ISSN: 0973-7510
E-ISSN: 2581-690X
Nipah virus (NiV) is an emerging zoonotic pathogen of global concern, causing serious life-threatening illness in humans. The first outbreak was reported from Malaysia in 1998. The virus is classified under the family Paramyxoviridae. Geographically, the deadly virus is known to be distributed in Southeast Asia. Its recent emergence in Kerala, India, indicates its public health emergency and necessitates the urgent proactive measures for the development of a safe and effective vaccine for the prevention and control of the disease. As a single-stranded RNA virus, there is no effective antiviral or vaccine available to fight this virus. Attempt on the development of vaccines had been hampered due to the highly infectious nature of the virus and the requirement of specialised bio-containment facility to handle the pathogen. Considering the potential advantages of reverse vaccinology approaches, the present study attempted to predict vaccine candidates targeting NiV virus genes encoding non-structural and structural proteins (specifically glycoprotein G, glycoprotein F, and W protein) circulating in Southeast Asia. The results of the analysis have suggested two potential vaccine candidates based on various parameters such as antigenicity, immunogenicity, non-toxicity, and non-allergenicity. In view of the global interest on urgent requirement of an effective vaccine, the present study predicted candidate vaccine antigens using bioinformatic tools and their promising usefulness as candidate peptides along with ongoing vaccine discovery efforts has been discussed.
Viral Zoonoses, Immunoinformatic, Structural and Non-structural Proteins, Emerging Infection, Reverse Vaccinology
The emergence and re-emergence of infectious diseases are very serious public health threats worldwide,1 and approximately 75% of these diseases are zoonotic and transmitted between humans and animals. Nipah virus is a zoonotic pathogen in humans that causes serious illnesses, such as encephalitis/respiratory diseases with fatal outcomes. It is a close relative of the Hendra virus and is classified under the family Paramyxoviridae, within the order: Mononegavirales and the subfamily Paramyxovirinae. It is transmitted to animals/humans by certain species of fruit bats (flying foxes), primarily Pteropus spp.2 and is now distributed in many of Southeast Asian countries, namely Cambodia, East Timor, Indonesia, Malaysia, Papua New Guinea, Vietnam, Thailand, Bangladesh, and India.3 In a recent episode of Nipah virus outbreak in India and Bangladesh, the virus was found to be transmitted between Pteropus bats and humans.3,4 Due to its highly infectious nature, ease of dissemination and increased rates of mortality and morbidity from the viral outbreak, NiV has been categorized as a BSL 4 pathogen.5
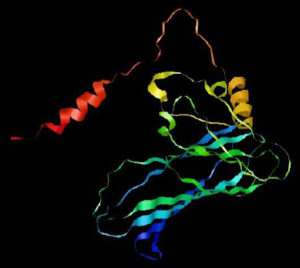
This virus has helical symmetry and is an RNA virus with an envelope, and a negative sense, non-segmented, single-stranded genome. The genetic material of the Nipah virus contains a succession of six genes, namely nucleocapsid protein (N), matrix protein (M), phosphoprotein (P), attachment glycoprotein (G), fusion glycoprotein (F), and long polymerase (L). The Nucleoprotein, Phosphoprotein, and long polymerase proteins attach to the viral nucleic acid forming the viral ribonucleoprotein (vRNP) (Figure 1). The attachment (G) and fusion (F) glycoproteins are the causes of virion adhesion to the susceptible host and subsequent entry into the cell.6 Gene P encodes four proteins, a phosphoprotein (P), and three non-structural proteins, namely, protein V, protein W, and protein C7 (Table 1).
