ISSN: 0973-7510
E-ISSN: 2581-690X
The swift and precise diagnosis of pyogenic infections is essential for efficient treatment, particularly in resource-constrained facilities. The present study assessed the effectiveness of chromogenic agar (HiCrome) in isolating pyogenic organisms in comparison to traditional culture techniques as periodic reassessment is crucial to ensure its continued diagnostic accuracy and cost-effectiveness in the face of evolving pathogens, changing laboratory conditions, and antimicrobial stewardship requirements. One hundred clinical samples suspected of pyogenic infections were analysed using HiCrome agar and traditional culture methods. Bacterial isolates were identified using colony morphology and biochemical assays. Antibiotic susceptibility was assessed via the disk diffusion method, and the turnaround time (TAT) for results was documented. HiCrome agar demonstrated a rapid isolation rate, with 57.55% of isolates documented within 24 h In contrast, conventional methods required 48 h for complete identification, achieving a 100% success rate at that time. On the antibiogram, Escherichia coli and Klebsiella pneumoniae exhibited high sensitivity to carbapenems, while Pseudomonas aeruginosa showed variable resistance patterns. The HiCrome agar effectively isolated the key pyogenic organisms, including Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Enterococcus faecium (all with 100% matching rate), Staphylococcus aureus (92.31% matching rate), Pseudomonas aeruginosa (85.71% matching rate) but had limitations with certain fastidious organisms, which showed lower matching rates. HiCrome agar is an efficient tool for the swift identification of pyogenic infections, offering considerable benefits in turnaround time. Nonetheless, traditional approaches are crucial for thorough identification, especially for demanding organisms. A synergistic approach employing both approaches may enhance diagnostic precision and elevate patient outcomes in clinical environments.
Pyogenic Infections, Turnaround Time, Klebsiella pneumoniae, Pseudomonas aeruginosa
In clinical microbiology, the rapid and accurate identification of pathogens is essential for effective patient management, especially in instances of pyogenic infections.1 Pyogenic infections, marked by pus formation resulting from bacterial invasion, could impact multiple tissues and organs, resulting in considerable morbidity and mortality. Prevalent causal pathogens comprised Streptococcus pyogenes, Staphylococcus aureus, Escherichia coli, Klebsiella spp., Proteus spp., and Pseudomonas spp.2 In India, the prevalence of infectious diseases was significant, with a crude mortality rate of almost 417 per 100,000 individuals, highlighting the urgent necessity for prompt identification and treatment of pyogenic infections.3,4
Conventional cultural techniques for identifying bacterial infections typically necessitate prolonged incubation periods and may not yield prompt results essential for successful clinical decision-making. In this context, chromogenic agar (Chrom agar) has surfaced as a viable option. Chrom agar is a differential medium that enables the presumptive identification and confirmation of microorganisms via discrete color changes in the colonies, which occur due to the interaction between specific enzymes produced by target bacteria and the chromogenic substrates included in the medium. This feature facilitates expedited identification relative to traditional approaches, as evidenced by numerous studies that underscore its efficacy in detecting a spectrum of diseases, including multidrug-resistant organisms and uropathogens.5,6
The benefits of Chrom agar surpass its swift identifying skills. In resource-constrained centers, where access to sophisticated diagnostic instruments may be limited, Chrom agar provides an economical and user-friendly alternative. The preparation technique entails basic boiling and transfer into Petri dishes, obviating the necessity for specialist apparatus or substantial technical proficiency.5,7
Notwithstanding these benefits, the efficacy of Chrom agar in isolating pyogenic organisms has not been well investigated. The study intended to address the gap in previous research, which had predominantly concentrated on the application of Chrom agar in identifying uropathogens and multidrug-resistant organisms, by assessing its efficacy for pyogenic infections. The rising incidence of antibiotic resistance in prevalent bacteria underscores the necessity for prompt detection and treatment techniques customized to individual patient requirements, particularly in at-risk groups such as individuals with co-morbidities or impaired immune systems. The present research aimed to offer significant insights into the efficacy of Chrom agar as a diagnostic measure in clinical microbiology by systematic analysis of these factors. The present study had two objectives: The first objective was to evaluate the efficacy of Chrom agar in isolating pyogenic pathogens and to compare its performance with traditional culture methods for bacteriological profiles in reducing TAT. The second objective was to observe the antibiotic susceptibility patterns of the isolates.
Study design and sample size
A prospective observational study was conducted among 100 consecutive pyogenic samples submitted for culture to the microbiology department of PSG Diagnostic Centre, PSGIMSR, located in Coimbatore, Tamil Nadu, India. The study was conducted after obtaining approval from the Institutional Human Ethical Committee (IHEC) with reference number PSG/IHEC/2023/Appr/Exp/430.
Samples were included in the present study, if they were pus samples or exudates obtained from abscess, wound and post-operative aspirates. Tissues samples were also included along with fluids from sterile areas: CSF, synovial joint fluid, ascetic fluid, pleural and pericardial fluid. The study was funded by the Indian Council of Medical Research (ICMR) under the short-term studentship program (Reference number: 2023-11849)
Sample collection
The received samples were cultured within two hours of collection and if any delay was expected, the samples were stored at 4-6 °C for up to four hours.
