developed globally to interpret the set of probe intensities into genotypes, following enhancement of the significance in preliminary microbial studies.26
Preliminary analyses of microbial populations employing molecular approaches focused on specific microbial groups inhabited in the various bodily fluids, as opposed to individual organisms, which enabled the precise differentiation of the mentioned body fluids. Various efficient approaches, including multiplex real-time PCR for the analysis of microflora DNA extracted from various samples, were found to be capable of determining specific and unique signals in vaginal, oral, and fecal fluids. These strategies were then proven to be successful for use with forensic samples and high-throughput automation in forensic laboratories was considered.27 However, due to the limited number of markers and working samples, investigations also noted a high incidence of false positive and negative results, which resulted in the research community choosing alternative efficient strategies and techniques.
Earlier, a plethora of methodologies described microbial populations according to their morphology, metabolism and pathogenicity. However, a paradigm shift occurred in 1960 along with the rapid advent of numerical taxonomy, which encompassed quantifying the G-C content of DNA. The aforesaid strategy suggested minimal G-C variance intraspecifically within 2%-3%. From 1960 to 1980, chemotaxonomy gained traction for its use in analyzing bacterial cell components; however, in the mid-1990s, 16S rRNA gene sequencing replaced the previous methods, as it could be used to identify strains as distinct species differing by more than 98.7% in their sequences.28,29 The popularity of this method stems from the accessibility, simplicity and inherent limitations constraining adaptive evolution.29,30
Studies demonstrated that the massively parallel pyrosequencing of eukaryotic V9 hypervariable regions of SSU rRNA genes enabled the assessment of species diversity within extensively sampled populations and the identification of novel species in various environments.31 Likewise, another protocol was designed for microbial eukaryotes using primers created for the v9 hypervariable regions of small subunit rRNA genes, or a combination of both the v4 and v9 hypervariable regions.32 Such microbial sequencing data undergoes processing, quality assessment, taxonomic categorization, and functional interpretation using data analysis pipelines such as QIIME, Mothur, and MG-RAST.33 A glut of statistical approaches, including multivariate analysis, linear regression, and machine learning algorithms, have been primarily implemented to distinguish temporal patterns, identify microbial indicators, and develop specific models for estimating the postmortem interval (PMI).4,34,35
The human fecal microbiome was sequenced using these approaches, and
194.1 × 106 reads were generated from a single sample.36 This study involved a thorough comparison between 16S rRNA amplicon and Whole Genome Shotgun Sequencing (WGS) methods; the use of Illumina HiSeq and MiSeq platforms; and analyses of reads versus de novo assembled contigs, as well as the effect of shorter and longer reads. Whole genome shotgun sequencing was found to offer several advantages over the 16S amplicon method, providing the better detection of bacterial species, improved diversity assessment, and enhanced gene prediction. In recent years, the application of next-generation sequencing (NGS) to analyze bacteria in the human skin found on regularly touched surfaces ruled out the possibility of these microbial consortia harboring extensive genetic information appropriate for forensic research.37 Due to the greater ability to detect low-abundance microbial consortia within a selected sample, terminal-restriction fragment length polymorphism (T-RFLP) is the chief approach used in microbial fingerprinting.
Recent forensic investigations continue to focus on Multilocus VNTR analysis (MLVA) combined with single-nucleotide polymorphism (SNP) marker detection to deliver reliable evidence and link suspects to a crime. However, more extensive tools, such as single-molecule real-time (SMRT) sequencing, which is capable of reading 500 million base pairs in less than two hours, have broader applications beneficial to microbial forensics.38 One of the key challenges allied with forensic research involves limited (often low-quality) sample availability, and contamination. To tackle this challenge, metagenomic profiling has recently been conducted; this involves sequencing all available nucleic acid traces within a sample and patching together the different variants of one specific agent. In this method, SMRT and HiFi sequencing, along with Divisive Amplicon Denoising Algorithm 2 (DADA2), help to clarify all the variants of full-length microbial genes with single-nucleotide resolution and a near-zero error rate.38 The rapid evolution and widespread adoption of next-generation sequencing (NGS) technology, owing to its cost-effectiveness and high throughput, have transformed it into a crucial tool for numerous genomics researchers. Exploiting the capabilities of NGS permits the simultaneous analysis of multiple forensic-relevant genetic loci across a plethora of genetic materials, thereby directing the way towards new avenues for forensic science research.39
Microbiome-linked practical implications in forensic research
Bio crimes
