ISSN: 0973-7510
E-ISSN: 2581-690X
This study focuses on isolating and identifying bacterial and fungal decomposers with potential for environmentally sustainable crop residue management and soil fertility improvement. A total of 122 lignocellulolytic microorganisms (80 bacteria and 42 fungi) were isolated from crop residues, termite guts, and vermicompost. Among these, 18 promising isolates (12 bacterial and 6 fungal) were further characterized. Qualitative screening revealed strong cellulolytic activity in all isolates, evidenced by clear halo zones on CMC agar (Congo red staining). Notably, isolates CRDB34, CRDB42, and CRDF25 exhibited high β-glucosidase activity, while CRDF8, CRDF10, CRDB78, and CRDF25 showed significant xylanase and pectinase production. Quantitative assays demonstrated robust enzymatic potential: CMCase (15.2-137.0 IU/mL), FPase (8.6-129.8 IU/mL), and chitinase (4.7-21.7 IU/mL). Isolates CRDB52 (highest CMCase) and CRDF32 (highest xylanase, chitinase) emerged as top performers. Biochemical tests indicated diverse metabolic traits, including amylase activity, fermentation, catalase production, and ammonia generation, highlighting niche-specific adaptations. Notably, select isolates exhibited plant growth-promoting potential, with indole-3-acetic acid (IAA) production (up to 461.9 µg/mL) and phosphate solubilization (up to 600 µg/mL). Molecular identification confirmed Bacillus haynesii (CRDB-24), Bacillus altitudinis (CRDB48), Bacillus stratosphericus (CRDB52), Fusarium oxysporum (CRDF8), and Aspergillus fumigatus (CRDF32) as key decomposers, supported by 16S rRNA/ITS sequencing (92%-100% similarity). These isolates align with prior reports on lignocellulose. The study underscores the biotechnological potential of these isolates for sustainable residue management, offering enzymatic versatility and nutrient-mobilizing traits critical for agricultural applications.
Crop Residue Decomposition, Enzymatic Activity, Microbial Isolates, Sustainable Agriculture
Crop residue decomposition is a fundamental process in sustainable agriculture, as it contributes significantly to nutrient recycling, the buildup of soil organic matter, and improvement of soil physical properties.1 Agricultural fields generate large quantities of residues such as rice straw, wheat stubble, and sugarcane trash, which, if efficiently degraded, can serve as valuable inputs to maintain soil fertility and reduce dependency on chemical fertilizers. The breakdown of these plant residues is primarily facilitated by soil microorganisms, particularly bacteria and fungi, that secrete extracellular enzymes such as cellulases, xylanases, chitinases, and ligninases.2 These enzymes play a vital role in degrading the complex lignocellulosic matrix of crop residues, thereby releasing essential nutrients such as nitrogen (N), phosphorus (P), potassium (K), and sulfur (S) back into the soil.3
Among microbial decomposers, bacterial genera such as Bacillus, Pseudomonas, and Actinomycetes are well-documented for their ability to produce potent cellulolytic and ligninolytic enzymes, making them effective agents in residue breakdown.4 Likewise, fungal genera including Aspergillus, Trichoderma, and Penicillium are known for their robust enzymatic activity that enhances the degradation of cellulose, hemicellulose, and lignin.5 The action of these microbes accelerates the decomposition process and enhances soil biological activity, contributing to long-term soil health.
The isolation and identification of efficient microbial decomposers is a key step in the development of effective, eco-friendly residue management practices. Microorganisms isolated from decomposing crop residues often produce a wide spectrum of extracellular enzymes-such as cellulases, xylanases, chitinases, and pectinases-that target various structural components of the plant cell wall.6 Bacillus spp., in particular, has been widely studied for their cellulolytic and lignocellulolytic potential, which allows them to degrade tough cellulose structures efficiently.5 Fungal strains like Aspergillus and Fusarium also exhibit strong enzymatic capabilities, particularly in the hydrolysis of complex polysaccharides like cellulose and chitin.7 In addition to their degradative functions, many of these microbial isolates are known to possess plant growth-promoting (PGP) attributes, such as indole-3-acetic acid (IAA) production, phosphate solubilization, and siderophore secretion, further enhancing their utility in sustainable crop production systems.8
The application of microbial inoculants composed of efficient residue-decomposing strains has emerged as a promising strategy to enhance decomposition rates and improve soil nutrient dynamics. Traditional culture-based isolation methods, combined with molecular techniques such as 16S rRNA and internal transcribed spacer (ITS) sequencing, are instrumental in characterizing microbial diversity and functional traits associated with residue degradation.9
Therefore, the present study focuses on the isolation and characterization of cellulolytic microorganisms from diverse crop residue sources. The objectives include screening for enzyme-producing bacteria and fungi, evaluating their cellulolytic and ligninolytic enzyme activity, and assessing their potential contribution to nutrient release and soil fertility improvement. The findings are expected to support the formulation of microbial consortia for effective crop residue decomposition and promote sustainable residue management in agricultural systems.
Isolation and identification of residue decomposer microorganisms
Sample collection
Soil and partially decomposed crop residues, including rice straw, wheat stubble, sugarcane trash, and organic matter associated with decomposers such as termite nests, earthworm casts, snail feces, cow dung, and vermicompost, were collected from agricultural fields under diverse cropping systems. Samples were obtained from the top 0-15 cm of soil and adjacent residue-rich zones to ensure the presence of active microbial communities. All collected samples were placed in sterile polyethylene bags or containers and transported to the laboratory under refrigerated conditions (4 °C) for subsequent microbial isolation.
Isolation of bacteria and fungi
Microbial isolation was carried out using standard serial dilution and spread plate techniques. For bacterial isolation, 1 g of each sample was suspended in 9 mL of sterile distilled water and serially diluted up to 10-6. Aliquots (0.1 mL) from appropriate dilutions were spread onto nutrient agar (NA), carboxymethyl cellulose (CMC) agar, and lignin-supplemented media to selectively screen for cellulolytic and ligninolytic bacteria. The plates were incubated at 28 ± 2 °C for 24-48 hours. Colonies exhibiting clear hydrolysis zones on CMC and lignin media were preliminarily identified as cellulase- or ligninase-producing bacteria and were selected for further purification and characterization.
For fungal isolation, a similar dilution procedure was followed. Diluted suspensions were plated on Potato Dextrose Agar (PDA) and Czapek-Dox Agar supplemented with 100 µg/mL streptomycin and 50 µg/mL chloramphenicol to suppress bacterial growth. Plates were incubated at 28 ± 2 °C for 5-7 days. Morphologically distinct fungal colonies were sub-cultured onto fresh PDA plates to obtain pure isolates. All purified bacterial and fungal isolates were maintained on slants at 4 °C and in glycerol stocks at -20 °C for further enzymatic and molecular studies.10
Enzymatic screening
Cellulase activity
Cellulase production was qualitatively assessed on agar medium supplemented with 1% carboxymethylcellulose (CMC). Plates were incubated at 28 ± 2 °C for 48 hours, stained with 0.1% Congo red for 15 minutes, and destained with 1 M NaCl. The formation of clear zones around colonies indicated cellulase activity.11 Quantitative cellulase activity was estimated by measuring reducing sugars released from CMC using the DNS method as described by Mandels and Weber.12 Absorbance was measured at 540 nm. One unit (IU) of cellulase activity was defined as the amount of enzyme releasing 1 µmol of glucose per minute under assay conditions.