Table (1):
Proteins of NiV and their functions
| Type of Protein | Name of Protein | Function |
|---|---|---|
| Structural proteins | G Glycoprotein | Promotes virion entry by clathrin mediated endocytosis |
| F Glycoprotein | Merge the viral and host cell membrane | |
| P Phosphoprotein | Helps for replication and transcription of virus | |
| N Nucleoprotein | Protects viral genome from nucleases | |
| M Matrix protein | Facilitates RNA progression | |
| L Protein | Catalyzes transcription of viral mRNA | |
| Non-structural proteins | C Protein | Counteracts with cellular IFN |
| V Protein | Inhibits host immune response | |
| W Protein | Blocks IFN alpha & beta |
Glycoproteins on the viral surface, such as the G and F proteins present on the NiV outer membrane, are regarded as prime immune system targets due to their ability to interact with host cell receptors.8,9 In earlier studies, Rebecca et al. reported that the Paramyxoviridae virus family possesses two glycoproteins anchored in the membrane, that serve as targets for neutralizing antibodies.10 Pratik et al. reported that because of the synchronized actions of fusion and attachment glycoproteins, it is quite natural for the Nipah virus to infiltrate the target cell following the binding process.11 According to Brent et al., the soluble recombinant G glycoprotein subunit has the potential to offer protection not only against identified strains of Henipaviruses such as NiV M and NiV B but also against HeV in four animal species. It was further confirmed that the NiV G glycoprotein is a strong immunogen for the construction of pre-F/G (HN/H) Chimera.10,11 As stated by Yoneda et al., the W-protein plays a vital role in inhibiting the synthesis of a/b interferon (IFN-a/b), thus eluding the innate immune response.12 The structural proteins F and G as well as the non-structural protein W are included in the scope of previous investigations aimed at identifying the immunodominant protein of NiV and these findings have been confirmed.13 Hence, we focused on the NiV structural and non-structural proteins F, G and W from the isolates from India and Malaysia, which have antigenic determinants and can confer an effective immune system defense against infection.14
According to the Centers for Disease Control and Prevention (CDC), the current treatment options are limited to supportive care, and approved vaccines or drugs for human use are still unavailable (http://www.cdc.gov/vhf/ nipah/prevention/index.html). More conclusive evidence on the effectiveness of antiviral drugs for treating NiV infections in humans is needed due to the limited number of ex vivo and in vivo studies conducted thus far. Animal model studies have evaluated a number of antiviral drugs for the control of NiV infection such as ribavirin, remdesivir and favipiravir15 (Table 2).
Table (2):
Status of currently available antiviral drugs for NiV infection
| Antiviral drug | Species | Remarks | Ref. |
|---|---|---|---|
| Ribavirin | Human (Malaysia outbreak) | 36% decrease in death rate | 15 |
| Syrian hamster | Not any advantageous impact | 15 | |
| Chloroquine | Syrian hamster | Not any advantageous impact | 15 |
| Ferret | 15 | ||
| Neutralizing antibodies | Syrian hamster | Pretreatment resulted in 100% survival; post treatment provided only partial protection | 16 |
| m102.4 antibody | Ferret | 100% existence was observed once treatment was administered one day post inoculation | 17 |
| Poly(I)-poly(C12U) | Syrian hamster | An 80% existence was observed once treatment was administered two hours post inoculation and continued for 9 more days | 18 |
| VIKI-PEG 4-chol | Syrian hamster | A 40% existence was observed when treatment was administered 2 days after inoculation | 19 |
| Remdesivir | African green monkey | Prevents viremia not viral replication | 20 |
| Acyclovir | Human (Singapore outbreak) | Acting as an analogue to deoxyguanosine triphosphate | 21 |
| Favipiravir | Hamster | Highest antiviral activity | 22 |
| Rintatolimid (Ampligen) | Hamster | Blocks viral replication | 23 |
NiV has been listed by the WHO as one of the top 10 emerging viruses for which urgent research and development are needed (World Health Organization. WHO R&D Nipah Baseline Situation Analysis; WHO: Geneva, Switzerland, 2018) (https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON490). Traditional vaccine development is quite difficult due to the extremely lethal nature of NiV, and the virus needs to be cultured in vitro. Several investigations are in progress for the development of vaccines against NiV infection, and majority of these investigations are based on a single protein /epitope of the protein (Table 3).