Converting toxins and pathogens into bioweapons enables their potential exploitation in bioterrorism and biocrime. Considering the feasibility and relatively uncomplicated nature of deploying such bioweapons, forensic science must be prepared to aid investigations, holding accountable those responsible and preventing similar future occurrences.40
Several reports of biocrimes have been documented in the United States, and one of the well-known instances involved an HIV-positive dentist in Florida. The dentist was accused of performing oral surgery on six patients while infected with HIV, and failing to disclose the associated risk to them. During the trial, a phylogenetic analysis of HIV strains from the dentist and some patients was used as evidence, together with the surgical records. Although the specific incident that resulted in HIV transmission to these individuals remained unidentified, the epidemiological data supported transmission from dentist to patient rather than from one patient to another.41 Similarly, a man in Canada was found guilty of intentional HIV transmission resulting in the death of two women, marking one of the first murder convictions associated with an HIV transmission incident in the country.42
Cause of death
In a well-documented case in the United Kingdom, during an investigation into the death of an 8 week-old infant, a pathologist who testified failed to disclose that Staphylococcus aureus, commonly found on the skin, was deemed a contaminant recovered from the infant’s cerebral fluid and other body parts. Although there seemed to be no immediate cause for the infant’s death, suspicions fell on the mother, as she had lost previous infant. The woman was later found guilty of causing the deaths of both infants, the first by suffocation and the second by shaking, resulting in a life sentence. However, later discovered microbiological evidence and the child’s white blood cell profile indicated the possibility of a bacterial illness, such as meningitis and staphylococcal septicemia. This revelation eventually led to the mother’s release after her conviction was considered unreliable.43
Determining the PMI
In 2009, the Tayside Police in the United Kingdom investigated a case in which a man had been repeatedly stabbed and killed in a closed apartment, which prevented fly infestation. The crime scene showed fungal growth on parts of the blood-soaked carpet and a couch. Therefore, forensic experts replicated the crime scene by replacing some clean carpet and fabric with bovine blood. They introduced fungi from the original crime scene into the blood-stained carpet samples and kept them under controlled laboratory conditions, mirroring the temperature and humidity of the apartment. Three Penicillium griseofulvum colonies were found on the cadaver’s abdomen. The study focused on fungi, including Mucor plumbeus, Penicillium citrinum, and Penicillium brevicompactum. Based on the results of a comparison of the sizes of the new colonies, both in culture and on the carpet, with those at the murder scene, a subsequent admission of guilt was compatible with the suggestion that the death had occurred approximately five days before the body was discovered.44
Geolocation
Fungi can provide evidence of disturbed or moved tree branches or logs, which assists in the location of bodies. The South Wales Police investigated a case in which the growth pattern of mushrooms was used to challenge the grave finder, who claimed to have not disturbed the site. The alignment of fungal growth on logs suggested prior disturbance, disproving the grave finder’s testimony. Simple experiments testing the time needed for fungal reorientation could therefore be crucial in time-sensitive investigations.44
Personal identification
In a criminal case concerning the forensic examination of items associated with the sexual assault of a minor, the authorities seized an object that was considered likely to have been used in the sexual assault. The object sheltered a combination of genetic material, specifically the DNA from both the suspect and the victim. The microbial analysis revealed that the vaginal bacterial community was highly represented on the object used in the rape. Another case involved digital penetration in a rape incident; human and microbial DNA from both the suspect and the victim was found on the suspect’s finger. The human DNA was used to establish a mixture of contributor identities, while the microbial DNA, originating from bacteria characteristic of the vaginal tract, was used as evidence to support digital penetration.45
Establishing time of death
When a body is recovered from a water reservoir, the presence of specific microbiological bioindicators such as Photobacterium, Pseudoalteromonas, Vibrio cholerae, Vibrio harveyi, Marinomonas, Aeromonas, Pseudomonas and Shewanella only in samples taken from lungs indicates that the body was placed into water after death.3 In an experimental study, pig heads were immersed in seawater to model postmortem submersion.46 The results showed dynamic microbial succession, with distinct changes in bacterial communities over time and across seasons. For example, Fusobacterium, Nevskia, Psychrobacter, and Vibrio were detected during the early stages of decomposition, but they later disappeared. Seasonal differences were also observed: Marinomonas species dominated in autumn, while Polaribacter and Tenacibaculum were more prevalent in winter.