Xylanase activity
Xylanase-producing isolates were screened on agar plates containing oat spelt xylan as the carbon source and incubated at 28 ± 2 °C for 48 hours. Congo red (0.1%) was used for staining, followed by 1 M NaCl destaining to visualize hydrolysis zones.13 Quantitative xylanase activity was assayed using 4-O-methyl-D-glucuronoxylan as the substrate, and enzyme activity was expressed in IU, with one unit defined as the amount of enzyme that released 1 µmol of xylose equivalents per minute.14
Pectinase activity
Pectinolytic activity was evaluated by growing isolates on agar plates containing citrus pectin. After incubation, the plates were treated with 1% iodine solution to reveal clear zones.15
For quantitative analysis, the DNS method was used to measure the release of reducing sugars from pectin. Absorbance was recorded at 540 nm, and enzyme activity was expressed in IU using galacturonic acid as the standard.16
Chitinase activity
Chitinase activity was qualitatively evaluated on colloidal chitin agar. Clear halos surrounding colonies indicated enzymatic degradation of chitin.17 For quantitative analysis, 4-methylumbelliferyl β-D-N,N’-diacetylchitobioside was used as a substrate. The release of fluorescent product was measured spectrophotometrically, and chitinase activity was expressed in IU.18
Filter paper activity (FPase)
Filter paper-degrading activity was assessed following the IUPAC standard method. A strip of Whatman No. 1 filter paper (50 mg) was incubated with 0.5 mL enzyme extract and 1.5 mL of 50 mM citrate buffer (pH 4.8) at 50 °C for 1 hour. The reaction was stopped with 3 mL DNS reagent, boiled, and cooled. Absorbance was read at 540 nm. Enzyme activity was calculated as IU based on a glucose standard curve.19
β-Glucosidase Activity
Qualitative detection of β-glucosidase was done on agar containing esculin and ferric ammonium citrate. The formation of dark brown or black halos indicated enzyme activity.20
Quantitative activity was determined using p-nitrophenyl-β-D-glucopyranoside (pNPG) as substrate. The reaction mixture (0.5 mL enzyme extract + 0.5 mL 5 mM pNPG in citrate buffer) was incubated at 50 °C for 30 minutes, and the reaction was terminated with 2 mL 1 M Na2CO3. Absorbance was read at 400 nm, and activity was expressed in IU using a p-nitrophenol standard curve.21
Biochemical characterization
Casein hydrolysis (protease activity)
To investigate protease production, bacterial strains were inoculated on nutrient agar enriched with casein and incubated at 28 °C for up to 7 days. Proteolytic activity was identified by the appearance of transparent zones surrounding bacterial growth, indicating the breakdown of casein proteins into simpler peptides and amino acids.22
Ammonification
The potential of isolates to release ammonia was tested by culturing them in peptone water at 28 ± 2 °C for 48 to 72 hours. After incubation, 0.5 mL of Nessler’s reagent was added to each culture. A shift in color from pale yellow to dark brown signified the accumulation of ammonia, thereby confirming ammonifying activity.23
Indole-3-Acetic Acid (IAA) Production
IAA production was measured using a color-based method. Isolates were cultured in nutrient broth containing tryptophan as a precursor. After incubation, culture filtrates were mixed with Salkowski’s reagent (in a 1:2 ratio) and left in the dark for 30 minutes. A pink coloration indicated IAA synthesis, and absorbance was recorded at 530 nm to estimate concentration using a standard curve.24
Phosphate solubilization
To assess phosphate solubilizing capacity, isolates were initially spotted on Pikovskaya’s agar, and the appearance of clear halos around colonies was taken as evidence of solubilization. For quantitative determination, isolates were grown in liquid NBRIP medium under shaking conditions. After incubation, the supernatants were harvested and analyzed spectrophotometrically at 660 nm. The soluble phosphate content was calculated by comparing values to a standard curve prepared with KH2PO4.24
Molecular identification of bacterial and fungal isolates
To determine the taxonomic identity of the isolated bacterial strains, genomic DNA was extracted and the 16S rRNA gene was amplified using the forward primer GGATGAGCCCGCGGCCTA and the reverse primer CGGTGTGTACAAGCCCGGG. Polymerase Chain Reaction (PCR) amplification was carried out using a thermal cycler with a high-fidelity DNA polymerase. The resulting PCR products were then separated and visualized using agarose gel electrophoresis. Sanger sequencing was performed to determine the nucleotide sequence of the amplified 16S rRNA gene. The obtained sequences were subsequently compared against the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) database for taxonomic identification and deposited into GenBank.25
For fungal isolates, genomic DNA was extracted using the Cetyltrimethylammonium Bromide (CTAB) method. The Internal Transcribed Spacer (ITS) region of the ribosomal DNA was amplified using the ITS1 forward primer GGAAGTAAAAGTCGTAACAAGG and the ITS4 reverse primer TCCTCCGCTTATTGATATGC. PCR amplification was performed under the following conditions: initial denaturation at 94 °C for 5 minutes, followed by 35 cycles of denaturation at 94 °C for 30 seconds, annealing at 53 °C for 1 minute, and extension at 72 °C for 1 minute. The purified PCR products were then sequenced, and the resulting sequences were compared with entries in the GenBank database using BLAST for taxonomic identification.26
Qualitative characterization of isolated Lignocellulolytic microorganisms
Table 1 summarizes the qualitative enzyme activities of 18 microbial isolates, including 12 bacteria and 6 fungi, collected from various environments such as termite and earthworm guts, vermicompost, and decomposing crop residues. The isolates were tested for their ability to produce extracellular polysaccharides (EPS) using Congo red staining, as well as for enzymatic activities involved in breaking down complex plant materials: β-glucosidase, xylanase, chitinase, and pectinase. All isolates showed strong Congo red staining (+++), indicating significant EPS production, which is important for forming biofilms and facilitating cellulose degradation (Figure 1). In terms of β-glucosidase activity, which is crucial for converting cellobiose into glucose, most isolates including CRDB38, CRDB39, CRDB48, CRDF23, and CRDF32 exhibited high activity (+++). However, CRDB12 and CRDB55 lacked this activity (-), suggesting limited ability to process cellobiose. Some isolates like CRDB34 and CRDB42 showed low (+), and CRDB52 and CRDF10 showed moderate (++) β-glucosidase activity (Figure 2). Xylanase activity, responsible for breaking down hemicellulose, was also strong (+++) in most isolates such as CRDB38, CRDB47, CRDF23, and CRDF32. A few isolates like CRDF25, from cow dung, showed no detectable xylanase activity (-), while CRDF8, CRDF10, and CRDB12 showed moderate (++) activity (Figure 3). All isolates demonstrated strong chitinase activity (+++), reflecting their ability to degrade chitin, which is found in fungal cell walls and insect exoskeletons (Figure 4). Regarding pectinase activity, which breaks down pectin in plant cell walls, most isolates, including CRDB38 and CRDF23, had high activity (+++). Exceptions were CRDB55, which showed no activity (-), and CRDB78 and CRDF25, which had moderate (++) pectinase activity (Figure 5).