Table (3):
Currently available NiV vaccines (Under development)
| Vaccine | Species | Remarks | Ref. |
|---|---|---|---|
| Vaccinia virus vector | Syrian hamster | One vaccination provided protection against the fatal challenge | 24 |
| Canarypox virus vector | Pig | Two vaccinations provided clinical disease protection | 25 |
| VSV G | Ferret | One vaccination provided protection against heterologous fatal challenge | 26 |
| Syrian hamster | One vaccination provided protection against fatal challenge | 27 | |
| African green monkey | Induced stable and robust humoral responses with moderate cellular response | 28 | |
| Adeno associated virus vector | Syrian hamster | One vaccination provided protection against fatal challenge | 29 |
| Measles virus | African green monkey | Two vaccinations provided protection against fatal challenge | 30 |
| After two vaccinations, there is protection from clinical disease | |||
| sG NiV | Cat | After three vaccinations, protection against clinical disease is achieved | 31 |
| All vaccinated animals were protected from disease | 32 | ||
| sG HeV | Ferret | Two vaccinations provided protection against fatal challenge for up to 1 year | 33 |
| African green monkey | Vaccination provided protection against fatal challenge | 34 |
To overcome the limitations of traditional vaccine design and development, modern computational approaches have emerged, including, reverse vaccinology, systems biology, next-generation sequencing technology, and the design of epitope-based vaccines.35,36 Singh et al. reported that Immunoinformatics, also known as computational immunology, combines computational capabilities with the vast amount of genetic and proteomic data collected from pathogens. This integration aims to comprehend their immune responses, and the acquired information is subsequently employed in the development of vaccines.37 A study conducted by Martinelli revealed that using in silico tools for designing approaches to treat specific pathogens allows researchers to adapt procedures according to their objectives at a minimal cost.38 Employing computational tools prior to conducting laboratory experiments is beneficial since it is a cost-effective and time-efficient approach. The term computational vaccinology refers to the utilization of computational and mathematical methods to condense extensive immunological data into a more concise and manageable format.39 Fadaka et al. reported that immunoinformatics approaches are gaining increasing acceptance as the primary strategy for developing potent vaccines against various microorganisms, especially viruses.40 According to Shawan et al., the application of computational vaccinology, bolstered by strategies such as vaccinomics and immunoinformatics, has positioned the world at an advantageous stage. This perspective allows for the economical and time-efficient screening and detection of antigens of interest, facilitating the development of vaccine candidates to combat emerging pathogenic invasions.41 Targeting the right protein or gene that can provide the necessary outcomes or immune response is essential for improving the development of vaccines and medicinal agents.42 Hence, in this study, we utilised two structural proteins (G and F) and one non-structural protein (W) for the designing of two vaccine constructs (Malaysian and Indian) using the Reverse vaccinology approach.
Protein sequences retrieval
The sequences of the two structural proteins attachment glycoprotein G (ID: CBM41034.1, QBQ56723.1), fusion glycoprotein F (ID: CBM41033.1, QBQ56722.1), and one non-structural protein W protein (ID: QHR78997.1, QBQ56719.1) of Indian and Malaysian isolates of NiV were obtained from the NCBI protein database (http://www.ncbi.nlm.nih.gov/protein) in the FASTA format.
Protein Variability Testing (PVS)
To analyse and characterize the differences/variations in the amino acid sequences /structures of proteins (the Indian and Malaysian NiV proteins), protein variability testing was performed using the Protein Variability Server (http://imed.med.ucm.es/PVS/). This server employs various variability metrics to calculate the variability of sequences within a multiple sequence alignment.
Prediction of CTL, HTL and linear B-Cell epitopes
The IEDB (https://www.iedb.org/) Immune Epitope Database, was utilized for the prediction of cytotoxic T lymphocytes (CTL) and helper T-cell epitopes from peptides/proteins. The Net MHC pan EL4.1 and IEDB recommended the 2.22 method for the prediction process.43 The IEDB selection process incorporates a consensus approach, which incorporates NN-align, SMM-align, CombLib, and Sturniolo, if there is an appropriate predictor for the molecule.44 The prediction of linear B-cell epitopes was carried out by employing the ABC Pred software tool, which is accessible at http://crdd.osdd.net/raghava/abcpred/. Notably, a B-cell epitope refers to a short peptide capable of cross-reacting with an antibody, that binds to a conformational epitope.
Toxicity, allergenicity, antigenicity and immunogenicity prediction of selected epitopes
To ensure safety, all epitopes were subjected to toxicity screening with the ToxinPred2 server (https://webs.iiitd.edu.in/raghava/toxinpred2/), and only non-toxic epitopes were considered for subsequent analysis. The server utilizes a wide range of techniques and information sources for prediction including machine learning techniques, BLAST, and MERCI. To predict the allergenicity of the epitopes, we utilized the AllerTOP v.2.0 server (https://www.ddg-pharmfac.net/AllerTOP/method.html), which is the initial server for alignment-free in silico prediction of allergens depending on the physical and chemical properties of protein sequences.
The VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) server was employed for the prediction of antigenicity of epitopes, and it is the first alignment-free method in which protein sequences are subjected to an autocross-covariance (ACC) transformation at a threshold of 0.4. To evaluate the immunogenicity of the predicted epitopes, the IEDB Immunogenicity (Class I) server (https://tools.iedb.org/immunogenicity/) was utilized to predict the immunogenicity of the peptide MHC (pMHC) complex. This tool utilizes the arrangement of amino acids within the peptide and their properties.
