ISSN: 0973-7510
E-ISSN: 2581-690X
Streptococcus agalactiae (group B Streptococcus, GBS) is a pathogen associated with severe diseases in newborn and immunocompromised patients. One of the commonly used approaches for GBS identification is the CAMP test. It represents enhance of hemolysis when co-cultivating GBS with a β-toxin producing strain of S. aureus. In recent years, in addition to false positive results observed in other bacterial species, CAMP-negative GBS isolates have also been reported, questioning the specificity and sensitivity of the test. CAMP-negative phenotype is characterized by a lack of expression or deletion of cfb gene. According to data, the CovR/S regulatory system, β-hemolysin/cytolysin (cylE), and C5a protease (scpB) genes are possibly involved in the expression of a CAMP-factor. In our study 14 strains out of 294 (4.8%) were tested phenotypically negative for CAMP-factor, but positive for cfb gene. Among the CAMP-negative isolates the antibiotic susceptibility testing revealed resistance rates of 71.4%, 42.9%, and 100.0% for macrolides, lincosamides, and tetracyclines, respectively. Multidrug-resistant (MDR) isolates accounted for 42.9%. Detected serotypes were Ia (35.7%), III (21.4%), V (21.4%), and IV (7.1%). Frequencies of the analyzed virulence factors were as follows: cylE (85.7%) and scpB (92.9%). There was no statistical significance regarding antibiotic resistance and the distribution of the examined virulence genes between strains with CAMP-positive and negative phenotypes. The current study indicated that although the CAMP-test serves as an effective screening diagnostic tool, it is crucial to combine it with additional methods to obtain a conclusive microbiological diagnosis of GBS.
S. agalactiae, GBS, CAMP-test, CAMP-negative Phenotype
Streptococcus agalactiae (also referred to as group B Streptococcus, GBS) is a Gram-positive encapsulated bacterium with a tendency to arrange in chains, non-motile, and non-spore-forming. It is a fast-growing, nutritionally fastidious, facultative anaerobe, most commonly beta-hemolytic on blood agar, but with significantly weaker and less pronounced hemolysis than Streptococcus pyogenes (group A Streptococcus, GAS). Biochemically, GBS is catalase negative, hydrolyzes sodium hippurate, and it is resistant to bacitracin.1 GBS is a pathogen that is of extreme importance in the pathology of pregnancy and delivery, the leading cause of meningitis and septicemia in the neonate.2 The carriage of GBS in the genital system of females varies between 11.0% and 30.0% and it is more common in pregnant women than in non-pregnant ones.3 Almost half of the colonized females pass the pathogen on to their newborns.4 It is also an emerging opportunistic agent in immunocompromised individuals.2,5 GBS possess a large number of virulence factors, which can be divided into adhesion and colonization factors, immune evasion factors, toxins, and enzymes.6
The CAMP test, named after the first researchers who observed this reaction (Christie, Atkins, and Munch-Petersen), is often used for microbiological identification of GBS. First described in 1944, it shows an arrowhead-shaped enhancement of beta-hemolysis when sphingomyelinidase-secreting S. aureus is cultivated on sheep blood agar in adjacent GBS, which produces CAMP factor (25 kDa protein). Sphingomyelinase converts the sphingomyelin of sheep erythrocytes into ceramide, which makes them susceptible to the effects of pore-forming CAMP factor toxin.7,8 CAMP factor, encoded by the cfb gene, is not essential for virulence and does not promote adhesion, invasion, or biofilm formation. It is considered an important confirmatory test for the identification of S. agalactiae that is low cost and easy to be performed. But its specificity and sensitivity have been questioned.9-11
On the one hand, Streptococcus porcinus, Streptococcus pseudoporcinus, Streptococcus iniae, Listeria monocytogenes, Listeria ivanovii, Rhodococcus equi, Pasteurella haemolytica, Aeromonas spp., Vibrio spp., Cutibacterium acnes, group G streptococci, and others test positive. The gene sequence encoding the CAMP factor of GAS (cfa) is homologous to the GBS cfb gene and it is highly expressed when GAS is incubated under 5.0% CO2 in the presence of CaCl2 or MgCl2. Analogous genes have been detected in other bacteria – Streptococcus uberis, Cutibacterium acnes, and Bartonella henselae.8,12-16
On the other hand, cases of CAMP-negative strains detected by polymerase chain reaction (PCR) and nucleoid sequencing have been reported. This may be due to a defect or absence of the cfb gene. In some cases, the gene may be intact, but there is low gene expression, transcription defects, or low CAMP factor activity.10,17-20