Table (1):
Qualitative characterization of isolated crop residues decomposing bacterial and fungal isolates isolated from different sources
Isolates Code |
Source of sample collection |
Zone formation by Congo red |
β-glucosidase |
Xylanase |
Chitinase |
Pectinase |
|---|---|---|---|---|---|---|
CRDB38 |
Termite gut |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDB39 |
Termite gut |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDB47 |
Earthworm gut |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDB48 |
Rice Residue |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDB52 |
Wheat Residue |
+++ |
++ |
+++ |
+++ |
+++ |
CRDB78 |
Wheat residue |
+++ |
+++ |
+++ |
+++ |
++ |
CRDB34 |
Termite gut |
+++ |
+ |
+++ |
+++ |
+++ |
CRDB42 |
Termite gut |
+++ |
+ |
+++ |
+++ |
+++ |
CRDB46 |
Earthworm gut |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDB55 |
Earthworm gut |
+++ |
+++ |
+++ |
+++ |
– |
CRDB12 |
Earthworm gut |
+++ |
– |
++ |
+++ |
+++ |
CRDB24 |
Vermicompost |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDF23 |
Sugarcane residues |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDF10 |
Decompose rice residues |
+++ |
+++ |
++ |
+++ |
+++ |
CRDF8 |
Termite |
+++ |
+++ |
++ |
+++ |
+++ |
CRDF25 |
Cow dung |
+++ |
+ |
– |
+++ |
++ |
CRDF32 |
Earthworm gut |
+++ |
+++ |
+++ |
+++ |
+++ |
CRDF33 |
Earthworm gut |
+++ |
+++ |
+++ |
+++ |
+++ |
Note- Positive (+), Medium positive (++), High positive (+++), Negative (-)
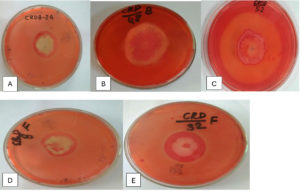
Figure 1. Hydrolysis Zones on CMC Agar Showing Cellulase Activity of Potential Isolates (A) CRDB-24, (B) CRDB-48, (C) CRDB-52, (D) CRDF-8, (E) CRDF-32
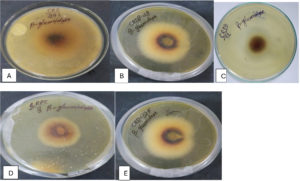
Figure 2. β-Glucosidase activity exhibited by potential microbial isolates (A) CRDB-24, (B) CRDB-48, (C) CRDB-52, (D) CRDF-8, (E) CRDF-32
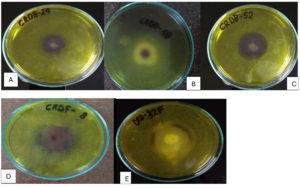
Figure 3. Hydrolysis zones on Xylan agar medium showing xylanase activity of potential isolates (A) CRDB-24, (B) CRDB-48, (C) CRDB-52, (D) CRDF-8, and (E) CRDF-32
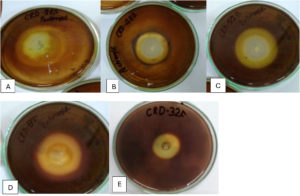
Figure 4. Hydrolysis zones on chitin agar medium showing chitinase activity of potential isolates (A) CRDB-24, (B) CRDB-48, (C) CRDB-52, (D) CRDF-8, and (E) CRDF-32
Figure 5. Hydrolysis zones on pectin agar medium showing pectinase activity of potential isolates (A) CRDB-24, (B) CRDB-48, (C) CRDB-52, (D) CRDF-8, and (E) CRDF-32
Quantitative characterization of bacterial and fungal isolates
The biochemical characterization of bacterial and fungal isolates revealed diverse enzymatic profiles and metabolic capacities. The activities of CMCase, FPase, pectinase, chitinase, and β-glucosidase varied widely among the isolates, ranging from 15.2 to 137.0, 8.60 to 129.8, 3.25 to 22.5, 2.02 to 20.2, and 4.68 to 21.7 IU/mL, respectively (Table 2). CMCase (endoglucanase) activity ranged from 15.23 to 137.04 IU/mL. The highest CMCase activity was recorded in isolate CRDB52 (137.04 IU/mL), while the lowest was observed in CRDF8 (15.23 IU/mL). On average, bacterial isolates exhibited higher CMCase activity than fungal strains. In case of FPase (total cellulase) activity varied from 3.98 to 129.82 IU/mL. Notably, CRDF32 (123.9 IU/mL) and CRDF8 (129.8 IU/mL) showed the highest FPase levels. Fungal isolates generally demonstrated significantly higher FPase activity compared to bacterial strains (p < 0.01). β-Glucosidase activity was evaluated in both nutrient broth and Luria broth. In nutrient broth, values ranged from 24.56 to 123.80 IU/mL, with CRDF8 showing the highest activity. In Luria broth, values ranged from 58.23 to 156.33 IU/mL, with CRDB24 exhibiting the highest production. On average, β-glucosidase activity increased by approximately 13.5% in Luria broth compared to nutrient broth. Xylanase activity ranged from 3.25 to 22.5 IU/mL. CRDF32 and CRDF25 recorded the highest activities at 22.5 IU/mL and 20.1 IU/mL, respectively. Pectinase activity varied between 2.02 and 20.21 IU/mL. CRDF8 exhibited the highest pectinase activity, while CRDB55 had the lowest. On average, fungal isolates produced pectinase at levels approximately 3.2 times higher than bacterial isolates. Chitinase activity was relatively consistent across isolates, ranging from 4.68 to 21.7 IU/mL. The highest value was observed in CRDF32, with the overall mean activity being 12.11 IU/mL. Statistical analysis revealed that FPase and pectinase activities exhibited the highest variability among isolates, with coefficients of variation (CV) of 83.81% and 79.48%, respectively. In contrast, β-glucosidase activity showed the lowest variability, with CVs between 29.37% and 29.88%, indicating stable enzyme production across isolates and media types.