Typically, the blood and cerebrospinal fluid of healthy individuals infrequently harbor microbial consortia. However, upon death, natural defences begin to deteriorate, which potentially allows bacteria and fungi existing in the skin and in the gut to distribute over the body.43 The primary phyla used to estimate the post-mortem interval (PMI) are Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria. One study found that microbial community richness (phyla, genera, and species) first declined and then increased, with a turning point at around day 24. Using a random forest algorithm, a model based on three bacterial genera (Atopostipes, Facklamia, and Cerasibacillus) was developed, providing valuable insights into long-term microbial succession and its potential application in estimating the PMI. Within the genera, Atopostipes and Facklamia were highlighted as major predictors, while Cerasibacillus showed a pronounced late-stage increase. At the species level, the preferred species were A. bizertensis, V. lutrae, B. fragilis, I. indica, E. faecalis, and P. mirabilis for this purpose.47 Certain studies have also validated the significance of the term “microbial clock” in forensic science research, and the developed “microbial clock” has been used to estimate the time of death through an analysis of changes in the microorganisms around a corpse. By comparing the body’s microbial composition to this clock, scientists can determine the time of death, which is crucial in criminal investigations when the body is found at a crime scene. While the microbial clock technique is still in its early stages, scientists and legal experts collaborate to apply and integrate this new technology into practice.48
Human microbiome as a biometric identifier
The microbial consortia inhabiting the human gut and skin are diverse and show prominent variations among selected individuals.49 Such microbial populations, especially skin bacteria, have very robust cell walls and can withstand external environmental pressures such as temperature variations, humidity, and UV radiation. As such, they can survive on any surface for extended periods.50,51
Recent microbiome studies have revealed notable diversity in both specific community structures and microbial activity and have found that the human microbiome can be individualized at lower taxonomic levels such as genus, species, and strains.52 Hence, employing Massively Parallel Sequencing (MPS)-based methods to compare the microbiomes of suspects allows for fine-resolution data, potentially leading to suspect identification or establishing connections between perpetrators and victims.53 The implementation of advanced and recent approaches, especially metagenomics, has made it possible to characterize the hundreds of thousands of bacteria that comprise a person’s microbial community (Table 1).54 Since microbial cells have a 1:1 to 10:1 ratio to human cells, the human microbiome is an example of a high-copy-number genetic marker.55,56 As such, it could be used to supplement partial or inconclusive STR profiles to improve resolution for human source attribution.57
Table (1):
Microbiome present at different body sites
| Body sites | Microbiome |
|---|---|
| Small intestine | Streptococcus, Escherichia coli, Clostridium |
| Oral cavity | Fusobacterium nucleatum (Most common) Streptococcus, Lactobacillus, Actinomyces, Neisseria and Veillonella (1st year of birth) |
| Streptococcus (Most dominant) Haemophilus (In buccal mucosa) Actinomyces (Supragingival plaque) Prevotella (Subgingival plaque) |
|
| Firmicutes, Proteobacteria, Actinobacteria, Bacteroides, Fusobacterium | |
| Pubic hair | Lactobacillus iners, Prevotella spp., Aggregatibacter segins |
| Vaginal secretions | Lactobacillus crispatus, Lactobacillus gasseri |
| Skin | Corynebacterium, Micrococcus, Propionibacterium, Pseudomonas, Rothia, Staphylococcus, Malassezia |
It has been demonstrated that the microbiota variation between different individuals is greater than that within the same individual, and humans have personalized microbiomes with a high degree of interpersonal diversity. Studies have also focused on determining whether such differences are significant enough to differentiate individuals.58 Linking a person’s microbiome to a specific body region can also serve as valuable forensic evidence. For instance, identifying a suspect’s microbiota on a victim could establish a link between a particular body area and sexual assault. In cases lacking DNA evidence, microbial fingerprints might connect specific body parts to crimes and individuals to criminal incidents (Table 2).59
Table (2):
Microbial markers present on hand, foot, manubrium for body site identification
| Body site | Microbial markers | Accuracy (%) |
|---|---|---|
| Hand | Propionibacterium acnes | 95.80~100 |
| Propionibacterium granulosum | ||
| Propionibacterium humerusii | ||
| Propionibacterium phage P1 1 | ||
| Propionibacterium phage P100 A | ||
| Foot | Propionibacterium phage PAD20 | 54.20~83.20 |
| Propionibacterium phage PAS50 | ||
| Propionibacterium sp. 434 HC2 | ||
| Propionibacterium sp. 5 U 42AFAA | ||
| Propionibacterium sp. HGH0353 | ||
| Manubrium | Propionibacterium sp. KPL1844 | 70.80~95.80 |
| Propionibacterium sp. KPL1854 | ||
| Propionibacterium sp. KPL2008 | ||
| Rothia dentocariosa | ||
| Corynebacterium jeikeium | ||
| Corynebacterium pseudogenitalium | ||
| Corynebacterium tuberculostearicum | ||
| Corynebactrium sp. HFH0082 | ||
| Corynebacterium sp. KPL1818 | ||
| Corynebacterium sp. KPL1824 |
The most temporally stable features of an individual’s microbiome have been identified as the single-nucleotide variation (SNV) profiles of Propionibacterium acnes derived from the skin and the gene signatures (including clade-specific markers and 1-kb genomic windows) from the gut microbiome, with stability lasting nearly three years.60,61
Microbiome forensics in cold cases: solving old crimes with new techniques
Robbery
Certain new approaches have had a significant impact on solving old crimes. The following story actually serves to show how the scientific community can assist in solving old crimes using advanced approaches. This crime involved a robbery in which a family was tied up, and the police found fresh footprints leading to car tire prints and a pile of feces covered with tissue.62 The tissue was sent to the forensic laboratory for investigation. Initially, human DNA profiling identified an unknown male, marked as “unknown male A”. Despite no matches in the DNA database, a later arrest of a suspect for other robberies led to a match with the DNA profile from the tissue. During a preliminary hearing, the defence questioned the recovered DNA, suggesting the tissue might have been unrelated to the crime. To address this, a second analysis was conducted, involving new samples from the tissue for both human DNA and microbial population profiling. The results showed a complete male DNA profile matching the suspect’s, with a random match probability of 1 in 1 billion. Microbial population analysis revealed similarities between the tissue and fecal samples. The overall microarray profile and the detection of (faecal) specific bacteria, Akkermansia, Bacteroides, Collinsella, Prevotella, Ruminococcus, Enterobacter, and Faecalibacterium, and the family Lachnospiraceae, were used to evaluate the evidence. The combined results, evaluated under hypotheses, strongly supported that the stain on the tissue contained faecal material from the suspect. This comprehensive analysis, including microbial profiling, proved crucial in court, leading to the suspect’s conviction. This case highlights the importance of microbial population analysis, especially when establishing the origin of biological material is critical in forensic investigations.