The production of toxins and adhesion factors is regulated by the two-component system known as GBS CovR/S (CsrR/S).21 The beta-hemolysin/cytolysin is expressed by almost all GBS strains. It is responsible for the formation of beta-hemolysis on blood agar and protection from phagocytes. Its activity is regulated by the cylE gene.22 Streptococcal C5a protease (ScpB) is a surface protein with enzymatic activity that belongs to the family of serine proteases. It is involved in the inactivation of one of the components of complement C5a (chemotactic factor for phagocytic cells), a product of the action of C5a convertase.23 Capsular polysaccharides promote the formation of biofilms while reducing the efficacy of phagocytosis and the complement system. Due to the different antigenicity of the capsular polysaccharides, 10 serotypes (Ia, Ib, II-IX) are known. While serotypes Ia, V, and III are more common in invasive infections in adults, serotype III is considered to be the most virulent, frequently identified as a prevalent cause of late-onset meningitis in neonates.24,25 Inactivation of the genes associated with CovR/S (CsrR/S) regulatory system leads to a significant increase in the production of beta-hemolysin/cytolysin and C5a protease, a significant decrease in the production of CAMP factor and no change in the production of capsular polysaccharides.26
The aim of the study was to identify the CAMP-negative GBS isolates, to determine the genetic determinants associated with the regulation of CAMP factor, as well as antibiotic susceptibility of these isolates. This will help in monitoring the emergence and spread of phenotypically CAMP-negative GBS isolates, leading to diagnostic difficulties.
Sample collection
As part of routine diagnostics, a total of 294 GBS isolates were obtained from inpatient and outpatient patients. The specimens were collected between September 2021 and November 2024 from three Bulgarian hospitals. Among these, 14 (n = 14) strains were presented with CAMP negative phenotype (4.8%). The age range of this group of patients was between 21 and 57 years. All the materials were vaginal swab samples, collected from pregnant (35.7%) and non-pregnant women (64.3%) with clinical signs of genital infection. The samples were delivered to the laboratory in transport nutrient media, accompanied by smears on glass slides, and subsequently microbiological analyses were performed systematically. Every patient provided written informed consent.
Bacterial strains
All strains suspected to be GBS were tested with conventional biochemical methods, CAMP-test, latex-agglutination test (PathoDxtra Strep Grouping Kit ThermoScintific, Oxoid, UK) and if necessary, with Crystal GP (Becton Dickinson, Kelberg, Germany). GBS strains were stored in skim milk at -70 °C and were sub-cultured three times on Columbia agar (Becton Dickinson, Kelberg, Germany) with 5.0% sheep blood for 18-24 h at 35 °C in 5.0% CO2 atmosphere before the antibiotic susceptibility testing and other tests. Streptococcus pneumoniae ATCC 49619, Staphylococcus aureus ATCC 25923, S. agalactiae ATCC 13813, and Enterococcus faecalis ATCC 29212 were used as control strains according to the EUCAST 2025 guidelines and previous studies.27
CAMP test
Initially, S. aureus (ATCC 25923) was streaked in the middle of the blood agar. Perpendicularly about 1 mm to the first strip, we inoculated S. agalactiae (ATCC 13813) and E. faecalis (ATCC 29212), which served as positive and negative controls. The tested samples were placed adjacent to S. aureus in the same manner. The blood agar was then incubated at 35-37 °C for 18-24 h. An arrowhead-shaped increase in beta-hemolysis between S. aureus and the sample indicated a positive CAMP reaction. The described phenomenon was not present in the CAMP-negative samples.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing to erythromycin, clindamycin, and tetracycline was performed by determining the minimum inhibitory concentration (MIC) using E-tests (Laboratories Pvt. Ltd., India). The MICs of penicillin G and vancomycin were determined using a broth microdilution test (MIKROLATEST® MIC, Erba Lachema s.r.o., Czech Republic). For interpretations of the results of antibiotic testing, EUCAST recommendations were used (EUCAST 2025).27
DNA extraction
DNA extraction was performed using an extraction kit (DNA-Sorb-A DNA extraction kit, Sacace Biotechnologies Srl, Italy) according to the manufacturer’s instructions. All DNA extracts were stored at -70 °C before being used in experiments.