Procedure
The collected samples were subjected to direct microscopy, conventional bacterial culture, and antimicrobial susceptibility test (AST). Direct microscopic examination was included to (1) confirm that the submitted specimen is adequate and truly purulent by visualizing inflammatory cells, pus cells and debris, (2) provide a rapid presumptive diagnosis by directly identifying microorganisms (e.g., Gram-positive cocci or Gram-negative bacilli) before culture results are available, and (3) guide the selection of appropriate culture media and downstream identification tests, thereby ensuring quality control and optimizing laboratory work flow. The conventional methods include inoculation onto blood agar, MacConkey agar and chocolate agar medium and incubating aerobically at 37 °C for 24 h and, followed by examination of the inoculated plates for culture characteristics. The organisms were identified to species level using biochemical tests and morphologically using direct microscopy. The specimens were simultaneously inoculated onto Urochrome agar (HIMEDIA Laboratories, India)and incubated at 35-37 °C for 18-24 h. The colour of the colonies on HiCrome UTI agar was noted. For quality control, cultures procured from ATCC were used. Colony characteristics observed on HiCrome UTI agar with reference cultures were: E. coli ATCC 25922- pink-purple colonies, Klebsiella pneumoniae ATCC 13883-blue to purple mucoid colonies, Pseudomonas aeruginosa ATCC 27853 colourless (greenish pigment may be observed), Proteus mirabilis ATCC 12453-light brown, S. aureus ATCC 25923- golden yellow, Enterococcus faecalis ATCC 29212- blue small, and Acinetobacter baumannii complex colourless colonies. The bacteria from the test samples were identified in comparison with the color produced with reference cultures (Figure 1). The antimicrobial susceptibility testing was done by disc diffusion method employing Mueller-Hinton agar plates and results were read 24 h of incubation at 35-37 °C.
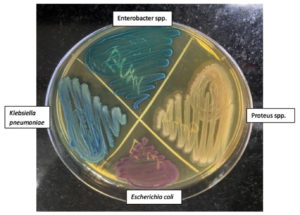
Figure 1. Bacterial growth on chromogenic agar shown with test sample colonies of different colours (Enterobacter spp., Proteus spp., Klebsiella pneumoniae and Escherichia coli)
All biological and hazardous materials were handled with all necessary precautions and stored in the laboratory till the study period. On completion of the study, they were segregated and disposed of in a proper way according to the hospital’s biomedical waste management protocol.
Detection of MRSA
The MRSA isolates were first identified by the Cefoxitin (30 µg) disc diffusion method. According to CLSI guidelines, a zone diameter of <22 mm was considered as an MRSA isolate.8
Detection of ESBL
This was done by phenotypic confirmation. The CLSI advocates use of cefotaxime (30 µg) or ceftazidime (30 µg) disks with or without clavulanate (10 µg) for phenotypic confirmation of the presence of ESBLs. A difference of >5 mm between the zone diameters of either of the cephalosporin disks and their respective cephalosporin/clavulanate disks is taken to be phenotypic confirmation of ESBL production.8
Detection of AmpC
The isolate was further tested for AmpC if zone size of cefoxitin <18 mm (Resistant) (Cefoxitin disc strength 30 µg). Phenotypic confirmation of AmpC by cefoxitin-cloxacillin double disk synergy test-A 0.5 McFarland’s suspension of test organism was inoculated on Muller-Hinton agar. Cefoxitin (30 µg) and cefoxitin-cloxacillin (30 µg/200 µg) disks were placed 30 mm apart. After 24 hours of incubation the zone diameter was measured. If there was increase in zone diameter between cefoxitin and cefoxitin-cloxacillin by >4 mm then the test was considered positive for AmpC production.8,9
Detection of CRE
For Enterobacterales zone size less than or equal to 19 for Imipenem and meropenem by disc diffusion is considered as resistant.8
For Pseudomonas
Zone size less than or equal to 15 for Imipenem and meropenem by disc diffusion is considered as resistant.8
For Acinetobacter
Zone size less than or equal to 18 for Imipenem and zone size less than or equal to 14 for meropenem by disc diffusion is considered as resistant.8
Statistical analysis
Quantitative data were expressed as the mean and standard deviation. Qualitative data were expressed as percentages. McNemar’s test was used to determine the association between related values, and the chi-square test was used for independent values. Statistical significance was set at p <0.05.
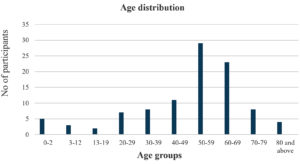
Of the study specimen, 53 percent samples were obtained from female participants and most specimen were obtained from participants aged between 50 to 59 years (29%) followed by 60 to 69 years (23%), 40 to 49 years (11%) (Figures 2 and 3).
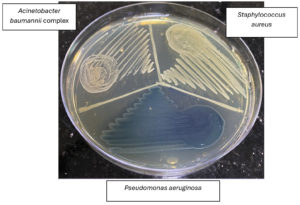
Figure 2. Bacterial growth on chromogenic agar shown with test sample colonies of different colours (S. aureus, Pseudomonas aeruginosa, Acinetobacter baumannii complex)
Table 1 depicts the distribution of sample type, antibiotics administered and gram stain reactions, the results showed that most samples obtained were pus (66%), followed by wound samples (21%), other exudates (6%), vaginal swabs (4%), tissue swabs (2%), and ear swabs (1%). Regarding the administration of antibiotics majority of the participants did not receive any antibiotics (71%) and when received, seven percent receive meropenem followed by amoxicillin-clavulanic acid five percent, ciprofloxacin 3 percent, clindamycin three percent, piperacillin-tazobactam three percent, and vancomycin (2%), while the remaining nine percent were treated with other antibiotics. With regard to Gram stain reaction, Gram-negative rods were the most prevalent documented, accounting for 53% of all samples followed by Gram-positive cocci in pairs (48%), Gram-positive rods (nine percent), Gram-positive cocci (seven percent), Gram-positive cocci in chains (six percent), Gram-positive cocci in clusters (three percent), Gram-negative coccobacilli (two percent), and yeast-like organisms (two percent). Notably, 22% samples exhibited no reaction to the gram staining.