Designing the multiepitope vaccine candidates-PNiV Vac-M and PNiV Vac-I
Antigenicity, immunogenicity, non-toxicity, and non-allergenicity play a vital role in new vaccine construction with epitopes. Two vaccine candidates were designed, PNiV Vac-M (Malaysia) and PNiV Vac-I (India). The CTL, HTL, and B-cell epitopes that passed the screening were chosen. HTL and linear B cell epitopes were linked by a GPGPG linker and the CTL epitopes were linked by an AAY linker.
Prediction of the quality and physiochemical properties of PNiV Vac-M and PNiV Vac-I
The quality attributes (toxicity, allergenicity and antigenicity) of PNiV Vac-M & PNiV Vac-I were assessed, Toxinpred 2.0 was used for toxicity prediction, and allergenicity was estimated via the Allertop 2.0 and VaxiJen 2.0 software tools to assess the antigenicity scores of the vaccine candidates. The Protparam tool (https://web.expasy.org/protparam/) available on the ExPASy server was used to predict physiochemical properties (molecular weight, amino acid number and composition, isoelectric point, instability and aliphatic index, extinction coefficient, net charge and hydropathicity value-GRAVY).
Two-dimensional & three-dimensional structure prediction
We used the PSIPRED (https://bioinf.cs.ucl.ac.uk/psipred/) tool to generate the secondary structure of PNiV Vac-I & PNiV Vac-M. PSIPRED is a PSI-blast based secondary structure prediction tool, that uses artificial neural network machine learning methods in its algorithm (https://en.wikipedia.org/wiki/PSIPRED). 3D models of the final vaccine construct were generated by employing the Alphafold Colab (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb).
Refinement, energy minimization & validation
For refinement of the 3D model of PNiV, the Vac-I and PNiV Vac-M Galaxy refine web server was used (https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). The Galaxy Refine server operates on a refinement methodology that underwent successful validation in refinement experiments conducted during CASP10. Top of Form To minimize the energy of PNiV Vac-I and PNiV Vac-M Forms, a Chiron server was used (https://dokhlab.med.psu.edu/chiron/login.php). Chiron swiftly minimizes the energy of protein molecules by employing discrete molecular dynamics, utilizing an all-atom representation for every residue within the protein. The PNiV Vac-I & PNiV Vac-M were validated using RAMPAGE (https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/).
Molecular docking of PNiV Vac-M & PNiV Vac-I with toll-like receptor 4
An effective method for assessing the cross-molecular interactions between proteins and proteins is in silico molecular docking. The Schrodinger module version 11.8 was used to perform the protein-protein docking of the TLR4 with PNiV Vac-M and PNiV Vac-I. The protein structure was generated using the protein preparation wizard within the Maestro 11.8 version of the Schrodinger suite. The protein was pre-processed by introducing hydrogen bonds to stabilize the atomic positions and by eliminating extraneous chains, excluding water molecules, and removing heteroatom ion compounds. Energy minimization was carried out using the force field of OPLS_3e (Optimized Potentials for Liquid Simulations, version 3e). Protein-protein docking was performed using the Schrodinger Bio luminate package with default parameters according to the manufacturer’s protocol.
MD simulation of the PNiV Vac-M & PNiV Vac-I -TLR4 complex
Molecular dynamics was used to simulate the docked complexes, and protein complex MD simulations were conducted using the Desmond program (Desmond 2018-4). Using the steepest descent method, the prepared systems underwent energy minimization across 2000 interactions. To equilibrate the system, Desmond’s default parameters were used. Then, using the NPT (normal pressure and temperature) ensemble with a time step of 2 fs, the 100 ns MD simulations were run with equilibrated systems at 300 K and constant pressure of 1 atm.
Protein selection and protein sequence retrieval
Among the NiV structural proteins F (Fusion protein), G (Attachment Glycoprotein), M (Matrix protein), L (RNA Polymerase), P (Phosphoprotein) and M (Matrix protein) and the non-structural proteins C, V and W, highly antigenic and immunogenic proteins i.e., glycoprotein F (fusion), glycoprotein G (attachment) and the W protein were selected and further subjected to epitope prediction. The F glycoprotein, G glycoprotein and W protein sequences of Indian and Malaysian isolates/strains of NiV were obtained from the NCBI database in the FASTA format and stored along with their accession numbers for further studies (Table 4).