Detection of genetic profiles
All collected GBS strains were confirmed by PCR using forward and reverse primers STRA-AgI and II, which target the 16S to 23S rRNA intergenic spacer region.28 We used previously described primer sequences for the identification of macrolide, lincosamide, and tetracycline resistance genes as well as the capsular serotypes.29 For cfb gene amplification, we used the methods described by Zhou et al.30 The primer sequences for the virulence factors examined in this study are listed in Table 1.30,31 The reaction conditions for conventional PCR were initial denaturation at 95 °C for 5 min, followed by 30-35 cycles consisting of denaturation at 95 °C for 30 sec, annealing for 30 sec, and elongation at 72 °C for 1 min; final elongation at 72 °C for 5-10 min. For the PCR reaction and interpretation of the result using gel electrophoresis (2.0% agarose), we used prime Taq premix 2x (Genet Bio, Daejeon, South Korea) and GelRed nucleic acid gel stain (Biotium, San Francisco, USA).
Table (1):
Primer sequences and amplification conditions for the detection of GBS virulence genes
| Primer sequence (5’→3’) | Product size (bp) | Annealing temp. (°C) | Ref. | |
|---|---|---|---|---|
| cfb | F TGGTAGTCGTGTAGAAGCCTTA | 370 | 58 | 30 |
| R TCCAACAGCATGTGTGATTGC | ||||
| cylE | F TGACATTTACAAGTGACGAAG | 268 | 55 | 31 |
| R TTGCCAGGAGGAGAATAGGA | ||||
| scpB | F ACAATGGAAGGCTCTACTGTTC | 255 | 60 | 31 |
| R ACCTGGTGTTTGACCTGAACTA |
F: Forward primer, R: reverse primer
Statistical analysis
Statistical analyses were carried out with IBM SPSS Statistics for Windows v19.0 (IBM Corp., Chicago, IL, USA). Fisher’s exact test was used. A p < 0.05 was considered statistically significant.
14 strains out of 294 (4.8%) tested phenotypically negative for the CAMP factor but positive for the cfb gene (Figures 1 and 2).
Figure 1. (A), (B) and (C) Examples of CAMP-negative GBS isolates. S. agalactiae (ATCC 13813) and E. faecalis (ATCC 29212) served as positive and negative controls. CAMP-positive isolates are also presented
Figure 2. Gel electrophoresis (2.0% agarose) of the amplified cfb gene from 14 CAMP-negative isolates. Molecular weight marker 100-1000 bp DNA Ladder (Meridian Bioscience, USA) was used, followed by one negative control and positive results for cfb gene of tested isolates
The frequencies of the other analyzed virulence factors in the CAMP-negative isolates were: cylE (85.7%) and scpB (92.9%), and three isolates testing positive for these two factors showed significant beta-hemolysis (Table 2). Detected serotypes were Ia (35.7%), III (21.4%), V (21.4%), and IV (7.1%). Two strains were non-typeable.