Table (1):
Distribution of samples based on type, antibiotics administered and reaction to gram staining
| Clinical Presentation | Count (%) | |
|---|---|---|
| Sample Type | Pus | 66 (66%) |
| Wound | 21 (21%) | |
| Others (exudates) | 6 (6%) | |
| Vaginal Swab | 4 (4%) | |
| Tissue Sample | 2 (2%) | |
| Ear Swab | 1 (1%) | |
| Antibiotics Given | None | 71 (71%) |
| Meropenem | 7 (7%) | |
| Amoxicillin – Clavulanic acid | 5 (5%) | |
| Ciprofloxacin | 3 (3%) | |
| Clindamycin | 3 (3%) | |
| Piperacillin + Tazobactam | 3 (3%) | |
| Vancomycin | 2 (2%) | |
| Others | 9 (9%) | |
| Gram’s Reaction | Gram Negative coccobacilli | 2 (2%) |
| Gram-negative rods | 53 (53%) | |
| Gram-positive cocci in chain | 6 (6%) | |
| Gram-positive cocci in clusters | 3 (3%) | |
| Gram-positive cocci in Pair | 48 (48%) | |
| Gram-positive cocci Pair & chain | 1 (1%) | |
| Gram-positive cocci Pair & cluster | 1 (1%) | |
| Gram-positive cocci Seen | 7 (7%) | |
| Gram-positive rods | 9 (9%) | |
| Yeast like organism | 2 (2%) | |
| No Bacteria Seen | 22 (22%) | |
Table 2 depicts the evaluation of culture media by comparing the results among conventional and HiCrome methods. The result revealed that HiCrome agar effectively isolated common pathogens such as E. coli and K. pneumoniae with 100 percent matching rates with that of conventional media, followed by S. aureus and P. aeruginosa demonstrating matching rates of 92.31% and 85.71% respectively; however, limitations were observed for Proteus vulgaris, Pseudomonas putida, Citrobacter koseri,
A. baumannii complex and Candida albicans, which showed 0% matching rates.
Table (2):
Evaluation of culture media by conventional and HiCrome
Gold Standard-Organism Isolated |
Total |
Matched (%) |
|---|---|---|
Nil/No Growth |
35 |
33 (94.29%) |
Escherichia coli |
21 |
21 (100%) |
Klebsiella pneumoniae |
20 |
20 (100%) |
Staphylococcus aureus |
13 |
12 (92.31%) |
Pseudomonas aeruginosa |
7 |
6 (85.71%) |
Enterococcus faecalis |
5 |
4 (80%) |
Proteus mirabilis |
4 |
4 (100%) |
Proteus vulgaris |
2 |
0 (0%) |
Citrobacter koseri |
2 |
0 (0%) |
Acinetobacter baumannii complex |
2 |
0 (0%) |
Enterococcus faecium |
1 |
1 (100%) |
Pseudomonas putida |
1 |
0 (0%) |
Candida albicans |
1 |
0 (0%) |
Morganella morganii |
1 |
0 (0%) |
Table 3 displays the distribution of the subjects based on bacterial isolates with most specimens reporting no bacterial counts (34%). The commonly detected bacterial isolate was E. coli (21%), followed by K. pneumoniae (20%), S. aureus (13%), and P. aeruginosa (7%). Other isolates observed included E. faecalis (5%), P. mirabilis (4%), and Acinetobacter baumannii complex (2%).
Table (3):
Distribution of samples based on the bacterial isolates obtained
Bacterial Isolates |
Count (%) |
|---|---|
Nil |
34% |
Escherichia coli |
21% |
Klebsiella pneumoniae |
20% |
Staphylococcus aureus |
13% |
Pseudomonas aeruginosa |
7% |
Enterococcus faecalis |
5% |
Proteus mirabilis |
4% |
Acinetobacter baumannii complex |
2% |
Citrobacter koseri |
2% |
Proteus vulgaris |
2% |
Others |
4% |
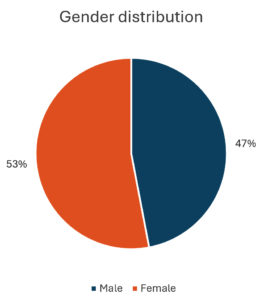
The pie chart illustrates the proportion of male and female specimens. It shows that 47% of the specimens are male, represented in blue, while 53% are female, shown in orange. This visual highlight a slightly higher representation of females in the dataset (Figure 4).
Table 4 shows the antimicrobial sensitivity patterns among the different Gram-negative isolates identified. Among the 21 E. coli isolates, 90.48% were susceptible to netilmicin, 85.71% were susceptible to tobramycin, imipenem, and meropenem respectively followed by amikacin (76.19%), while 71.43% were susceptible to cefepime, gentamicin and tigecycline. Out of the 20 K. pneumoniae isolates, 90% were susceptible to gentamicin and tigecycline followed by netilmicin (80%), tobramycin and amikacin (75% each), imipenem and meropenem (70%). Out of the P. aeruginosa isolates, 86.71% were susceptible to tobramycin followed by imipenem, meropenem, cefepime, and ciprofloxacin (71.43% each).