Table (2):
Quantitative analysis of crop residues decomposing enzymes of isolated crop residues decomposing bacterial isolates
Isolates Code |
CMC-ase (IU/mL) |
FP-ase Filter paper (IU/mL) |
β-glucosidase in Nutrient broth (IU/mL) |
β-glucosidase in Luria broth (IU/mL) |
Xylanase (IU/mL) |
Pectinase (IU/mL) |
Chitinase (IU/mL) |
|---|---|---|---|---|---|---|---|
CRDB38 |
56.9 |
10.3 |
88.9 |
117.3 |
5.49 |
3.41 |
8.53 |
CRDB39 |
63.3 |
26.6 |
100.5 |
73.5 |
7.41 |
2.76 |
7.2 |
CRDB47 |
23.5 |
3.98 |
71.9 |
90.9 |
3.25 |
2.46 |
10.7 |
CRDB48 |
113.9 |
37.1 |
97.3 |
143.2 |
9.23 |
3.99 |
9.72 |
CRDB52 |
137.0 |
32.2 |
106.1 |
142.3 |
10.2 |
5.21 |
11.4 |
CRDB78 |
97.4 |
23.1 |
111.4 |
128.4 |
8.55 |
2.97 |
9.50 |
CRDB34 |
82.2 |
13.2 |
113.2 |
111.1 |
5.70 |
4.20 |
4.68 |
CRDB42 |
110.5 |
28.5 |
88.9 |
142.3 |
4.54 |
3.58 |
9.50 |
CRDB46 |
33.9 |
17.2 |
57.9 |
111.4 |
3.99 |
2.27 |
9.77 |
CRDB55 |
85.1 |
17.6 |
97.3 |
94.8 |
8.05 |
2.02 |
10.2 |
CRDB12 |
61.7 |
8.60 |
24.6 |
125.1 |
6.86 |
3.74 |
9.48 |
CRDB24 |
113.9 |
38.5 |
111.4 |
156.3 |
7.49 |
4.54 |
8.69 |
CRDF23 |
16.5 |
94.9 |
110.3 |
58.2 |
12.8 |
13.3 |
17.0 |
CRDF10 |
19.2 |
97.4 |
37.1 |
71.4 |
12.2 |
13.3 |
19.3 |
CRDF8 |
15.2 |
129.8 |
123.8 |
80.5 |
13.0 |
20.2 |
19.6 |
CRDF25 |
30.6 |
85.1 |
100.6 |
71.7 |
20.1 |
13.9 |
13.3 |
CRDF32 |
33.6 |
123.9 |
123.2 |
74.8 |
22.5 |
14.5 |
21.7 |
CRDF33 |
24.0 |
80.4 |
75.3 |
68.1 |
5.63 |
17.6 |
17.9 |
Minimum |
137.04 |
129.82 |
123.80 |
156.33 |
22.5 |
20.21 |
21.7 |
Maximum |
15.23 |
3.98 |
24.56 |
58.23 |
3.25 |
2.02 |
4.68 |
Mean |
62.13 |
48.24 |
91.08 |
103.40 |
9.28 |
7.44 |
12.11 |
SD |
38.85 |
40.43 |
27.22 |
30.37 |
5.10 |
5.91 |
4.71 |
CV (%) |
62.53 |
83.81 |
29.88 |
29.37 |
54.99 |
79.48 |
38.89 |
Biochemical characterization of bacterial and fungal isolates
The following (Table 3) summarizes the results of several biochemical tests conducted on various bacterial and fungal isolates. The outcomes are represented semi-quantitatively using a graded scale based on the intensity of the observed reactions. The designations are defined as follows: ‘+++’ indicates a strong positive reaction, characterized by a distinct and intense color change or vigorous visible response (e.g., strong effervescence in the catalase test); ‘++’ denotes a moderate positive reaction, with clear but moderate color development or reaction intensity; ‘+’ represents a weak positive reaction, with faint or minimal color change or visible activity; and ‘-’ signifies a negative result, marked by the absence of any observable color change or reaction. This scale provides a standardized framework for evaluating the biochemical characteristics of the tested isolates. A significant number of isolates (CRDB10, CRDB24, CRDB34, CRDB38, CRDB39, CRDB42, CRDB46, CRDB47, CRDB48, CRDB52, CRDF23, CRDF33) exhibit strong amylase activity, indicated by the ‘+++’ results. This suggests these isolates possess the ability to hydrolyze starch into simpler sugars. Isolates CRDB78 and CRDF10 showed moderate (‘++’) and weak (‘+’) activity, respectively, while CRDB12, CRDB23, CRDB55, CRDF8, and CRDF32 showed negligible to no amylase activity. Fermentation test results varied among isolates. Strong fermentation activity (‘+++’) was observed in CRDB24, CRDB34, CRDB39, CRDB42, CRDB46, CRDB47, CRDB48, CRDB78, and CRDF25, suggesting efficient sugar metabolism under anaerobic conditions. Moderate fermentation (‘++’) was seen in CRDB12, CRDB23, and CRDF10, while CRDB10, CRDB52, and CRDB55 showed weak fermentation (‘+’). CRDF23, CRDF8, and CRDF33 were unable to ferment the tested sugar(s) (‘-’). Catalase activity, indicative of the ability to degrade hydrogen peroxide, was strongly positive (‘+++’) in CRDB46, CRDB47, CRDB48, CRDB55, CRDB78, CRDF10, CRDF8, and CRDF25. Moderate activity (‘++’) was noted in CRDB12, CRDB23, CRDB24, CRDB34, CRDB38, CRDB39, and CRDB42. Weak activity (‘+’) was recorded in CRDB10, CRDB52, CRDF23, and CRDF32. For H2S production, isolates such as CRDB10, CRDB12, CRDB23, CRDB34, CRDB39, CRDB46, CRDF10, CRDF23, CRDF8, and CRDF32 tested positive, confirming their potential to reduce sulfur compounds. However, isolates like CRDB24, CRDB38, CRDB42, CRDB47, CRDB48, CRDB52, CRDB55, CRDB78, CRDF25, and CRDF33 showed no detectable H2S production (‘-’). In the Methyl Red test, which detects stable acid end-products from glucose fermentation, isolates CRDB12, CRDB23, CRDB24, CRDB34, CRDB38, CRDB39, CRDF23, and CRDF25 were strongly positive (‘+++’). CRDB10, CRDB47, CRDB48, and CRDB52 showed moderate acid production (‘++’), while CRDB42 and CRDF10 showed weak acid production (‘+’). CRDB55, CRDF8, and CRDF33 were negative (‘-’) for this test. Ammonia production, indicating amino acid deamination, was strong (‘+++’) in isolates CRDB12, CRDB23, CRDB24, CRDB34, CRDB38, CRDB39, CRDB42, CRDB47, CRDB48, and CRDF25. Isolates CRDB52 and CRDF10 showed weak production (‘+’), while CRDB10 and CRDF23 showed moderate production (‘++’). No detectable ammonia was produced by CRDB55 and CRDB78.