Table (2):
Distribution of tested virulence factors among CAMP-negative and CAMP-positive GBS isolates
Factors of virulence |
CAMP-negative (n = 14) |
CAMP-positive (n = 280) |
Total number (n = 294) |
p-value* (CAMP-negative/CAMP positive) |
|---|---|---|---|---|
cfb |
14 (100.0%) |
280 (100.0%) |
294 (100.0%) |
|
cylE |
12 (85.7%) |
250 (89.3%) |
262 (89.1%) |
0.655 |
scpB |
13 (92.9%) |
261 (93.2%) |
274 (93.2%) |
1 |
*a p-value <0.05 is considered statistically significant
Table (3):
Distribution of antibiotic resistance among CAMP-negative and CAMP-positive GBS isolates
Antibiotics |
CAMP-negative (n = 14) |
CAMP-positive (n = 280) |
Total number (n = 294) |
p-value* (CAMP-negative/CAMP positive) |
|---|---|---|---|---|
Penicillin |
0 |
0 |
0 |
|
Erythromycin |
10 (71.4%) |
169 (60.4%) |
179 (60.9%) |
0.577 |
Clindamycin |
6 (42.9%) |
70 (25.0%) |
76 (25.9%) |
0.206 |
Tetracycline |
14 (100.0%) |
246 (87.9%) |
260 (88.4%) |
0.383 |
*a p-value <0.05 is considered statistically significant
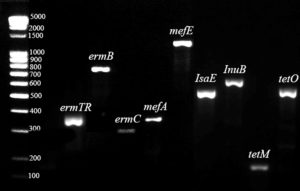
All CAMP-negative strains were susceptible to penicillin and vancomycin, while resistance to macrolides, lincosamides, and tetracyclines was 71.4%, 42.9%, and 100.0% respectively (Table 3). Multidrug-resistance (MDR) strains that were not susceptible to three or more classes of antibiotics accounted for 42.9%, and all CAMP-negative serotype III isolates belonged to this group. The main genetic profiles of the tested antibiotic resistance genes were: ermA/TR+tetM, ermA/TR+ermC+tetM, ermB+tetM and mefA+msrD+tetM (Table S1 and Figure 3).
Figure 3. Gel electrophoresis (2.0% agarose) of the amplified gene determinants for the identification of macrolide, lincosamide, and tetracycline resistance. Molecular weight marker 100-5000 bp DNA Ladder Extended (Carl Roth GmbH, Germany) was used
There was no statistical significance regarding antibiotic resistance and the distribution of the examined virulence genes between strains with CAMP-positive and negative phenotypes
(p > 0.05) (Tables 2 and 3).
In our study, all CAMP-negative isolates were positive for the cfb gene but showed no phenotypic expression during testing. Similar results have been reported by other authors.10,18 In contrast, in other studies, most isolates examined showed a deletion of the entire gene, while others reported deletion of parts of the gene.19,20,30,32 The common characteristic in the aforementioned cases is that there was no increase in beta-hemolysis between the control strain S. aureus and the isolates that were examined, leading to negative CAMP results.
According to previous studies CAMP-test was reported as a reliable method for the identification of GBS and even amplification of the cfb gene was selected for rapid identification of GBS in clinical samples.33-36 In recent years, there have been increasing reports of CAMP-negative GBS isolates, which contradicts the generally assumed ubiquitous distribution of the CAMP factor.10 In the present study, 4.8% of examined strains were CAMP-negative. Zhou et al., whose amplification protocol was used in the current study, found that 7.9% of samples had no gene expression.30 In a recently published study by Lai et al., the CAMP-negative rate was found to be 3.6%. Although this rate is lower than our own findings, it is still considerably higher than the 1.0% previously documented in the literature.32 This indicated that the CAMP test method and primers that target the cfb gene shouldn’t be the only presumptive test used to identify GBS.30
Not all S. aureus strains are suitable for the CAMP test, leading to false negative results. The production of β-toxin encoding sphingomyelinase is observed in almost all strains of S. aureus; however, this toxin becomes inactive due to the integration of mobile genetic elements into its encoding gene. As a result, it has been reported that most strains do not produce β-toxin, and its synthesis is specifically identified in particular lineages.37,38 In the present study, we used a control strain of S. aureus, which produces β-toxin and thereby exhibits a positive CAMP test when co-cultivated with GBS. Some authors have proposed that Staphylococcus pseudintermedius serves as a reliable alternative of S. aureus in CAMP-test, as it produces β-toxin.39,40 Some bacterial species also produce CAMP or CAMP-like factors, leading to false positive tests. Important examples of these are GAS and L. monocytogenes. In differential consideration, L. monocytogenes is the third most common cause of neonatal meningitis after GBS and E. coli.41,42 Importantly, L. monocytogenes is not susceptible to cephalosporins, a common choice of empirical treatment for bacterial infections.42 GAS produced stronger and more pronounced hemolysis than GBS.1 However, in the current study, three CAMP-negative isolates showed significant beta-hemolysis with positive cylE and scpB genes in accordance to the results reported by Jiang et al.26