Table (4):
NiV protein details
| No. | Name of protein | Country from which isolated | NCBI Accession number | No. of amino acids | Mol. wt (kD) | Amino acid composition (%) | |
|---|---|---|---|---|---|---|---|
| Aliphatic | Aromatic | ||||||
| 1 | Attachment glycoprotein G | India | QBQ56723.1 | 602 | 67179.06 | 39.2 | 9.1 |
| 2 | Fusion glycoprotein F | QBQ56722.1 | 546 | 60280.90 | 43 | 7.2 | |
| 3 | W Protein | QBQ56719.1 | 450 | 48917.63 | 37.7 | 4.9 | |
| 4 | Attachment glycoprotein G | Malaysia | CBM41034.1 | 602 | 67056.91 | 40.2 | 8.9 |
| 5 | Fusion glycoprotein F | CBM41033.1 | 546 | 60254.87 | 42.6 | 7.0 | |
| 6 | W Protein | QHR78997.1 | 450 | 49521.72 | 36.8 | 4.7 | |
Evaluation of protein variability
The protein variability between structural and non-structural proteins of Indian and Malaysian isolates/strains was analysed using the protein variability server, which is based on Shannon entropy analysis.45 The Shannon entropy (H) for every position is as follows
Here, Pi signifies the proportion of residues of amino acid type i, while M represents the total number of amino acid types. The range of H spans from 0 indicating the presence of only one residue at that position, to 4.322 where each of the 20 residues is equally represented at that position. In general, positions with H >2.0 are regarded as variable, while those with H <2 are deemed conserved. Positions are deemed highly conserved when H <1.0.
The Shannon entropy of the G, F and W proteins of structural and non-structural proteins from Indian and Malaysian isolates/strains were found to be 1. These proteins remain highly conserved without any significant variations (Figure 2).
(c)
Figure 2. Protein variability analysis results. (a) G glycoprotein, (b) F glycoprotein, (c) W protein
Prediction of CTL, HTL and linear B-cell epitopes
To predict T-cell epitopes, which are peptides identified by T-cell receptors and presented on antigen presenting cells (APCs) through class I or II molecules, the prediction tool for T-cell epitopes of the Immune Epitope Database and Analysis Resource (IEDB) was utilized. Ten epitopes were selected for each protein according to their percentile rank and good binding was indicated by a low percentile rank. These selected epitopes were further tested against MHC I supertype alleles, specifically the human leucocyte antigens of India and Malaysia HLA-A*24:02 and HLA-A*24:07, respectively.
Prediction of the HTL epitopes was carried out using the IEDB server. The default mode peptide length of 15 residues was adjusted, and the epitopes were then screened against DRB1*15 and DRB1*12:02 MHC II alleles found in India and Malaysia, respectively. The NetMHCII pan method was employed for prediction, and epitopes were chosen depending on their percentile rank, with low ranks indicating good binding properties. The humoral immune system relies heavily on the crucial participation of B lymphocytes which produce targeted antibodies against pathogens. Ten linear B-cell epitopes for each protein were predicted using the ABC web server. The epitope that received the highest score was chosen for vaccine construction (Table 5-12).
Table (5):
TCL Epitopes of G Glycoproteins from the Indian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4*) | Immunogenicity |
|---|---|---|---|---|---|
| G Glycoprotein India | AFLIDRINW | Non-toxin | Probable | 0.9509 | 0.3091 |
| YFPAVGFLV | Non-allergen | 0.8143 | 0.2008 |
*Threshold value
Table (6):
HTL epitopes of G Glycoproteins from the Indian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) | Immunogenicity |
|---|---|---|---|---|---|
| G Glycoprotein India | TLYFPAVGFLVRTEF | Non-toxin | Probable Non-allergen | 1.0010 | 91.0893 |
| DTLYFPAVGFLVRTE | 0.8064 | 92.2596 | |||