Sexual assault
Similar to the above-mentioned incident, a female patient reported sexual assault by a male nurse administering rectal medication during an epileptic seizure. Swab samples were taken from the nurse’s genital area (1-neck, 2-Glans, 3-penis shaft, 4-scrotum) and subjected to DNA analysis (swabs 1-4) and microbial population profiling (swabs 1-3) using PCR and microarray techniques. The victim claimed rape both vaginally and anally. The microarray patterns of the swabs showed a similarity to faecal and vaginal samples. Human DNA profiles were generated, indicating that the victim was the main contributor and the suspect an additional contributor. The Bayesian framework was used to report evaluative forensic evidence, and the microbial analysis supported the hypothesis of bacteria similar to gut/faecal populations. The mixed STR profiles had a random match probability less than 1 in 1 billion. The report proved crucial in court, leading to the suspect’s conviction. This case illustrates the importance of microbial population analysis, especially in complex situations where determining the origin (faecal or vaginal) is critical. It also highlights the utility of microbial profiling when traditional techniques such as mRNA profiling yield no results, providing valuable information for cell type identification in forensic investigations. It can also be useful in cases with only DNA evidence, especially in old or cold cases.62
Legal and ethical considerations
The admissibility of forensic microbiology evidence in court is contingent on compliance with the legal standards of the specific jurisdiction. In the United States, adherence to the “Daubert standard” is crucial, requiring evidence collected through validated, standardized, protocols with known error rates. Additionally, the procedures must undergo peer review, be published, and be considered generally acceptable within the relevant scientific community, following the guidelines outlined by the Scientific Working Group on Microbial Genetics and Forensics (SWGMGF).63
To ensure the acceptability of forensic microbiology evidence in a legal context, it is imperative to establish standard operating procedures (SOPs) governing the collection, analysis, and interpretation of microbial samples. Furthermore, the implementation of quality control (QC) and quality assurance (QA) measures, consistent with those employed in diagnostic testing laboratories, are essential for substantiating the precision of bacteriological analysis outcomes.64
Currently, no specific regulations or acts governing the use of post-mortem microbiome evidence in court cases exist, as noted in previous studies.57,65 The introduction of postmortem microbiome evidence into legal proceedings underscores the significance of validating microbiological evidence and the meticulous collection of samples.3 The presented postmortem microbiome, having undergone thorough validation and widely accepted regulatory pathways, can serve as expert testimony with recognized accuracy. However, challenges remain with respect to statistical analysis complexity, and associated approval is based on factors such as acuity, precision, reliability, quantitation limit, and interpretability.66
Potential utility and advantages of human microbiome profiling in forensic science
Unlike human DNA, which is generally stable over time, human microbiomes are dynamic, evolving with lifestyle and environmental changes. However, this very individuality of the microbiome presents a unique potential for personal identification in forensic investigations, especially when conventional DNA is degraded or absent. One of the major advantages of microbial forensics is the high abundance of human-associated microbial cells, which often outnumber human somatic cells by a significant margin.55 This makes microbial DNA a potentially more readily available resource than human DNA in some forensic contexts, particularly in touch or trace evidence where traditional nuclear DNA is limited or degraded. Furthermore, microbiomes can be retrieved from low-biomass samples such as skin cells, saliva traces, or even residual sweat on surfaces, enabling investigations in scenarios where DNA yield would otherwise be insufficient.61
Another advantage lies in the variation of microbial communities across different body sites (e.g., skin, oral cavity, gut), which can help identify the anatomical origin of a biological sample. For instance, the presence of skin-associated microbiota on a weapon might suggest direct handling, while gut-associated microbes could indicate fecal contamination. In addition, microbial evidence may help establish indirect or trace contact between individuals or between a person and a location or object, offering investigative leads when other forms of evidence are lacking.67
Moreover, microbial communities may persist longer than human DNA in certain environmental conditions. While human DNA degrades with time and environmental stressors, some microbial communities can remain relatively stable or even thrive, potentially serving as a supplementary or alternative forensic marker when traditional DNA evidence has deteriorated.2
Limitations and challenges associated with human microbiome profiling in forensics
Despite these promising aspects, microbiome profiling in forensic science is fraught with certain limitations and challenges. First and foremost, the human microbiome is highly dynamic. Unlike human DNA, which remains genetically consistent throughout a person’s life, microbial communities fluctuate with time, health, hygiene, antibiotic use, and environment. This temporal variability makes it difficult to establish consistent microbial signatures for long-term identification or comparison.68 Therefore, any significant lapse in time between sample collection from a crime scene and that from a suspect can hinder meaningful comparison.