In two studies conducted in China involving CAMP-negative isolates, the rates of antibiotic resistance were found to be lower in CAMP-negative isolates compared to CAMP-positive isolates; however, there was no statistical significance, except for the distribution of tetracyclines in one of them.30,32 Additionally, all strains reported by Lai et al. were identified as serotype III.32 In the current study, despite the lack of statistical significance, the resistance rates to the antibiotics examined was higher in the CAMP-negative compared to CAMP-positive isolates by 11.0%, 17.9%, and 12.1%, respectively. Furthermore, all CAMP-negative serotype III strains were MDR, which is substantial because this serotype is the most virulent and a common cause of neonatal meningitis.24 This result highlights the emergence of resistance among isolates, which could be incorrectly diagnosed, leading to difficulties in management of severe infections. The probable explanation for the lack of significance in the distribution of CAMP-negative isolates is that the CAMP factor is not essential for virulence. These observations are supported by studies conducted on CAMP-negative isolates.9,30,32
Rosa-Fraile et al. presented a comparison between different methods for GBS detection according to relative sensitivity and specificity.36 DNA sequencing was rated with the highest results. Unfortunately, this method is time-consuming and not available for routine diagnosis. Another method with high specificity is the use of Granada-type medium. It differentiates based on the hemolysis of GBS and cylE gene expression.36 In our study, two CAMP-negative strains were nonhemolytic and cylE-negative. This leads to additional difficulties in the microbiological interpretation of the results. The CAMP test is an effective screening tool and often serves as a first line of diagnosis, however, only a combination of all routine tests can definitively establish the final diagnosis.
In the present study, CAMP-negative isolates accounted for 4.8%, including serotype III MDR strains. Moreover, some of those isolates exhibited atypical hemolysis, which posed further diagnostic challenge. We examined the genetic determinants associated with the regulation of CAMP factor, as well as antibiotic resistance, which would facilitate the monitoring of phenotypically CAMP-negative GBS isolates. We concluded that the CAMP test is exclusively a screening tool and should be combined with other tests to achieve a definitive diagnosis of GBS, which is especially important for pregnant women.
Additional file: Table S1.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the Medical University of Sofia (Council of Medical Science), Contract number D-298/18.12.2023.
DATA AVAILABILITY
All datasets generated or analyzed during the study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Gizachew M, Tiruneh M, Moges F, Tessema B. Streptococcus agalactiae maternal colonization, antibiotic resistance and serotype profiles in Africa: a meta-analysis. Ann clin microbiol antimicrob. 2019;18(1):14.
Crossref - Gergova R, Boyanov V, Muhtarova A, Alexandrova A. A Review of the impact of Streptococcal infections and antimicrobial resistance on human health. Antibiotics (Basel). 2024;13(4):360.
Crossref - Sadeh M, Salehi-Abargouei A, Azartoos N, Mirzaei F, Khalili MB. Distribution of Streptococcus agalactiae among Iranian women from 1992 to 2018: A systematic review and meta-analysis. Jundishapur J Microbiol. 2020;13(7):e102314.
Crossref - Rao GG, Khanna P. To screen or not to screen women for Group B Streptococcus (Streptococcus agalactiae) to prevent early onset sepsis in newborns: recent advances in the unresolved debate. Ther Adv Infect Dis. 2020;7:2049936120942424.
Crossref - Saad EJ, Baenas DF, Boisseau CS, et al. Streptococcus agalactiae bacteremia in non-pregnant adult patients at two teaching hospitals. Rev Argent Microbiol. 2018;50(3):280-284.