| GVAIGIATAAQITAG | 1.0404 | 96.1663 | |||
| AGVAIGIATAAQITA | 0.9497 | 93.4635 | |||
| RGSPPYQEGKSVNAE | 1.0176 | 98.5261 | |||
| GSPPYQEGKSVNAEN | 0.9395 | 96.8956 |
Table (7):
B-Cell epitopes of G Glycoproteins from Indian and Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) |
|---|---|---|---|---|
| G Glycoprotein India | SFSWDTMIKFGDVQTV | Non-toxin | Probable Non-allergen | 1.0016 |
| SGSLMMTRLAVKPKNN | 1.1763 | |||
| G Glycoprotein Malaysia | SFSWDTMIKFGDVQTV | 1.0016 | ||
| KKVRFENTTSDKGKNP | 0.8723 |
Table (8):
HTL epitopes of F Glycoproteins from the Indian & Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) | Immunogenicity |
|---|---|---|---|---|---|
| F Glycoprotein India | SEWISIVPNFILVRN | Non-toxin | Probable Non-allergen | 0.5788 | 65.948 |
| F Glycoprotein Malaysia | ITFISFIIVEKKRNT | 1.7597 | 82.1445 | ||
| LITFISFIIVEKKRN | 1.5214 | 76.5204 | |||
| TFISFIIVEKKRNTY | 1.4822 | 81.2113 | |||
| FISFIIVEKKRNTYS | 1.4463 | 77.8806 | |||
| ISFIIVEKKRNTYSR | 1.2290 | 76.7376 |
Table (9):
BCL epitopes of F Glycoproteins from the Indian and Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) |
|---|---|---|---|---|
| F Glycoprotein India | LGSVNYNSEGIAIGPP | Non-toxin | Probable Non-allergen | 1.1587 |
| EGIAIGPPVFTDKVDI | 0.7407 | |||
| F Glycoprotein Malaysia | MVVILDKRCYSNLLIL | 0.7160 |
Table (10):
TCL epitopes of W Proteins from the Indian & Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) | Immunogenicity |
|---|---|---|---|---|---|
| W Protein India | GLNPTAIPF | Non-toxin | Probable
Non-allergen |
1.8141 | 0.16721 |
| DQLEFEDEF | 1.033 | 0.38262 | |||
| W Protein Malaysia | AQPPYHWSI | 0.9115 | 0.10237 |
Table (11):
HTL epitopes of W Proteins from the Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) | Immunogenicity |
|---|---|---|---|---|---|
| W Protein Malaysia | NVCLVSDAKMLSYAP | Non-toxin | Probable
Non-allergen |
0.8356 | 91.4996 |
| GNVCLVSDAKMLSYA | 0.7717 | 91.4531 |
Table (12):
BCL epitopes of W Proteins from the Indian & Malaysian isolates
| Protein & Origin | Epitopes | Toxicity | Allergenicity | Antigenicity score (Th value: 0.4) |
|---|---|---|---|---|
| W Protein India | SSTSPTDGTIGKRVSN | Non-toxin | Probable
Non-allergen |
1.1938 |
| HVRGSPPYQEGKSVNA | 0.9715 | |||
| W Protein Malaysia | HWSIERSISPDKTEIV | 0.6304 | ||
| GRTIEGQSIRDNLQAK | 0.4629 |
Assessment of the toxicity, allergenicity, antigenicity, and immunogenicity of the selected epitopes
The chosen epitopes were assessed for their quality traits, i.e. toxicity, allergenicity, antigenicity, and immunogenicity. Epitopes exhibiting high antigenicity and immunogenicity, along with nontoxic and nonallergic properties, were preferred for further investigation. Sixteen and 13 epitopes were selected from the 60 epitopes of the Indian and Malaysian isolates, respectively.
Construction of the multiepitopic vaccine candidate- PNiV Vac-M & PNiV Vac-I
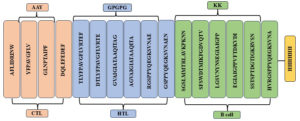
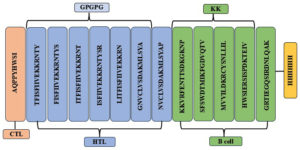
The development of multiepitopic vaccine candidate involves the fusion of various categories of epitopes. Two vaccine candidates were designed; one representing the Malaysian isolate, PNiV Vac-M and the other representing the Indian isolate PNiV Vac-I. PNiV Vac-M was constructed using 1 CTL, 7 HTL and 5 B-cell epitopes, and PNiV Vac-I was constructed using 4 CTL, 6 HTL and 6 B-cell epitopes. The selected immunogenic epitopes were further linked together using a KK linker (B cell), an AAY linker (HTL), and a GPGPG linker (CTL). T-helper cells targeted epitopes using GPGPG linkers, while AAY linkers were employed to associate B-cell linear epitopes. Additionally, for easy downstream processing, a 6xHis tag made of polyhistidine was appended at the C-terminus (Figure 3).