Additionally, microbial transfer between individuals can occur through direct (skin-to-skin) or indirect (via shared surfaces) contact, complicating source attribution. This makes it difficult to definitively associate a microbial sample with a single individual, particularly in public or shared environments.69 Further, microbial profiles often comprise mixed-origin samples, especially on shared objects or in communal spaces, posing another obstacle for accurate interpretation when no suspects are yet identified.
Contamination presents another significant challenge. In particular, low-biomass microbial samples are susceptible to exogenous contamination from laboratory reagents, handling tools, or environmental surfaces. Negative extraction controls often yield non-zero microbial reads, underscoring the importance of rigorous contamination controls and interpretation criteria.70 As such, establishing the forensic relevance of a microbial profile necessitates demonstrating its distinction from background or control samples, which is often a difficult and technical task.
Standard operating procedures (SOPs) currently used for traditional forensic DNA analysis are not optimized for microbiome sampling, storage, and extraction. The protocols for microbiome analysis must consider microbial viability, anaerobic sensitivity, and storage conditions, which may conflict with existing forensic workflows. Furthermore, microbial DNA extraction and sequencing protocols lack standardization, making inter-laboratory reproducibility and validation a major hurdle.71
The complexity of microbial data also demands using advanced bioinformatics tools for interpretation. However, different analysis pipelines can yield varying results from the same raw data due to discrepancies in algorithms, reference databases, and filtering thresholds. This variation limits the ability to compare results across labs or studies, thereby reducing forensic reliability.72 Moreover, microbial community structures do not yet have standardized population frequency databases analogous to those used in STR-based human DNA profiling, which hinders statistical modeling and inference.
Furthermore, implementing microbial forensic capabilities in routine practice would require substantial investment in infrastructure, training, and legal validation. The admissibility of microbial evidence in courts is yet to be firmly established, and the forensic community would need robust guidelines and accreditation standards before microbiome evidence can be routinely accepted.
A significant barrier to using microbial forensics in real-world cases is that most forensic professionals currently lack the necessary expertise, particularly in advanced bioinformatics and the specialized interpretation required for microbiome analysis. This skills gap means forensic practitioners may not be able to reliably interpret or testify about microbiome evidence, which can threaten the credibility and legal acceptance of such evidence in courtrooms. To overcome this limitation, there is a strong need for targeted training initiatives, interdisciplinary collaboration, and the development of standardized protocols and guidelines to ensure consistent, reliable use of microbial forensic methods and their admissibility in legal proceedings.73,74
The use of metagenomics in forensics is in its early stages, and it faces significant challenges. Microbiomes are not yet approved as evidence for individual identification, geolocation inference, and PMI estimation. The specificity and stability of individual microbes compared to human DNA markers require validation, and there is a need for well-established error rates. Quantitative machine learning methods (including K-nearest neighbors, random forests, and neural networks) show promise, but they require a sufficiently large sample size. The current evaluation criteria for microbiome evidence differ from traditional DNA likelihood calculations, requiring further research for acceptance in the forensic science community.75 Addressing ethical considerations, such as sample collection procedures and privacy concerns, is also crucial to ensure the responsible and ethical application of microbial forensics in PMI estimation.76
Future prospects
Metagenomic integration
To achieve a comprehensive under-standing of the functional activities within postmortem microbial communities, it is important to integrate metagenomic sequencing with metatranscriptomics, metaproteomics, and metabolomics.
Standardization
To ensure the reproducibility and comparability of results across different studies and forensic laboratories, standardized methods, protocols, and data analysis techniques are required.
Comprehensive databases
To enhance the accuracy of PMI estimation, it is vital to establish curated databases that can serve as valuable references.
Confounding factors
Additionally, confounding factors (such as environmental conditions, personal care products, and variations in microbial communities among individuals) must be thoroughly investigated and accounted for.