Table (4):
Antimicrobial susceptibility among the Gram-negative isolates
Antibiotics |
Escherichia coli (n = 21) |
Klebsiella pneumoniae (n = 20) |
Pseudomonas aeruginosa (n = 7) |
Proteus mirabilis (n = 4) |
Acinetobacter baumannii complex (n = 2) |
Citrobacter koseri (n = 2) |
Proteus vulgaris (n = 2) |
Morganellam organii (n = 1) |
Pseudomonas putida (n = 1) |
|---|---|---|---|---|---|---|---|---|---|
Tobramycin |
18 (85.71%) |
15 (75%) |
6 (85.71%) |
1 (25%) |
1 (50%) |
2 (100%) |
1 (50%) |
1 (100%) |
1 (100%) |
Imipenem |
18 (85.71%) |
14 (70%) |
5 (71.43%) |
3 (75%) |
– |
– |
1 (50%) |
1 (100%) |
– |
Meropenem |
18 (85.71%) |
14 (70%) |
5 (71.43%) |
3 (75%) |
– |
2 (100%) |
1 (50%) |
1 (100%) |
1 (100%) |
Amikacin |
16 (76.19%) |
15 (75%) |
4 (57.14%) |
1 (25%) |
– |
2 (100%) |
1 (50%) |
1 (100%) |
– |
Cefepime |
15 (71.43%) |
13 (65%) |
5 (71.43%) |
4 (100%) |
– |
2 (100%) |
2 (100%) |
1 (100%) |
– |
Gentamicin |
15 (71.43%) |
18 (90%) |
3 (42.86%) |
1 (25%) |
1 (50%) |
2 (100%) |
1 (50%) |
1 (100%) |
1 (100%) |
Tigecycline |
15 (71.43%) |
18 (90%) |
– |
1 (25%) |
1 (50%) |
– |
– |
– |
– |
Ertapenem |
14 (66.67%) |
12 (60%) |
– |
3 (75%) |
– |
– |
1 (50%) |
1 (100%) |
– |
Cefoperazone+ Sulbactam |
13 (61.9%) |
9 (45%) |
– |
2 (50%) |
– |
1 (50%) |
1 (50%) |
– |
– |
Netilmicin |
19 (90.48%) |
16 (80%) |
4 (57.14%) |
1 (25%) |
1 (50%) |
2 (100%) |
1 (50%) |
1 (100%) |
1 (100%) |
Piperacillin+ Tazobactam |
11 (52.38%) |
9 (45%) |
1 (14.29%) |
2 (50%) |
– |
1 (50%) |
– |
– |
– |
Doxycycline |
10 (47.62%) |
12 (60%) |
1 (14.29%) |
– |
2 (100%) |
1 (50%) |
– |
– |
– |
Co-Trimoxazole |
10 (47.62%) |
7 (35%) |
1 (14.29%) |
2 (50%) |
1 (50%) |
– |
1 (50%) |
1 (100%) |
1 (100%) |
Ceftriaxone |
9 (42.86%) |
6 (30%) |
– |
1 (25%) |
– |
1 (50%) |
– |
1 (100%) |
– |
Ciprofloxacin |
9 (42.86%) |
10 (50%) |
5 (71.43%) |
1 (25%) |
– |
2 (100%) |
– |
– |
– |
Cefotaxime |
8 (38.1%) |
7 (35%) |
– |
1 (25%) |
– |
1 (50%) |
– |
1 (100%) |
– |
Ceftazidime |
7 (33.33%) |
8 (40%) |
1 (14.29%) |
– |
– |
1 (50%) |
– |
1 (100%) |
1 (100%) |
Cefuroxime |
7 (33.33%) |
6 (30%) |
– |
1 (25%) |
– |
1 (50%) |
– |
1 (100%) |
– |
Ampicillin |
– |
1 (5%) |
– |
– |
– |
– |
– |
– |
– |
Cefazolin |
– |
– |
– |
– |
– |
– |
– |
– |
– |
Table 5 shows the results regarding the type of resistance encountered among the bacterial isolates and the results indicated that out of the 81 isolated organisms, S. aureus was present in 14 samples, with 4 isolates (4.93%) tested positive for Methicillin-resistant S. aureus (MRSA). Resistance to Extended Spectrum Beta-Lactamase (ESBL) was detected in 18 isolates (22.22%), including 10 isolates of E. coli, six isolates of K. pneumoniae, and two isolates of P. mirabilis. AmpC resistance was identified in 10 isolates (12.34%), comprising three isolates of E. coli, five isolates of K. pneumoniae, one isolate of P. mirabilis, and one isolate of Morganella morganii. P. aeruginosa was the sole bacterium demonstrating inducible AmpC resistance, with five isolates (6.17%) displaying AmpC resistance. Carbapenem resistance was detected in eight isolates (9.87%), comprising three isolates of A.baumannii, four isolates of K. pneumoniae, and one isolate of P. mirabilis. Additionally, five isolates exhibited resistance to various drug categories: one isolates each from E. coli and K. pneumoniae displayed AmpC + ESBL resistance; two K. pneumoniae isolates showed AmpC + Carbapenem resistance; and one P. mirabilis isolate revealed ESBL + AmpC + Carbapenem resistance. K. pneumoniae was the predominant bacterium exhibiting multidrug-resistance (n = 18), followed by E. coli, which had 14 samples.
Table (5):
Antimicrobial susceptibility among the Gram-positive isolates
Staphylococcus aureus (n = 13) |
Enterococcus faecalis (n = 5) |
Enterococcus faecium (n = 1) |
|
|---|---|---|---|
Ampicillin |
– |
5 (100%) |
1 (100%) |
Gentamicin |
12 (92.31%) |
5 (100%) |
1 (100%) |
Doxycycline |
10 (76.92%) |
– |
– |
Co-Trimoxazole |
12 (92.31%) |
– |
– |
Ciprofloxacin |
12 (92.31%) |
– |
– |
Tetracycline |
– |
4 (80%) |
1 (100%) |
Linezolid |
13 (100%) |
4 (80%) |
1 (100%) |
Clindamycin |
9 (69.23%) |
– |
– |
Vancomycin |
9 (69.23%) |
4 (80%) |
1 (100%) |
Erythromycin |
5 (38.46%) |
3 (60%) |
– |
Nil |
1 (7.69%) |
Table 6 shows the antimicrobial sensitivity patterns among the different Gram-positive isolates identified. Among the 13 S. aureus isolates, 100% were susceptible to linezolid followed by gentamicin, ciprofloxacin and cotrimoxazole (92.31% each). Out of the five E. faecalis isolates, 100% were susceptible to gentamicin as well as ampicillin followed by tetracycline, linezolid, and vancomycin (80% each). Out of seven Enterococcus faecium isolates, 100% were susceptible to ampicillin, gentamicin, tetracycline, linezolid, and vancomycin.