(b)
Figure 3. (a) Multiepitope vaccine construct- Indian Isolate, (b) Multiepitope vaccine construct- Malaysian Isolate
a) PNiV Vac-I
b) PNiV Vac-M
Assessment of the quality and physiochemical properties of PNiV Vac-M & PNiV Vac-I
The quality attributes (toxicity, allergenicity and antigenicity) and physiochemical properties (molecular weight, isoelectric point, amino acid number and composition, instability index, aliphatic index, extinction coefficient, net charge and hydropathicity value) of PNiV Vac-M and PNiV Vac-I were assessed. Both candidates were found to be nontoxic, nonallergenic and antigenic. The molecular weights of the vaccine candidates were 28982.80 and 27201.24 D for PNiV Vac-I and PNiV Vac-M, respectively. The amino acid residues of PNiV Vac-I and PNiV Vac-M were 279 and 245, respectively. PNiV Vac-I has a hydropathicity index of -0.292, and PNiV Vac-M has a value of -0.426. The aliphatic indices of PNiV Vac I and PNiV Vac-M were 67.53 and 77.51, respectively. PNiV Vac-I has an instability index of 26.41 and PNiV Vac-M has an instability index of 35.24. The isoelectric point of PNiV Vac-I was determined to be 9.30 and that of PNiV Vac-M was 10.08. The extinction coefficients were 25900 and 27055 for PNiV Vac-I and PNiV Vac-M, respectively (Table 13).
Table (13):
Physiochemical properties of the PNiV Vac-M & PNiV Vac-I vaccine candidates
Vaccine construct |
Amino acid |
Molecular weight (D) composition |
Aliphatic index |
Theoretical pI |
Instability index |
GRAVY |
Extinction coefficient |
Toxigenicity |
Allergenicity |
Antigenicity |
|---|---|---|---|---|---|---|---|---|---|---|
PNiV Vac-I |
279 |
28982.80 |
67.53 |
9.30 |
26.41 |
-0.292 |
25900 |
Non-toxin |
Probable Non-allergen |
0.7997 |
PNiV Vac-M |
245 |
27201.24 |
77.51 |
10.08 |
35.24 |
-0.426 |
27055 |
Non-toxin |
Probable Non-allergen |
0.8002 |
Prediction of the 2D and 3D structures of PNiV Vac-M and PNiV Vac-I: refinement, energy minimization & validation
Prediction of the 2D structure was performed using PSIPRED which revealed that PNiV Vac-M had 6.0% helices, 31.6% strands, 4.8% beta strands and 57.6% random coils, whereas PNiV Vac-I had 9.6% helices, 31.1% strands, 8.9% beta strands and 50.1% random coils (Figure 4). 3D structure prediction was conducted on the designed vaccine candidates using and Alphafold software tool (Figure 5). The resulting 3D structures were refined, subjected to energy minimization and further validated via a Ramachandran plot. The resulting data indicate that the structures exhibit high-quality resolution, and atomic clashes between the residues upon steric confirmation.
Molecular docking interaction between PNiV Vac-M & PNiV Vac-I and TLR 4
The protein database provided the crystal structure of the human TLR 4 (PDB ID: 4g8a) complex. The refined PNiV Vac-I & PNiV Vac-M underwent molecular docking with the monomeric form of human TLR4 immune receptors. The crystallographic structure of the human TLR 4 complex consists of 10 chains labelled A through H (A-H), where the A and B chains represent the human TLR 4 receptor, the C and D chains correspond to lymphocyte antigen 96, and the E-H chains are associated with various forms of glucopyranose.
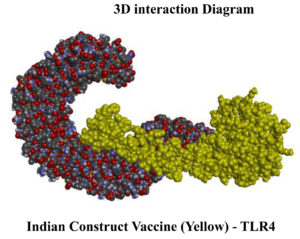
The protein complexes were further subjected to protein and protein docking as per the Schrodinger Bio luminate protocol with their respective target proteins. For the TLR4- PNiV Vac-I complex a Piper score of -316.054 kcal/mol and a Piper energy of -587.658 kcal/mol were obtained with 9 hydrogen bond interactions. For TLR4-PNiV, the Vac-M complex had a Piper score of -451.548 and a Piper energy of -634.281 with 9 hydrogen bond interactions (Table 14 and 15). The results revealed that the interactions were thermodynamically stable and graphical interaction analysis was carried out through LigPlot+ and PDBsum.