Emerging field potential
With great promise for PMI estimation, the discipline of Microbiological Forensics is still in its infancy. While more study and evaluation of the effectiveness of forensic microbiological analysis techniques is necessary, criminals are now leaving more than just fingerprints at crime scenes: blood, saliva, and semen can also be used to track DNA. Forensic microbiological analysis plays a critical role in the examination of microorganisms that can function as microscopic evidence in cases when persons seek to conceal other body fluids. It is anticipated that the increasing importance of such evidence can settle outstanding issues and protect human rights, and it can also be used to satisfy the need for proof beyond a reasonable doubt in court cases.
Validation and refinement
Further research is essential to validate findings across diverse environmental contexts and refine the use of predictive models for accurately estimating the PMI.
Microbial succession patterns and functional understanding
The continued exploration of microbial succession patterns and their correlation with PMI, along with the shifting of focus from the presence of specific microbes to understanding the potential of microbial communities, holds promise for enhancing forensic investigations and providing valuable information for the justice system.
Forensic science practitioner training
Integrating microbial forensics into routine casework necessitates practitioner training. Most forensic scientists are not currently equipped with the specialized knowledge needed to interpret microbiome data, which often relies on advanced molecular biology techniques, next-generation sequencing, and complex bioinformatics. Targeted training programs should therefore focus on core areas such as microbial ecology, sequencing technologies, data analysis pipelines, and the legal implications of presenting microbiome evidence. Interdisciplinary collaboration with microbiologists, data scientists, and legal experts is essential. Furthermore, accreditation frameworks and continuing education initiatives will help to ensure that practitioners are adequately competent to provide reliable interpretations and withstand scrutiny in judicial proceedings.73,75
The applications of microbiome research have expanded significantly, allowing for a range of forensic uses, such as estimating post-mortem intervals to unraveling hidden burials. The Human Microbiome Project enhanced our understanding of the complex relationship between microorganisms and human health, leading to explorations of the “Thanatomicrobiome” and its potential in forensic science. As technology continues to advance, incorporating microbiome techniques into forensic research promises to deepen our understanding of crime scenes, post-mortem processes, and the complex interplay between microbial communities and human biology.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MRS, ASAS, LJ, CVP, and MSSP wrote the original draft. MSSP designed the study. MRS and MSSP wrote, reviewed, and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Speruda M, Piecuch A, Borzecka J, Kadej M, Ogorek R. Microbial traces and their role in forensic science. J Appl Microbiol. 2022;132(4):2547-2557.
Crossref - Metcalf JL, Xu ZZ, Weiss S, et al. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science. 2016;351(6269):158-162.
Crossref - Spagnolo EV, Stassi C, Mondello C, Zerbo S, Milone L, Argo A. Forensic microbiology applications: a systematic review. Leg Med. 2019;36:73-80.
Crossref - Javan GT, Finley SJ, Can I, Wilkinson JE, Hanson JD, Tarone AM. Human thanatomicrobiome succession and time since death. Sci Rep. 2016;6(1):29598.
Crossref - Hachem M, Ahmad H, Pilankar I, Abdelrahim I, Alfalasi F, Asif Z. Advances in Human Microbiome as an Emerging Tool in Forensics. Int J Emerg Technol. 2020;11(3):70–76. Accessed August 2, 2025.
- Dash HR, Das S. Microbial degradation of forensic samples of biological origin: potential threat to human DNA typing. Mol Biotechnol. 2018;60(2):141-153.
Crossref - Carter DO, Yellowlees D, Tibbett M. Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften. 2007;94(1):12-24.
Crossref - Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genom Hum Genet. 2012;13:151-170.
Crossref - Tuomisto S, Karhunen PJ, Vuento R, Aittoniemi J, Pessi T. Evaluation of postmortem bacterial migration using culturing and real time quantitative PCR. J Forensic Sci. 2013;58(4):910-916.
Crossref - Can I, Javan GT, Pozhitkov AE, Noble PA. Distinctive thanatomicrobiome signatures found in the blood and internal organs of humans. J Microbiol Methods. 2014;106:1-7.
Crossref - Hyde ER, Haarmann DP, Petrosino JF, Lynne AM, Bucheli SR. Initial insights into bacterial succession during human decomposition. Int J Legal Med. 2015;129(3):661-671.
Crossref - Carter DO, Metcalf JL, Bibat A, Knight R. Seasonal variation of postmortem microbial communities. Forensic Sci Med Pathol. 2015;11(2):202-207.
Crossref - Parkinson RA, Dias K-R, Horswell J, et al. Microbial Community Analysis of Human Decomposition on Soil. In: Ritz K, Dawson L, Miller D. (eds) Criminal and Environmental Soil Forensics. Springer, Dordrecht. 2009:379-394.
Crossref - Pechal JL, Crippen TL, Tarone AM, Lewis AJ, Tomberlin JK, Benbow ME. Microbial community functional change during vertebrate carrion decomposition. PloS One. 2013;8(11):e79035.