Table (6):
Type of resistance observed among the bacterial isolates
| Organism | Total samples | Type of resistance | Positive samples | Percent |
|---|---|---|---|---|
| Staphylococcus aureus | 100 | ESBL | 4 | 4.93% |
| Escherichia coli | 81 | ESBL | 10 | 12.34% |
| Klebsiella pneumoniae | 81 | ESBL | 6 | 7.41% |
| Proteus mirabilis | 81 | ESBL | 2 | 2.47% |
| Escherichia coli | 81 | AmpC Resistance | 3 | 3.70% |
| Klebsiella pneumoniae | 81 | AmpC Resistance | 5 | 6.17% |
| Proteus mirabilis | 81 | AmpC Resistance | 1 | 1.23% |
| Morganella morganii | 81 | AmpC Resistance | 1 | 1.23% |
| Pseudomonas aeruginosa | 81 | Inducible AmpC Resistance | 5 | 6.17% |
| Acinetobacter baumannii | 81 | Carbapenem Resistance | 3 | 3.70% |
| Klebsiella pneumoniae | 81 | Carbapenem Resistance | 4 | 4.93% |
| Proteus mirabilis | 81 | Carbapenem Resistance | 1 | 1.23% |
| Multidrug-resistant Organisms | ||||
| Escherichia coli | AmpC + ESBL Resistance | 1 | ||
| Klebsiella pneumoniae | ESBL + AmpC Resistance | 1 | ||
| Klebsiella pneumoniae | AmpC + Carbapenem Resistance | 2 | ||
| Proteus mirabilis | ESBL + AmpC + Carbapenem Resistance | 1 | ||
From Table 7, it was observed that by conventional method, TAT for all bacterial identification was 48 h, whereas, using chrome agar, 57.55% isolates were reported within 24 h.
Table (7):
Turnaround time bacterial identification between Conventional and HiCrome
TAT |
Conventional |
Chrome |
|---|---|---|
24 Hours |
0 (0%) |
46 (57.5%) |
48 Hours |
80 (100%) |
34 (42.5%) |
The increasing prevalence of antimicrobial resistance in pathogens, especially among the hospitalized patients, requires swift and precise identification of infectious agents. The necessity for prompt intervention is emphasized, as suitable antibiotic therapy can markedly enhance patient outcomes.10 The establishment of new microbiological and the use of novel microbial identification media are essential for timely disease diagnosis and monitoring of antimicrobial resistance. This study investigates the effectiveness of Chrom agar for identifying the bacteria associated with pyogenic infections, emphasizing bacteriological profiles, antibiotic susceptibility patterns, and comparing turnaround times with traditional culture methods.
The specimen collected in the present study were mostly from the female participants (53 percent), with the mean age of all the participants being 49.55 ± 20.72 years. The higher proportion of specimen obtained from females suggested a greater prevalence of pyogenic infections, such as those affecting skin and tissues. For instance, studies had shown that S. aureus is a common isolate in wound infections, and its prevalence could vary based on gender demographics.11,12 Understanding this distribution is crucial for tailoring appropriate treatment protocols and antibiotic stewardship programs.
Regarding the clinical presentation of pyogenic infections among the participants, the majority of samples (66%) were pus, highlighting the actual nature of these infections, followed by wound samples (21%). The results were in contrast with a study conducted by Kalita et al., where the majority samples were wound swabs (75.53%), followed by pus aspirate (24.47%), which highlights the variability in sample types with pyogenic infections across different groups and study settings.13
Available evidence had shown that prior antibiotic exposure could significantly influence the outcomes of microbial cultures. Studies had shown that administering antibiotics before collecting samples could lead to negative results in culture. For instance, researches had indicated that samples from the patients who received antibiotics prior to sample collection often exhibited lower isolation rate due to suppression of bacterial growth or selection for resistant strains.14,15 In the present study, the lack of prior antibiotic treatment among 71 percent of the participants enhanced the likelihood of obtaining reliable culture results, as these samples were less likely to be affected by antibiotic-induced alterations in microbial flora.
Regarding the Gram’s reaction, Gram-negative rods were predominant 53 percent, highlighting their role in these infections, while Gram-positive cocci in pairs were also present (48 percent). Results of a study conducted by Kalita et al. reported that Gram-negative organisms were predominantly isolated followed by Gram-positive cocci, which was in line with the findings of the present study.11 Many studies had reported the prevalence of Gram-negative organisms in causing pyogenic wound and skin-and-soft-tissue infections, encompassing abscesses, cellulitis, furunculosis, folliculitis, and surgical-site or other purulent wound infections.13,16-18 The results suggested that Gram-negative organisms play a major role in the aetiology of pyogenic infections, followed by Gram-positive cocci. Notably, the observation of 22 samples showing no bacterial growth suggested the need to identify potential non-bacterial aetiologies and necessitating the use of advanced identification techniques.