Crossref - Schindler Y, Rahav G, Nissan I, et al. Group B streptococcus virulence factors associated with different clinical syndromes: Asymptomatic carriage in pregnant women and early-onset disease in the newborn. Front Microbiol. 2023;14:1093288.
Crossref - Lang S, Palmer M. Characterization of Streptococcus agalactiae CAMP factor as a pore-forming toxin. J Biol Chem. 2003;278(40):38167-38173.
Crossref - Jin T, Brefo-Mensah E, Fan W, et al. Crystal structure of the Streptococcus agalactiae CAMP factor provides insights into its membrane-permeabilizing activity. J Biol Chem. 2018;293(30):11867-11877.
Crossref - Hensler ME, Quach D, Hsieh CJ, Doran KS, Nizet V. CAMP factor is not essential for systemic virulence of Group B Streptococcus. Microb Pathog. 2008;44(1):84-88.
Crossref - Guo D, Xi Y, Wang S, Wang Z. Is a positive Christie-Atkinson-Munch-Peterson (CAMP) test sensitive enough for the identification of Streptococcus agalactiae? BMC Infect Dis. 2019;19(1):7.
Crossref - Ballard MB, Mercado-Evans V, Marunde MG, Nwanosike H, Zulk J, Patras KA. Group B Streptococcus CAMP factor does not contribute to interactions with the vaginal epithelium and is dispensable for vaginal colonization in mice. Microbiol. Spectr. 2021;9(3):e0105821.
Crossref - Gase K, Ferretti JJ, Primeaux C, McShan WM. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect Immun. 1999;67(9):4725-4731.
Crossref - Kurosawa M, Oda M, Domon H, Saitoh I, Hayasaki H, Terao Y. Streptococcus pyogenes CAMP factor attenuates phagocytic activity of RAW 264.7 cells. Microbes Infect. 2016;18(2):118-127.
Crossref - Liatsos GD, Tsiriga A, Dourakis SP. Fatal Streptococcus pseudoporcinus disseminated infection in decompensated liver cirrhosis: a case report. J Med Case Reports. 2021;15:240.
Crossref - Siewert LK, Korotaev A, Sedzicki J, Fromm K, Pinschewer DD, Dehio C. Identification of the Bartonella autotransporter CFA as a protective antigen and hypervariable target of neutralizing antibodies in mice. Proc Natl Acad Sci U S A. 2022;119(25):e2202059119.
Crossref - Thomas TSM, Thomas J, le Roux K, Duze ST, Mkhwanazi F, Duse A. Diagnostic challenges with accurate identification of Listeria monocytogenes isolates from food and environmental samples in South Africa. Afr J Lab Med. 2022;11(1):1482.
Crossref - Podbielski A, Blankenstein O, Lutticken R. Molecular characterization of the cfb gene encoding group B streptococcal CAMP-factor. Med Microbiol Immunol. 1994;183(5):239-256.
Crossref - Hassan AA, Akineden O, Lammler C, Huber-Schlenstedt R. Molecular characterization of phenotypically CAMP-negative Streptococcus agalactiae isolated from bovine mastitis. J Vet Med B Infect Dis Vet Public Health, 2002;49(5):257-259.
Crossref - Tickler IA, Tenover FC, Dewell S, et al. Streptococcus agalactiae Strains with chromosomal deletions evade detection with molecular methods. J Clin Microbiol. 2019;57(4):e02040-18.
Crossref - Creti R, Imperi M, Stanziale A, Giuliani G, Fazii P, Savini V. Group B streptococci (GBS) strains evading molecular diagnostics showed novel chromosomal deletions encompassing the CAMP-factor (cfb) encoding gene. Eur J Clin Microbiol Infect Dis. 2023;42(7):913-916.
Crossref - Patras KA, Wang NY, Fletcher EM, et al. Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol. 2013;15(7):1154-1167.
Crossref - Armistead B, Herrero-Foncubierta P, Coleman M, et al. Lipid analogs reveal features critical for hemolysis and diminish granadaene mediated Group B Streptococcus infection. Nat. Commun. 2020;11(1):1502.