Crossref - Pechal JL, Crippen TL, Benbow ME, Tarone AM, Dowd S, Tomberlin JK. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int J Legal Med. 2014;128(1):193-205.
Crossref - Howard GT, Duos B, Watson-Horzelski EJ. Characterization of the soil microbial community associated with the decomposition of a swine carcass. Int Biodeterior Biodegradation. 2010;64(4):300-304.
Crossref - DeBruyn JM, Hauther KA. Postmortem succession of gut microbial communities in deceased human subjects. Peer J. 2017;5:e3437.
Crossref - Junkins EN, Speck M, Carter DO. The microbiology, pH, and oxidation reduction potential of larval masses in decomposing carcasses on Oahu, Hawaii. J Forensic Legal Med. 2019;67:37-48.
Crossref - Hyde ER, Haarmann DP, Lynne AM, Bucheli SR, Petrosino JF. The living dead: bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PloS one. 2013;8(10):e77733.
Crossref - Janaway RC, Percival SL, Wilson AS. Decomposition of Human Remains. In: Percival, S.L. (eds) Microbiology and Aging. Humana Press. 2009:313-334.
Crossref - Erquiaga MJ, Fabbri MR, Kowalczyk MF, et al. Changes in the microbiome at the onset and end of decomposition. J Biotechnol Bioinforma Res. 2020;2(4):1-5.
Crossref - Zhang J, Wang M, Qi X, et al. Predicting the postmortem interval of burial cadavers based on microbial community succession. Forensic Sci Int Genet. 2021;52:102488.
Crossref - Dmitrijs F, Guo J, Huang Y, et al. Bacterial succession in microbial biofilm as a potential indicator for postmortem submersion interval estimation. Front Microbiol. 2022;13:951707.
Crossref - Petrisor IG, Parkinson RA, Horswell J, et al. Microbial forensics. In: Morrison RD, Murphy BL (eds). Environmental Forensics. 1964:227-257.
Crossref - Vergnaud G, Pourcel C. Multiple Locus Variable Number of Tandem Repeats Analysis. In: Caugant D. (eds) Molecular Epidemiology of Microorganisms. Methods in Molecular Biology™, vol 551. Humana Press, Totowa, NJ. 2009;551:141-158.
Crossref - LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Research. 2009;37(13):4181-4193.
Crossref - Giampaoli S, Berti A, Valeriani F, et al. Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet. 2012;6(5):559-564.
Crossref - Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45(9):2761-2764.
Crossref - Janda JM. Taxonomic Classification of Bacteria. In: Greenc LH, Goldman E (eds). Practical Handbook of Microbiology. 2021:161-166.
Crossref - Haarkotter C, Saiz M, Galvez X, Medina-Lozano MI, Alvarez JC, Lorente JA. Usefulness of microbiome for forensic geolocation: a review. Life. 2021;11(12):1322.
Crossref - Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PloS One. 2009;4(7):e6372.
Crossref - Stoeck T, Bass D, Nebel M, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19(Suppl 1):21-31.
Crossref - D’Argenio V, Casaburi G, Precone V, Salvatore F. Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. BioMed Res Int. 2014;2014(1):325340
Crossref - Metcalf JL. Estimating the postmortem interval using microbes: Knowledge gaps and a path to technology adoption. Forensic Sci Int Genet. 2019;38:211-218.
Crossref - Belk A, Xu ZZ, Carter DO, et al. Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes. 2018;9(2):104.
Crossref - Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967-977.
Crossref - Rawal R. Microbial Fingerprinting-An Emerging Tool in Forensics: A Review. Review. Int J Innov Sci Res Technol. 2022;7(5):1037-1046.
Crossref - Gautier L. Microbial forensics: what we’ve learned from Amerithrax and beyond. BioTechniques. 2023;75(4):129-132.
Crossref - Yuguda YM. Application of Next Generation Sequencing (NGS) technology in forensic science: A review. GSC Biol Pharm Sci. 2023;23(2):155-159.
Crossref - Budowle B, Murch R, Chakraborty R. Microbial forensics: the next forensic challenge. Int J Legal Med. 2005;119(6):317-330.
Crossref - Ciesielski C, Marianos D, Ou C-Y, et al. Transmission of human immunodeficiency virus in a dental practice. Ann Intern Med. 1992;116(10):798-805.
Crossref - Huber J, Crawford T. Murder verdict in HIV case sets off alarms; first in Canada. National Post (Canada). April 6, 2009. Accessed August 2, 2025. https://nationalpost.com/news/murder-verdict-in-hiv-case-sets-off-alarms-first-in-canada
- Gunn A, Pitt SJ. Microbes as forensic indicators. Trop Biomed. 2012;29(3):311-30
- Hawksworth DL, Wiltshire PE. Forensic mycology: the use of fungi in criminal investigations. Forensic Sci Int. 2011;206(1-3):1-11.