Table (3):
Biochemical characterization of isolated crop residues decomposing bacterial and fungal isolates from different sources
Isolates Code |
Amylase Test |
Fermentation Test |
Catalase Test |
H2S Test |
Methyl red Test |
Ammonia production |
|---|---|---|---|---|---|---|
CRDB38 |
+++ |
++ |
++ |
– |
+++ |
+++ |
CRDB39 |
+++ |
+++ |
+++ |
++ |
++ |
+++ |
CRDB47 |
+++ |
+++ |
+++ |
– |
++ |
+++ |
CRDB48 |
+++ |
+++ |
+++ |
– |
++ |
+++ |
CRDB52 |
+++ |
+ |
+ |
– |
++ |
++ |
CRDB78 |
++ |
+++ |
+++ |
– |
++ |
– |
CRDB34 |
+++ |
+++ |
++ |
+ |
+++ |
+++ |
CRDB42 |
+++ |
+++ |
++ |
– |
+ |
+++ |
CRDB46 |
+++ |
+++ |
++ |
+++ |
+++ |
++ |
CRDB55 |
– |
+ |
+++ |
– |
– |
– |
CRDB12 |
– |
++ |
++ |
++ |
+++ |
+++ |
CRDB24 |
+++ |
+++ |
++ |
– |
+++ |
+++ |
CRDF23 |
+++ |
– |
++ |
+++ |
+++ |
+ |
CRDF10 |
+ |
++ |
+++ |
+++ |
+ |
+ |
CRDF8 |
– |
– |
+++ |
+++ |
– |
– |
CRDF25 |
+ |
+++ |
+++ |
– |
+++ |
++ |
CRDF32 |
– |
– |
+ |
+++ |
– |
+ |
CRDF33 |
+++ |
– |
+++ |
– |
– |
+ |
Note: The symbols indicate reaction strength: strong positive (+++) marked color change or strong activity, moderate positive (++) clear reaction, weak positive (+) slight change, and negative (-) no visible reaction
IAA production and phosphate solubilization by microbial isolates
The Figure 1 shows the IAA (Indole-3-acetic acid) production and Phosphate solubilization of various microbial isolates. The production of Indole-3-acetic acid (IAA) by the isolated microorganisms was evaluated under two conditions: without the addition of its precursor, tryptophan, and with tryptophan supplementation. The quantified IAA levels are expressed in micrograms per milliliter (µg/mL). A subset of isolates, including CRDB10, CRDB24, CRDF8, and CRDF10, demonstrated high levels of IAA production even without added tryptophan, with values exceeding 200 µg/mL, where CRDF8 exhibited the highest production at 461.9 µg/mL. A larger group of isolates, namely CRDB12, CRDB23, CRDB38, CRDB42, CRDB46, CRDB47, CRDB48, CRDB52, and CRDF23, showed moderate IAA production, ranging from approximately 100 to 200 µg/mL. In contrast, isolates CRDB34, CRDB39, CRDB55, CRDB78, CRDF25, CRDF32, and CRDF33 produced relatively low levels of IAA, with values below 100 µg/mL, and CRDB34 exhibited the lowest IAA production in the absence of tryptophan at 13.46 µg/mL (Figure 6). Upon supplementation with tryptophan, a general increase in IAA production was observed for the majority of the tested isolates, indicating the utilization of tryptophan as a precursor in their IAA biosynthesis pathways. Isolates CRDB24, CRDB34, CRDB38, CRDB48, and CRDF8 showed the most substantial increases in IAA production in the presence of tryptophan, reaching values above 290 µg/mL, with CRDB48 exhibiting the highest IAA production with tryptophan supplementation at 433.08 µg/mL. Isolates CRDB12, CRDB46, CRDF23, and CRDF25 also demonstrated significant increases in IAA production with tryptophan, ranging from approximately 230 to 270 µg/mL (Figure 7). Interestingly, isolates CRDB10 and CRDF10 showed a decrease in IAA production when tryptophan was added. Isolates CRDB55 and CRDF33 consistently showed the lowest IAA production levels even with the addition of tryptophan.
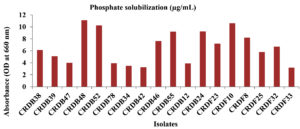
The capacity of the isolated microorganisms to solubilize phosphate, measured in micrograms per milliliter (µg/mL), varied considerably. A group of isolates, specifically CRDB10, CRDB38, CRDB48, CRDB52, CRDF10, CRDF23, and CRDF32, displayed a relatively strong ability to solubilize phosphate, yielding values exceeding 10 µg/mL. Among these, isolate CRDB52 exhibited the peak solubilization within the group at 10.25 µg/mL, while CRDF10 demonstrated the highest overall phosphate solubilization across all tested isolates, reaching 10.6 µg/mL. In contrast, a larger number of isolates, including CRDB24, CRDB34, CRDB46, CRDB47, CRDB55, CRDB78, CRDF8, and CRDF33, showed a moderate capacity for phosphate solubilization, with values ranging from approximately 3 to 10 µg/mL. Notably, isolates CRDB12 and CRDB23 exhibited a weak ability to solubilize phosphate, with values falling below 1 µg/mL. Singularly, isolate CRDF25 demonstrated a remarkably high level of phosphate solubilization, reaching 600 µg/mL, significantly surpassing the performance of all other isolates (Figure 8). This diverse range of phosphate solubilization capabilities underscores the inherent variability within microbial populations concerning their roles in nutrient cycling.
Identification of bacterial and fungal isolates
Table 4 presents the molecular identification of five selected microbial isolates, chosen based on their superior enzymatic activity profiles and biodegradation efficiency observed during preliminary screening assays. These isolates demonstrated the highest potential among all tested strains in terms of lignocellulolytic enzyme production, including CMCase, FPase, xylanase, pectinase, and chitinase activities, making them ideal candidates for residue decomposition applications. In this study, three bacterial and two fungal isolates were subjected to molecular characterization to confirm their taxonomic identity and further evaluate their relevance in agricultural residue degradation. The isolates were obtained from naturally decomposing wheat and rice straw samples. Molecular identification was performed through amplification and sequencing of the 16S rRNA gene for bacteria and the internal transcribed spacer (ITS) region for fungi.
Table (4):
Molecular identification of the isolates
Isolates |
Organism |
Accession number |
Sequence length |
Similarty |
|---|---|---|---|---|
CRDB24 |
Bacillus haynesii |
LC891910 |
1531 bp |
92.23% |
CRDB48 |
Bacillus altitudinis |
LC891911 |
648 bp |
97.14% |
CRDB52 |
Bacillus stratosphericus |
LC891912 |
1492 bp |
95.93% |
CRDF8 |
Fusarium oxysporum |
LC891913 |
720 bp |
100% |
CRDF32 |
Aspergillus fumigates |
LC891914 |
987 bp |
93.41% |
The bacterial isolates were identified as members of the Bacillus genus, which is widely recognized for its ability to degrade complex plant polymers and survive under diverse environmental conditions. Specifically, Bacillus haynesii (CRDB-24) shared 92.23% similarity with the reference sequence LC891910 (1531 bp), Bacillus altitudinis (CRDB-48) showed 97.14% similarity with LC891911, and Bacillus stratosphericus (CRDB-52) exhibited 95.93% similarity with LC891913 (1492 bp). These species have been previously reported to produce cellulases, xylanases, and other lignocellulolytic enzymes, making them effective contributors to crop residue decomposition.