The finding that 22 of the sample cultures showed no bacterial growth indicates the possibility of non-infectious causes or the existence of fastidious organisms that standard culture methods may not have identified. This observation was significant as it underscores the limitations of HiChrome agar and conventional culture techniques, which might not identify certain pathogens, especially those necessitating specialized growth conditions. The results of the present study regarding the bacterial organisms isolated aligned with the reports of Trojan et al. which identified E. coli (51.2%) as the most prevalent organisms from pus samples, followed by S. aureus (21%), K. pneumoniae (11.6%), P. aeruginosa (5.8%), Citrobacter spp. (3.5%), A. baumannii (2.3%), P. mirabilis (2.3%), and Streptococcus spp. (2.3%).19
This emphasizes the diagnostic value of chromogenic agar in identifying bacteria from purulent infections. The present study examined purulent infections other than UTIs; however, the recurrent isolation of E. coli across various infection types indicated a wider pathogenic capacity. S. aureus was detected in 13% of participants, highlighting the persistent involvement of Gram-positive cocci in pyogenic infections. S. aureus is a significant pathogen associated with skin and soft tissue infections, such as abscesses and cellulitis. The isolation rate was in accordance with findings corresponds with findings of Sudhaharan et al., which identified S. aureus as the predominant Gram-positive isolate in comparable contexts, accounting for 91.7% of Gram-positive isolates in purulent samples.16 The consistency observed in various studies underscores the significance of S. aureus in pyogenic infections and its implications for clinical practice. The result of the present study aligned with those of Poolman et al., who identified E. coli and S. aureus as key pathogens linked to bacteraemia and nosocomial infections, especially in older populations.20 The repeated identification of these organisms in multiple studies indicated that E. coli and S. aureus are the most significant causative organisms for infections, underscoring their importance as primary targets for antibiotic therapy.
In antibiogram, variations in antibiotic susceptibility could be observed among different isolates, underscoring the necessity for tailored antibiotic therapy. E. coli and K. pneumoniae exhibited significant sensitivity to netilmicin, tobramycin, imipenem, amikacin, and meropenem, suggesting the efficacy of carbapenems against these Gram-negative bacteria, consistent with the findings of Swain et al., P. aeruginosa demonstrated 85.71% sensitivity to tobramycin and the susceptibility to carbapenems and ciprofloxacin.21 However, it exhibited reduced susceptibility to gentamicin (42.86%) and piperacillin-tazobactam (14.29%), highlighting the necessity for judicious antibiotic selection. K. pneumoniae exhibited sensitivity to gentamicin; however, other studies had documented varying susceptibility patterns among Gram-negative isolates.21 All tested strains of S. aureus were susceptible to linezolid, aligning with the findings of Swain et al., indicating that linezolid is an effective treatment option for S. aureus infections.21 E. faecalis and E. faecium demonstrated susceptibility to gentamicin, ampicillin, and vancomycin; however, the limited number of isolates might constrain the generalisation of these findings. Continuous monitoring of antibiotic resistance patterns is crucial for effective infection control.
The results of the current study underscore a troubling trend in antimicrobial resistance (AMR), especially in pathogens like S. aureus and K. pneumoniae. As per the study, K. pneumoniae was a significant pathogen associated with pyogenic infections, consistent with the earlier research which showed a rising prevalence of multidrug-resistant strains in both community and hospital environments.22 The rising incidence of MRSA, associated with a notable increase in global mortality, highlights the critical need to tackle AMR. The existence of (ESBL) possessing pathogens, especially E. coli, often hampers the treatment options due to their resistance to various antibiotic classes.23 In addition, the rise in multidrug-resistant pathogens highlights a global challenge of antimicrobial resistance, fuelled by inadequate stewardship and the widespread misuse of antibiotics in both clinical and agricultural practices.23 A recent analysis from The Lancet indicated that antibiotic-resistant illnesses might result in almost 8 million fatalities each year by 2050 if prevailing trends persist, underscoring the urgent necessity for enhanced healthcare systems and antibiotic stewardship initiatives.24
A matching rate of 100 percent was observed with HiCrome agar regarding the isolation of E. coli and K. pneumoniae in comparison with conventional culture media, which highlighted the effectiveness of HiCrome agar in isolating these organisms in clinical settings. Similarly, 100 percent matching rate was observed with respect to the isolation of P. mirabilis and E. faecium, suggesting HiCrome agar as a reliable medium for the isolation of these pathogens as well.
In contrast, S. aureus showed a matching rate of 92.31%, while the detection rate for P. aeruginosa was 85.71%, and 80% for E. faecalis. Significant drawbacks were noted for P. vulgaris, P. putida, and C. albicans, which exhibited 0% matching rates. The results indicated that even though the medium is effective in general, it might not consistently detect all microorganisms because of the varied utilisation of media components.25
Previous studies had reported the use of chromogenic agar for detecting uropathogens, MRSA, and Shiga-toxin-producing E. coli, among others.17,26-33 The results of a study by Jangla et al. were similar to that of the present study with the colony characteristics of E. coli, K. pneumoniae, P. aeruginosa, S. aureus in HiCrome being similar to identification by Vitek 2 Compact.34 A study by Nahar et al., reported that both HiCrome UTI agar and Blood agar media supported the growth of all the bacteria under study, while MacConkey agar yielded an isolation rate of 91.72%, which showed the efficacy of HiCrome agar in identifying the uropathogens.35 The present study represented the first report of using chromogenic agar specifically for detecting the microorganisms responsible for pyogenic infections, further validating its application in diverse clinical contexts.
The comparison of (TAT) for bacterial identification between conventional culture methods and chromogenic agar revealed significant differences in efficiency and effectiveness. In the study, the conventional method required 48 h for bacterial identification, while Chrom agar demonstrated a remarkable capability, with 57.55% of isolates reported within 24 h. This difference was statistically significant (p <0.0001), indicating that Chrom agar was effective for rapid identification of pathogens. However, while Chrom agar showed a 42.5% success rate at 48 h, the conventional method achieved a 100% success rate at the same time point. This difference could be explained by the fact that the reference culture on chrome agar had shown 100 percent match, however, identification of certain organisms required further biochemical tests, which caused a delay in TAT. The results suggested that Chrom agar was particularly useful for immediate outcomes, making it advantageous in clinical settings, where rapid diagnosis is critical. In addition, the findings also aligned with recent studies that emphasised the utility of chromogenic media in reducing TAT for bacterial identification. For instance, research had shown that Chrom agar could facilitate quicker identification of various pathogens, including E. coli and S. aureus, compared to traditional methods.36-38 However, the conventional method remains reliable for comprehensive identification over longer periods, as it consistently yields accurate results across all tested organisms.