Crossref - Gouello A, Dunyach-Remy C, Siatka C, Lavigne J. Analysis of microbial communities: an emerging tool in forensic sciences. Diagnostics (Basel).2021;12(1):1.
Crossref - Dickson GC, Poulter RT, Maas EW, Probert PK, Kieser JA. Marine bacterial succession as a potential indicator of postmortem submersion interval. Forensic Sci Int. 2011;209(1-3):1-10.
Crossref - Zhao X, Zhong Z, Hua Z. Estimation of the post mortem interval by modelling the changes in oral bacterial diversity during decomposition. J Appl Microbiol. 2022;133(6):3451-3464.
Crossref - Kumari P, Yadav S. Investigating the Post-Mortem Interval (PMI) with Forensically Important Necrobiomes. Indian J Forensic Med Toxicol. 2023;17(3):97-103.
Crossref - Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694-1697.
Crossref - Smith SM, Eng R, Padberg Jr FT. Survival of nosocomial pathogenic bacteria at ambient temperature. J Med. 1996;27(5-6):293-302.
- Brooke JS, Annand JW, Hammer A, Dembkowski K, Shulman ST. Investigation of bacterial pathogens on 70 frequently used environmental surfaces in a large urban US university. J Environ Health. 2009;71(6):17-23.
- Blaser MJ, Dominguez-Bello MG. The human microbiome before birth. Cell Host & Microbe. 2016;20(5):558-560.
Crossref - Oliveira M, Amorim A. Microbial forensics: new breakthroughs and future prospects. Appl Microbiol Biotechnol. 2018;102(24):10377-10391.
Crossref - Sabunciyan S, Aryee MJ, Irizarry RA, et al. Genome-wide DNA methylation scan in major depressive disorder. PloS One. 2012;7(4):e34451.
Crossref - Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533.
Crossref - Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31(1):107-133.
Crossref - Schmedes SE, Woerner AE, Budowle B. Forensic human identification using skin microbiomes. Appl Environ Microbiol. 2017;83(22):e01672-17.
Crossref - Williams DW, Gibson G. Classification of individuals and the potential to detect sexual contact using the microbiome of the pubic region. Forensic Sci Int Genet. 2019;41:177-187.
Crossref - Eom Y-B. Microbial forensics: human identification. Biomedical Science Letters. 2018;24(4):292-304.
Crossref - Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854-866.
Crossref - Franzosa EA, Huang K, Meadow JF, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A. 2015;112(22):E2930-E2938.
Crossref - Quaak FC, van de Wal Y, Maaskant-van Wijk PA, Kuiper I. Combining human STR and microbial population profiling: Two case reports. Forensic Sci Int Genet. 2018;37:196-199.
Crossref - Budowle B, Harmon R. HIV legal precedent useful for microbial forensics. Croat Med J. 2005;46(4):514-521.
- Cunningham J, Pitt SJ. An introduction to biomedical science in professional and clinical practice. John Wiley & Sons. 2022.
- Liu Y, Hao B, Shi Y, et al. Violent offences of methamphetamine users and dilemmas of forensic psychiatric assessment. Forensic Sci Res. 2017;2(1):11-17.
Crossref - Gautam K, Rawal R. Microbial Clock: A review on forensic microbiology for crime scene investigations. J Fore Res. 2022;3(1):112.
Crossref - Meadow JF, Altrichter AE, Bateman AC, et al. Humans differ in their personal microbial cloud. Peer J. 2015;3:e1258.
Crossref - Jovel J, Patterson J, Wang W, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol. 2016;7:459.
Crossref - Lax S, Smith DP, Hampton-Marcell J, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048-1052.
Crossref - Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12(1):87.
Crossref - Quaak FC, van Duijn T, Hoogenboom J, Kloosterman AD, Kuiper I. Human-associated microbial populations as evidence in forensic casework. Forensic Sci Int Genet. 2018;36:176-185.
Crossref - Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16(1):276.
Crossref - Lee SB, Mills DK, Morse SA, Schutzer SE, Budowle B, Keim P. Education and training in microbial forensics. Microbial Forensics. 2020:473-495.
Crossref - Lopez CD, Vidaki A, Kayser M. Integrating the human microbiome in the forensic toolkit: Current bottlenecks and future solutions. Forensic Sci Int Genet. 2022;56:102627.
Crossref - Zhang J, Liu W, Simayijiang H, Hu P, Yan J. Application of microbiome in forensics. Genomics, Proteomics Bioinformatics. 2023;21(1):97-107.
Crossref - Jangid C, Dalal J. Exploring the Potential of Microbial Communities: Understanding Their Role in PMI Estimation. Unlocking the Mysteries of Death-New Perspectives for Post-mortem Examination. IntechOpen. 2023.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.