Crossref - Dobrut A, Brzychczy-Wloch M. Immunogenic proteins of Group B Streptococcus—potential antigens in immunodiagnostic assay for GBS detection. Pathogens, 2022;11(1):43.
Crossref - Shabayek S, Spellerberg B. Group B Streptococcal colonization, molecular characteristics, and epidemiology. Front. Microbiol. 2018;9:437.
Crossref - Ali MM, Asrat D, Fenta DA, Chaka T, Woldeamanuel Y. Group B Streptococcus colonization rate and serotype distribution among pregnant women and their newborns at Adama Hospital Medical College, Ethiopia. Sci Rep. 2020;10(1):9301.
Crossref - Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in group B Streptococcus. J Bacteriol. 2005;187(3):1105-1113.
Crossref - The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 15.0. 2025. Accessed on September 1, 2025. http://www.eucast.org
- Delannoy CM, Crumlish M, Fontaine MC, et al. Human Streptococcus agalactiae strains in aquatic mammals and fish. BMC Microbiol. 2013;13:41.
Crossref - Boyanov VS, Alexandrova AS, Hristova PM, Hitkova HY, Gergova RT. Antibiotic resistance and serotypes distribution in Streptococcus agalactiae Bulgarian clinical isolates during the years of 2021–2024. Pol J. Microbiol. 2024;73(4):505-514.
Crossref - Zhou J, Zhang L, Zhang Y, et al. Analysis of molecular characteristics of CAMP-negative Streptococcus agalactiae strains. Front Microbiol. 2023;14:1189093.
Crossref - Lopez Y, Parra E, Cepas V, et al. Serotype, virulence profile, antimicrobial resistance and macrolide-resistance determinants in Streptococcus agalactiae isolates in pregnant women and neonates in Catalonia, Spain. Enferm Infecc Microbiol Clin (Engl Ed). 2018;36(8):472-477.
Crossref - Lai X, Chen M, Wang J, et al. CAMP-negative Streptococcus agalactiaestrains exhibited complete or partial chromosomal deletions of the CAMP-factor encoding gene cfb. Microbiol Spectr. 2025;13(5):e0325724.
Crossref - Ke D, Menard C, Picard FJ, et al. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46(3):324-331.
Crossref - Gosiewski T, Brzychczy-Wloch M, Heczko PB. The application of multiplex PCR to detect seven different DNA targets in group B streptococci. Folia Microbiol (Praha). 2012;57(3):163-167.
Crossref - Park JS, Cho DH, Yang JH, et al. Usefulness of a rapid real-time PCR assay in prenatal screening for group B streptococcus colonization. Ann Lab Med. 2013;33(1):39-44.
Crossref - Rosa-Fraile M, Spellerberg B. Reliable Detection of Group B Streptococcus in the Clinical Laboratory. J Clin Microbiol. 2017;55(9):2590-2598.
Crossref - Salgado-Pabon W, Herrera A, Vu BG, et al. Staphylococcus aureus β-toxin production is common in strains with the β-toxin gene inactivated by bacteriophage. J Infect Dis. 2014;210(5):784-792.
Crossref - Poupel O, Kenanian G, Touqui L, Abrial C, Msadek T, Dubrac S. Timely excision of prophage Φ13 is essential for the Staphylococcus aureus infectious process. Infect Immun. 2025:e0031425.
Crossref - Savini V, Paparella A, Serio A, Marrollo R, Carretto E, Fazii P. Staphylococcus pseudintermedius for CAMP-test. Int J Clin Exp Pathol. 2014;7(4):1733-1734.
- Kmieciak W, Szewczyk EM, Ciszewski M. Searching for beta-haemolysin hlb gene in Staphylococcus pseudintermedius with species-specific primers. Curr Microbiol. 2016;73(1):148-152.
Crossref - Tavares T, Pinho L, Andrade EB. Group B Streptococcal Neonatal Meningitis. Clin Microbiol Rev. 2022;35(2):e0007921.
Crossref - Wu F, Nizar S, Zhang L, Wang F, Lin X, Zhou X. Clinical features and antibiotic treatment of early-onset neonatal listeriosis. J Int Med Res. 2022;50(8):3000605221117207.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.