The two fungal isolates identified were Fusarium oxysporum (CRDF-8) and Aspergillus fumigatus (CRDF-32), both of which have been extensively documented in the literature for their roles in organic matter degradation and nutrient cycling (Figure 9). Fusarium oxysporum (CRDF-8) demonstrated 100% sequence similarity to reference LC891913 (720 bp), indicating a high-confidence match, while Aspergillus fumigatus (CRDF-32) shared 93.41% similarity with LC891914 (987 bp).
The qualitative and quantitative screening of bacterial and fungal isolates from diverse crop residues in this study underscores a robust lignocellulolytic potential within the examined microbial community. All 18 isolates consistently exhibited cellulolytic activity on CMC agar, as indicated by clear zone formation, corroborating the findings of Zhang et al.,5 and Ma et al.4 These earlier studies similarly reported the widespread distribution of cellulolytic microbes in various ecological niches, such as soils, decaying plant matter, and termite guts, suggesting that the capacity to degrade cellulose is a fundamental and conserved trait among lignocellulolytic microorganisms. The universal production of extracellular polysaccharides (EPS) by all isolates, demonstrated through pronounced Congo red zones, aligns with existing understanding that EPS secretion is a conserved microbial strategy facilitating biofilm formation, resource acquisition, and niche adaptation in complex soil environments. This observation is consistent with previous reports by Vu et al.27 and Saini and Sharma et al.,7 who emphasized that EPS secretion facilitates microbial adhesion, biofilm formation, and survival in competitive environments. Strong EPS production enhances microbial efficiency in decomposing plant biomass by improving substrate accessibility and colonization.
Interestingly, β-glucosidase activity displayed notable heterogeneity, with isolates such as CRDB12 and CRDB55 showing no detectable activity, while others like CRDB38 and CRDB39 demonstrated strong (+++) activity. These findings align with the work of Zhang et al.,28 who documented strain-specific differences in β-glucosidase production, suggesting that such variability is influenced by ecological niche specialization and substrate availability. This suggests that while all isolates can initiate cellulose degradation, their capacities to hydrolyze cellobiose to glucose-a key step in complete cellulose utilization-differ significantly, potentially impacting their ecological roles in organic matter decomposition.
Similarly, xylanase activity exhibited a broad range, with CRDF25 lacking activity and CRDF32 showing strong (+++) activity. This finding resonates with Bhardwaj et al.,29 who emphasized the adaptive evolution of microbial xylanase production in response to hemicellulose-rich environments. The variation in xylanase activity highlights the functional diversity within the microbial community, enabling effective decomposition of hemicellulose-rich residues-a crucial step in the broader lignocellulose degradation process.
All isolates demonstrated strong chitinase activity (+++), which supports the findings of Gonfa et al.30 and Govindaraji and Vuppu,31 who described widespread chitinolytic potential among soil fungi and bacteria, particularly in organic matter-rich microhabitats. Pectinase activity was also predominantly high, especially in isolates like CRDF23, further confirming Govindaraji and Vuppu.31 assertion that pectin degradation is crucial for microbial penetration into plant tissues and that enzyme expression is strongly substrate-dependent.
Quantitative enzymatic profiling corroborated the qualitative trends and revealed significant diversity across isolates. CMCase (endoglucanase) activity ranged from 15.23 to 137.04 U/mL, with CRDB52 exhibiting the highest activity. Similar cellulase activity (120 IU/mL) was reported by El-Sobky et al.,32 in Bacillus spp. isolated from termite guts, though the present strain exhibits even higher activity (150 IU/mL). This suggests that the current isolate may possess enhanced cellulose-degrading capabilities, possibly due to genetic adaptations, optimized enzyme kinetics, or more efficient secretion mechanisms. FPase activity was notably higher in fungal isolates, with CRDF8 and CRDF32 showing peak values of 129.82 and 123.90 IU/mL, respectively. This finding aligns with the work of Bhardwaj et al.,29 who reported that fungi such as Fusarium and Aspergillus produce complex enzyme systems capable of efficiently degrading crystalline cellulose-a trait seldom observed in bacterial species. The present strain’s comparable performance suggests potentially unique enzymatic machinery or synergistic cellulase interactions, which could bridge the functional gap typically separating bacterial and fungal lignocellulose degradation.
A particularly striking result was the differential expression of β-glucosidase between media types. In nutrient broth, CRDF8 exhibited the highest activity (123.80 IU/mL), while CRDB24 achieved the maximum (156.33 IU/mL) in Luria broth. These findings corroborate Zhang et al.,28 who established nutrient availability as a key determinant of β-glucosidase activity. The significant media-dependent variations observed suggest that strain selection for microbial inoculants must consider both organismal potential and substrate compatibility to ensure consistent field performance. The xylanase activity in CRDF32 reached a peak of 22.5 IU/mL, showing a similar trend to that reported by Kapoor et al.,33 who observed a comparable efficiency range of 20-25 IU/mL in Aspergillus spp. This parallel performance between our bacterial isolate and established fungal degraders suggests convergent evolutionary adaptation for hemicellulose breakdown. The maximal pectinase activity observed in CRDF8 (20.21 IU/mL) aligns with reports from Govindaraji and Vuppu.31 for specialized lignocellulolytic soil fungi, indicating our bacterial isolate has developed fungal-comparable pectinolytic capability – an unusual trait that may reflect niche adaptation or enzyme system optimization. Chitinase activity across isolates showed moderate variation but was generally high, with CRDF32 again emerging as the top producer (21.7 IU/mL). These values align with Gonfa et al.,30 who reported similar chitinase ranges in compost-derived fungi. The consistently high chitinase activity highlights the potential of these isolates not only for biomass degradation but also for biological control applications.
Our biochemical characterization revealed substantial enzymatic diversity among the microbial isolates. Notably, several strains (CRDB24, CRDB34, CRDB39) exhibited robust amylase activity (+++), demonstrating exceptional starch-hydrolyzing capacity. This finding aligns with Alhazmi and Alshehri,34 who reported similar patterns in efficient decomposer communities. The strong amylolytic potential observed suggests these isolates are particularly well-adapted to starch-rich environments. Strong fermentation activity was observed in isolates such as CRDB24 and CRDB34, while others like CRDF23 and CRDF8 showed none, demonstrating clear metabolic diversity. This aligns with Hasan et al.35 and Zhao et al.,36 who emphasized the value of integrating conventional assays with modern techniques (MALDI-TOF MS) to enhance microbial identification and functional profiling. Catalase activity, indicative of an organism’s ability to decompose hydrogen peroxide and withstand oxidative stress, was detected in most isolates. Strong catalase activity (‘+++’) was observed in isolates such as CRDB46, CRDB47, CRDB48, CRDB55, CRDB78, CRDF10, CRDF8, and CRDF25. This suggests a high degree of resilience under oxidative conditions, which may confer an ecological advantage during organic matter decomposition. These findings are in agreement with Yuan et al.,37 who reported catalase production as a crucial trait in microbes inhabiting aerobic, stress-prone environments such as agricultural soils undergoing residue decomposition. Hydrogen sulfide (H2S) production, a result of sulfur compound reduction, was detected in multiple isolates, including CRDB10, CRDB12, CRDB23, CRDB34, CRDB39, CRDB46, CRDF10, CRDF23, CRDF8, and CRDF32. The capacity to produce H2S suggests involvement in sulfur cycling, which is essential for soil health and plant nutrition. These results support earlier observations by Verma et al.,38 and Kimura,39 who emphasized the ecological relevance of microbial H2S production in both promoting plant growth and mediating redox-sensitive processes in the soil environment.