The effectiveness of Chrom agar in short-term identification is crucial in acute care settings, where timely intervention can significantly impact patient outcomes. Nevertheless, the conventional method’s reliability over extended periods underscores the importance of evaluating both short-term and long-term effectiveness while selecting diagnostic methods. This dual approach ensures that clinicians can make informed decisions based on immediate needs while maintaining a high standard of accuracy in pathogen identification.
The present study recognized considerable limitations, notably its limited sample size, which constrained the generalisation of the results. The data from a restricted sample size might not reliably reflect the broader population dynamics, particularly concerning various diseases and resistance patterns in different situations. Although chromogenic agar facilitated expedited bacterial identification within 24 h it failed to yield exhaustive results for all isolates. Conversely, traditional approaches attained a 100% identification rate within 48 h indicating that chromogenic agar might not reliably expedite identification for rare or fastidious species, thus jeopardizing diagnostic accuracy when timely identification is essential.
This was the first investigation regarding the efficacy of chromogenic media in isolating and identifying pyogenic organisms, juxtaposing its findings with traditional approaches to elucidate their different advantages and disadvantages. By enabling visual, colour-based presumptive identification directly from the primary plate, HiCrome reduced the need for secondary media and cut the average reporting time from 48 h to approximately 24 h, translating into a measurable decrease in technician workload and overall laboratory costs. The clinical implications are substantial: swift identification facilitates expedited therapy decisions in acute care environments. Clinicians must acknowledge the constraints of chromogenic agar with certain organisms, while appreciating the dependability of traditional approaches. Combining both methodologies enhances diagnosis, hence improving patient care through prompt interventions and increased precision.
The present study demonstrated the efficacy of chromogenic agar for the swift identification of bacteria, particularly pyogenic species, while acknowledging its limitations in reliably detecting all pathogens, especially rare and fastidious ones. The results indicated that although chromogenic media could expedite therapeutic decisions, traditional approaches are crucial for thorough identification. An integrated strategy employing both approaches might enhance diagnostic precision and facilitate faster patient diagnosis in clinical environments, facilitating prompt treatment and better patient outcomes.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MMR conceptualized the study and applied methodology. MMR collected resources. MMR, LS and SS performed project administration, supervised the study, performed software work and performed formal analysis and data validation. GM and MMR performed visualization, investigation, data curation and wrote the original draft. MMR wrote, reviwed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by the Indian Council of Medical Research (ICMR), vide grant number ICMR-STS ID 2023-11849.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Human Ethical Committee, PSG Institute of Medical Sciences and Research, Peelamedu, Coimbatore, Tamil Nadu, India (Proposal No. 23/305; Ref No. PSG/IHEC/2023/Appr/Exp/430).
- Peri AM, Stewart A, Hume A, Irwin A, Harris PNA. New microbiological techniques for the diagnosis of bacterial infections and sepsis in ICU including point of care. Curr Infect Dis Rep. 2021;23(8):12.
Crossref - Umemura Y, Ogura H, Takuma K, et al. Current spectrum of causative pathogens in sepsis: A prospective nationwide cohort study in Japan. Int J Infect Dis. 2021;103:343-351.
Crossref - Lahiry S, Ramalingam R, Dalal K, et al. Tackling AMR crisis in India: Changing paradigm. Asian Journal of Medical Sciences. 2020;11(6):129-37.
- Ram B, Thakur R. Epidemiology and Economic Burden of Continuing Challenge of Infectious Diseases in India: Analysis of Socio-Demographic Differentials. Front Public Health. 2022;10:901276.
Crossref - Samra Z, Heifetz M, Talmor J, Bain E, Bahar J. Evaluation of use of a new chromogenic agar in detection of urinary tract pathogens. J Clin Microbiol. 1998;36(4):990-994.
Crossref - Truong TV, Twist A, Zaytsev A, et al. Evaluation of a novel chromogenic medium for the detection of Pseudomonas aeruginosa in respiratory samples from patients with cystic fibrosis. Microorganisms. 2022;10(5):1004.
Crossref - Sekyere JO, Sephofane AK, Mbelle NM. Comparative Evaluation of CHROMagar COL-APSE, MicroScan Walkaway, ComASP Colistin, and Colistin MAC Test in Detecting Colistin-resistant Gram-Negative Bacteria. Sci Rep. 2020;10(1):6221.
Crossref - Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 35th ed. CLSI supplement M100. Wayne, PA: CLSI; 2025. Available from: https://clsi.org/shop/standards/m100/
- Khari FIM, Karunakaran R, Rosli R, Tay ST. Genotypic and phenotypic detection of AmpC β-lactamases in Enterobacter spp. Isolated from a teaching hospital in Malaysia. PLoS One. 2016;113:e0150643.
Crossref - Bayot ML, Bragg BN. Antimicrobial Susceptibility Testing. In: StatPearls. StatPearls Publishing. 2024.
- Wadekar MD, Sathish JV, Jayashree, Pooja C. Bacteriological profile of pus samples and their antibiotic susceptibility pattern. Indian J Microbiol Res. 2020;7(1):43-47.
Crossref - Deboral A, Bhosale NK, Umadevi S. Aerobic Bacteriological and Antibiotic Susceptibility Profile of Pus Isolates from A Tertiary Care Hospital, Puducherry. J Pure Appl Microbiol. 2020;14(3):1961-1966.
Crossref - Kalita JM, Nag VL, Kombade S, Yedale K. Multidrug resistant superbugs in pyogenic infections: a study from Western Rajasthan, India. Pan Afr Med J. 2021;38:409.
Crossref - Gramberg MCTT, Van Hattem JM, Dijkstra JA, et al. Effect of prior antibiotic use on culture results in people with diabetes and foot osteomyelitis. Antibiotics (Basel). 2023;12(4):684.
Crossref - Li L, Xu L, Zhu R, Song J, Wang X. Effect of prior receipt of antibiotics on the pathogen distribution: a retrospective observational cohort study on 27,792 patients. BMC Infect Dis. 2020;20(1):8.
Crossref - Sudhaharan S, Kanne P, Chavali P, Vemu L. Aerobic bacteriological profile and antimicrobial susceptibility pattern of pus isolates from tertiary care hospital in India. J Infect Dev Ctries. 2018;12(10):842-848.
Crossref - Mueller M, Tainter CR. Escherichia coli infection. [Updated 2023 Jul 13]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. 2024.
- Wani FA, Bandy A, Alenzi MJS, et al. Resistance patterns of gram-negative bacteria recovered from clinical specimens of intensive care patients. Microorganisms. 2021;9(11):2246.
Crossref - Trojan R, Razdan L, Singh N. Antibiotic Susceptibility Patterns of Bacterial Isolates from Pus Samples in a Tertiary Care Hospital of Punjab, India. Int J Microbiol. 2016;2016:9302692.
Crossref - Poolman JT, Anderson AS. Escherichia coli and Staphylococcus aureus: leading bacterial pathogens of healthcare associated infections and bacteremia in older-age populations. Expert Rev Vaccines. 2018;17(7):607-618.
Crossref - Swain B, Samal D, Agrawal SN. Microorganisms causing wound infection and their antibiotic susceptibility pattern. Int J Res Med Sci. 2022;10(11):2617.
Crossref - Bajare B, Dhangar A, Tankhiwale S, Shrikhande S. Bacteriological profile of pyogenic infections and their antimicrobial susceptibility in a tertiary care hospital in central India. Med Lab J. 2024;18(2):16-18.
Crossref - Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10(S12):S122-9.
Crossref - GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990-2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404(10459):1199-1226.
Crossref - Carricajo A, Boiste S, Thore J, Aubert G, Gille Y, Freydière AM. Comparative evaluation of five chromogenic media for detection, enumeration and identification of urinary tract pathogens. Eur J Clin Microbiol Infect Dis [Internet]. Available from: https://link.springer.com/content/pdf/10.1007/s100960050403
- Marathe A, Zhu Y, Chaturvedi V, Chaturvedi S. Utility of CHROMagar™ Candida Plus for presumptive identification of Candida auris from surveillance samples. Mycopathologia. 2022;187(5-6):527-534.
Crossref - Ariza-Miguel J, Oniciuc EA, Sanz I, Fernandez-Natal I, Hernandez M, Rodriguez-Lazaro D. Evaluation of two commercially available chromogenic media for confirmation of methicillin-resistant Staphylococcus aureus from human, animal, and food samples. Int J Food Microbiol. 2015;209:26-28.
Crossref - Bayona JVM, Garcia CS, Palop NT, Cardona CG. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagar™ Candida for the detection of Candida auris in surveillance samples. Diagn Microbiol Infect Dis. 2020;98(4):115168.
Crossref - Lewis GL, Cernicchiaro N, Moxley RA. Performance of Chromogenic Agar Media for Isolation of Shiga Toxin-Producing Escherichia coli from Ground Beef. J Food Prot. 2020;83(7):1149-54.
Crossref - Palavecino EL. Rapid Methods for Detection of MRSA in Clinical Specimens. In: Ji Y. (eds) Methicillin-Resistant Staphylococcus Aureus (MRSA) Protocols. Methods in Molecular Biology, vol 1085. Humana Press, Totowa, NJ 2014;1085:71-83.
Crossref - Nerurkar V, Khan S, Kattungal S, Bhatia S. Identifying Candida and other yeast-like fungi: utility of an identification algorithm in a resource-limited setting. J Clin Diagn Res. 2014;8(12):DC01-DC04.
Crossref - Bentz ML, Le N, Min B, et al. Evaluation of CHROMagar Candida Plus for the detection of C. auris with a panel of 206 fungal isolates and 83 colonization screening skin-swabs. Microbiol Spectr. 2024;12(4):e0356423.
Crossref - Khan T, Faysal NI, Hossain MM, et al. Emergence of the novel sixth Candida auris Clade VI in Bangladesh. Microbiol Spectr. 2024;12(7):e0354023.
Crossref - Jangla SM, Naidu R, Patel SC, Gami UK, Machhi BS. Bacteriological Profile of Wound Swab and Pus Samples Using Conventional Media and Chromogenic Medium. J Krishna Inst Med Sci Univ. 2020;9(3):56-64.
- Khutade K, Shah H, Patil S, Patel H. Validation of an assessment of chromogenic media against conventional culture techniques for isolation, identification, and direct antibiotic susceptibility testing of uropathogens in resource-poor settings. Int J Pharm Biol Sci. 2024;13:116-122.
Crossref - Singh AK, Bhunia AK. Optical scatter patterns facilitate rapid differentiation of Enterobacteriaceae on CHROMagar™ Orientation medium. Microb Biotechnol. 2016;9(1):127-135.
Crossref - Robberts FJL, Owusu-Ofori A, Oduro G, et al. Rapid, low-complexity, simultaneous bacterial group identification and antimicrobial susceptibility testing performed directly on positive blood culture bottles using chromogenic agar. Am J Trop Med Hyg. 2022;107(6):1302-1307.
Crossref - Murray MP, Zinchuk R, Larone DH. CHROMagar Candida as the sole primary medium for isolation of yeasts and as a source medium for the rapid-assimilation-of-trehalose test. J Clin Microbiol. 2005;43(3):1210-2.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.