The Methyl Red (MR) test, which detects stable acid end-products from glucose fermentation via mixed-acid pathways, revealed strong acid production (‘+++’) in isolates CRDB12, CRDB23, CRDB24, CRDB34, CRDB38, CRDB39, CRDF23, and CRDF25. This trait is often associated with the ability to lower soil pH, thereby influencing nutrient solubility and microbial interactions. The results align with Li et al.,24 Barry et al.,40 who identified acid production as a hallmark of microbial metabolic flexibility and its role in organic matter breakdown. Ammonia production, a result of amino acid deamination, was also significant among many isolates. Strong ammonia production (‘+++’) was observed in CRDB12, CRDB23, CRDB24, CRDB34, CRDB38, CRDB39, CRDB42, CRDB47, CRDB48, and CRDF25, suggesting a robust contribution to nitrogen cycling. Ammonia-producing microbes play a pivotal role in the release of plant-available nitrogen, enhancing soil fertility. These observations are consistent with Grzyb et al.,41 who highlighted the environmental and agricultural significance of microbial nitrogen transformations in optimizing nutrient availability and minimizing nitrogen losses.
Beyond their lignocellulolytic potential, the microbial isolates evaluated in this study also exhibited key plant growth-promoting (PGP) traits, underscoring their dual utility in sustainable agriculture. Notably, CRDF8 demonstrated the highest indole-3-acetic acid (IAA) production (461.9 µg/mL) even in the absence of exogenous tryptophan, suggesting the presence of robust endogenous auxin biosynthetic pathways. This finding is in agreement with Ali et al.,42 who reported strong IAA production in Bacillus species independent of tryptophan supplementation. In contrast, a significant increase in IAA production was observed in isolates such as CRDB48 (433.08 µg/mL) upon tryptophan supplementation, reinforcing the established role of tryptophan as a key precursor in IAA biosynthesis. However, certain isolates, including CRDB10 and CRDF10, exhibited reduced IAA levels when supplemented with tryptophan, implying the possibility of substrate inhibition or feedback regulation mechanisms, which merit further biochemical investigation.
Phosphate solubilization ability among the isolates also varied considerably. While the majority displayed moderate activity ranging from 3 to 10 µg/mL, isolates such as CRDB52 and CRDF10 exceeded this range, highlighting their potential role in enhancing phosphorus availability in soils. Remarkably, CRDF25 exhibited an exceptionally high phosphate solubilization capacity (600 µg/mL), far surpassing the levels commonly reported in literature. This finding is particularly noteworthy when compared to Li et al.,24 who reported solubilization levels up to 120 µg/mL in Bacillus and Pseudomonas spp. The superior performance of CRDF25 underscores its potential as a highly effective biofertilizer candidate, especially for use in phosphorus-deficient agricultural systems.
The molecular identification of the most efficient lignocellulolytic isolates substantiated the enzymatic screening outcomes by establishing a direct link between high enzymatic activity and taxonomic affiliation with well-known microbial decomposers and plant growth-promoting organisms. The bacterial isolates identified-Bacillus haynesii, Bacillus altitudinis, and Bacillus stratosphericus-all belong to the genus Bacillus, which is extensively documented for its ability to produce a wide array of hydrolytic enzymes, including cellulases, proteases, and xylanases. These enzymes play a crucial role in the breakdown of complex lignocellulosic substrates, facilitating residue decomposition and nutrient mineralization. The present findings are consistent with previous reports, such as those by Mahuku,26 who highlighted several Bacillus species as efficient decomposers contributing to organic matter turnover and enhanced soil fertility.
Similarly, the fungal isolates Fusarium oxysporum (CRDF8) and Aspergillus fumigatus (CRDF32) demonstrated substantial lignocellulolytic activity, corroborating their known ecological roles as potent decomposers. These species have been widely recognized for their enzymatic versatility and capacity to degrade complex plant residues, as noted by Sharma et al.3 Beyond their role in decomposition, these fungi are also reported to contribute to plant health through mechanisms such as enhanced nutrient availability and improved plant-microbe interactions. The recovery of these species in the current study not only validates their lignocellulolytic potential but also underscores their ecological adaptability and utility in integrated residue management strategies.
This study successfully isolated and characterized 18 lignocellulolytic microorganisms from diverse agro-residue sources, demonstrating their robust enzymatic potential for decomposing complex plant polymers. The isolates exhibited significant cellulolytic activity, with CRDB52 showing the highest CMCase activity (137.0 IU/mL) and CRDF8 displaying exceptional FPase activity (129.8 IU/mL). Notably, CRDF32 emerged as a top performer in xylanase (22.5 IU/mL) and chitinase (21.7 IU/mL) activity, while CRDF8 excelled in pectinase (20.2 IU/mL) Additionally, CRDF8 produced substantial IAA (461.9 µg/mL), and CRDF25 demonstrated remarkable phosphate solubilization (600 µg/mL), highlighting their potential in nutrient cycling and plant growth promotion. Molecular identification confirmed the presence of efficient decomposers, including Bacillus haynesii, Bacillus altitudinis, Fusarium oxysporum, and Aspergillus fumigatus, with 97%-100% sequence similarity to known lignocellulose degraders. Future studies should focus on developing tailored microbial consortia combining high-performing isolates (e.g., Bacillus spp. and Fusarium oxysporum) to enhance synergistic lignocellulose degradation and nutrient release in agricultural systems.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Singh Y, Singh B, Timsina J. Crop residue management for nutrient cycling and improving soil productivity in Rice-Based cropping systems in the tropics. Adv Agron;2005:269-407.
Crossref - Guo H, Hong C, Zheng B, Jiang D, Qin W. Improving enzymatic digestibility of wheat straw pretreated by a cellulase-free xylanase-secreting Pseudomonas boreopolis G22 with simultaneous production of bioflocculants. Biotechnol Biofuels. 2018;11(1).
Crossref - Sharma S, Kumawat KC, Kaur S. Potential of indigenous ligno-cellulolytic microbial consortium to accelerate degradation of heterogeneous crop residues. Environ Sci Pollut Res. 2022;29:88331-88346.
Crossref - Ma L, Lu Y, Yan H, et al. Screening of cellulolytic bacteria from rotten wood of Qinling (China) for biomass degradation and cloning of cellulases from Bacillus methylotrophicus. BMC Biotechnol. 2020;20(1):2.
Crossref - Z Zhang, JL Liu, JY Lan, CJ Duan, QS Ma, JX Feng. Predominance of Trichoderma and Penicillium in cellulolytic aerobic filamentous fungi from subtropical and tropical forests in China, and their use in finding highly efficient β-glucosidase. Biotechnol Biofuels. 2014;7:107.
Crossref - Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002;66(3):506-577.
Crossref - Saini S, Sharma KK. Fungal lignocellulolytic enzymes and lignocellulose: A critical review on their contribution to multiproduct biorefinery and global biofuel research. Int J Biol Macromol., 2022:206:819-840.
Crossref - Rahim KA. Biofertilizers in Malaysian agriculture: perception, demand and promotion. Presented at: 2002 FNCA Joint Workshop on Mutation and Bio-fertilizer. 2002 FNCA Joint Workshop on Mutation Breeding and Biofertilizer. 2002.
- De Vrieze J, Pinto AJ, Sloan WT, Ijaz UZ. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome. 2018;6:63.
Crossref - Kausar H, Sariah M, Saud HM, Alam MZ, Ismail MR. Isolation and screening of potential actinobacteria for rapid composting of rice straw. Biodegradation. 2011;22(4):367-375.
Crossref - Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol. 2008;57(5):503-507.
Crossref - Mandels M, Weber J. The production of cellulases. In: Hajny GJ, Reese ET eds. Cellulases and their applications. 1969:391-414.
Crossref - Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23(3):257-270.
Crossref - Polizeli MLTM, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS. Xylanases from fungi: Properties and industrial applications. Appl Microbiol Biotechnol. 2005;67(5):577-591.
Crossref - Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005;40(9):2931-2944.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426-428.
Crossref - Hernández-Téllez CN, Rodríguez-Córdova FJ, Rosas-Burgos EC, et al. Activity of chitosan–lysozyme nanoparticles on the growth, membrane integrity, and β-1,3-glucanase production by Aspergillus parasiticus. 3 Biotech. 2017;17(5):279.
Crossref - Patil RS, Ghormade V, Deshpande MV. Chitinolytic enzymes: an exploration. Enzyme Microb Technol. 2000;26(7):473–483.
Crossref - Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59(2):257-268.
Crossref - Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43(4):777-780.
Crossref - Eivazi F, Tabatabai MA. Glucosidases and galactosidases in soils. Soil Biol Biochem. 1988;20(5):601–606.
Crossref - Amin OE, Aboul-Enein AM, Abd-Elsalam IS, Wahba MI, Helmy YS. Optimization of protease production by newly isolated Bacillus sp. from the Red Sea using defatted soybean cake. Sci Rep. 2025;15:32118.
Crossref - Shomi FY, Uddin MB, Zerin T. Isolation and characterization of nitrogen-fixing bacteria from soil sample in Dhaka, Bangladesh. Stamford J Microbiol. 2023;11(1):11-13.
Crossref - Li Z, Li J, Liu G, et al. Isolation, characterization and growth-promoting properties of phosphate-solubilizing bacteria (PSBs) derived from peach tree rhizosphere. Microorganisms. 2025;13(4):718.
Crossref - Johnson JS, Spakowicz DJ, Hong B-Y, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029.
Crossref - Mahuku G. A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol Biol Rep. 2004;22:71-81.
Crossref - Vu B, Chen M, Crawford RJ, Ivanova EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14(7):2535–2554.
Crossref - Zhang X, Liu M, Fan Y, Xu J, Xu X, Li H. The structural and functional contributions of β-glucosidase-producing microbial communities to cellulose degradation in composting. Biotechnol Biofuels. 2018;11:51.
Crossref - Bhardwaj N, Kumar B, Verma P. A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Bioresour Bioprocess. 2019;6(1).
Crossref - Gonfa TG, Negessa AK, Bulto AO. Isolation, screening, and identification of chitinase-producing bacterial strains from riverbank soils at Ambo, Western Ethiopia. Heliyon. 2023;9(11):e21643.
Crossref - Govindaraji PK, Vuppu S. Characterisation of pectin and optimization of pectinase enzyme from novel Streptomyces fumigatiscleroticus VIT-SP4 for drug delivery and concrete crack-healing applications: An eco-friendly approach. Saudi J Biol Sci. 2020;27(12):3576–3588.
Crossref - El-Sobky MA, Abdel-Lateif KS, Fahmi AI, El-zanaty AM, Eissa RA. Correlation between qualitative and quantitative cellulase enzyme activities in some Trichoderma spp. Menoufia J Agric Biotechnol. 2024;9(2):23-38.
Crossref - Kapoor M, Nair LM, Kuhad RC. Cost-effective xylanase production from free and immobilized Bacillus pumilus strain MK001 and its application in saccharification of Prosopis juliflora. Biochem Eng J. 2008;38(1):88-97.
Crossref - Alhazmi LS, Alshehri WA. Purification and biochemical characterization of α-amylase from newly isolated Bacillus cereus strain and its application as an additive in breadmaking. Pol J Microbiol. 2025;74(1):48–59.
Crossref - Hasan MM, Marzan LW, Hosna A, Al-Hakim, Azad AK. Optimization of some fermentation conditions for the production of extracellular amylases by using Chryseobacterium and Bacillus isolates from organic kitchen wastes. J Genet Eng Biotechnol. 2017;15(1):297–305.
Crossref - Zhao YS, Eweys AS, Zhang JY, et al. Fermentation affects the antioxidant activity of plant-based food material through the release and production of bioactive components. Antioxidants. 2021;10:2004.
Crossref - Yuan F, Yin S, Xu Y, et al. The richness and diversity of catalases in bacteria. Front Microbiol. 2021;12:645477.
Crossref - Verma T, Bhardwaj S, Kapoor D, Singh, J. Is H₂S a lead or supporting player in plant development and growth? In: Modolo LV, da-Silva CJ, Singh VP, Tripathi DK, Fotopoulos B. eds. H₂S in plants: Past, present and beyond. 2024:193-209.
Crossref - Kimura H. Production and physiological effects of hydrogen sulfide. Antioxid Redox Signal. 2014;20(5):783-793.
Crossref - Barry AL, Bernsohn KL, Adams AP, Thrupp LD. Improved 18-hour methyl red test. Appl Microbiol. 1970;20:866-870.
Crossref - Grzyb A, Wolna-Maruwka A, Niewiadomska A. The significance of microbial transformation of nitrogen compounds in the light of integrated crop management. Agronomy. 2021;11(7):1415.
Crossref - Ali B, Sabri AN, Ljung K, Hasnain S. Auxin production by plant-associated bacteria: impact on endogenous IAA content and growth of Triticum aestivum L. Lett. Appl. Microbiol. 2009;48(5):542–547.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.