Table (14):
Molecular Docking
No. |
Construct vaccine |
Piper Score |
Piper Energy |
|---|---|---|---|
1. |
Indian construct |
-316.054 |
-587.658 |
2. |
Malaysian construct |
-451.548 |
-634.281 |
Table (15):
Interaction of the Indian and Malaysian vaccine constructs with TLR4
| Indian vaccine construct vs TLR 4 | Malaysian vaccine construct vs TLR 4 | ||||||
|---|---|---|---|---|---|---|---|
| No. | Protein | Protein | Distance | No. | Protein | Protein | Distance |
| 1 | A: ASN339 | B: GLN38 | 2.6064 | 1 | A: GLN562 | B: SER54 | 1.9647 |
| 2 | A: LYS560 | B: TYR74 | 1.8231 | 2 | A: GLN565 | B: PRO36 | 2.372 |
| 3 | A: GLN578 | B: LEU52 | 2.6818 | 3 | A: HIS566 | B: GLY35 | 2.231 |
| 4 | A: THR584 | B: THR72 | 2.332 | 4 | A: SER589 | B: ARG94 | 1.7474 |
| 5 | A: HIS587 | B: THR72 | 2.2306 | 5 | A: GLN592 | B: ASN73 | 2.038 |
| 6 | A: ASN531 | B: ARG6 | 1.7476 | 6 | A: ASP596 | B: ARG72 | 1.6266 |
| 7 | A: GLU608 | B: TYR53 | 1.5927 | 7 | A: THR626 | B: ARG89 | 2.649 |
| 8 | A: GLU586 | B: THR72 | 1.634 | 8 | A: GLN588 | B: ARG94 | 1.7333 |
| 9 | A: THR626 | B: ARG252 | 2.1978 | 9 | A: CYS585 | B: ARG94 | 2.0301 |
Molecular dynamics simulation
Molecular dynamic simulations of the protein-protein complexes were performed for 100 ns of simulation time, and subsequent analyses were performed on the basis of the simulation trajectory. The root means square deviation (RMSD) acts as a metric for the average alteration in the structural displacement of atoms within a specific frame compared to the reference frame. The root mean square fluctuation (RMSF) is valuable for characterizing the localized variations along the protein chain.
Based on the simulation’s trajectory, the RMSD of the TLR4 protein backbone initially increased gradually until reaching 4.0 Å and then stabilized at 7.0 Å until 100 ns, and the RMSF of the complex stabilized at 3.0 Åuntil 100 ns. The RMSD of the PNiV Vac-I backbone increased gradually until reaching 5.0 Åand then stabilized at 7.0 to 8.0 Å 100 ns, and the RMSF of the complex stabilized at 4.5 Å until 100 ns. Both of the RMSD values were stable at 7.0 Å during the simulation period. In TLR4-PNiV Vac-M, the RMSD of the TLR4 protein backbone initially increased gradually until reaching 2.0 Å and then stabilized at 5.0 Å until 100 ns, and the RMSF of the complex stabilized at 1.5 Å until 100 ns. The RMSD of the PNiV Vac-M backbone increased gradually until reaching 6.0 Å and then stabilized at 10.5 Å 100 ns, and the RMSF of the complex stabilized at 6.0 Å until the 100 ns. Both of the RDDs show a stable constant value of 4.5 Å during the simulation period (Figure 6 and 7).
(b)
Figure 6. (a) 3D interaction diagram for PNiV Vac-I with TLR4, (b) 3D interaction diagram for PNiV Vac-M with TLR4
(b)
Figure 7. Results of Molecular Dynamics simulation of vaccine candidates. Root mean square deviation (RMSD) plots reflects the stability between the vaccine and TLR 4 and Root mean square fluctuation (RMSF) reflects the flexibility and fluctuation of the amino acids residues in the docked complexes. a) PNiV Vac-I b) PNiV Vac-M
Currently, there is no approved drug/vaccine available for the prevention and control of the highly infectious NiV infection (Table 2 and 3). Conventional vaccine development approaches need highly specialised containment facility (BSL-3 & BSL-4) for handling live or infectious NiV. At this juncture, reverse vaccinology approaches are found very promising in design and development of effective vaccine candidate based on their quality attributes such as toxicity, allergenicity, antigenicity, immunogenicity. In this study, amino acid sequences of the structural and non-structural proteins of Indian and Malaysian isolates of NiV were retrieved from the NCBI database and stored in the FASTA format for further analysis (Table 4).
Protein variability testing is a process of analysing and characterizing the differences/ variations in the amino acid sequences or structures of proteins. This type of testing is often used to determine the extent of variation between numerous protein samples or between proteins from different organisms to identify potential variations that may affect protein functions and behaviour. The present research utilized a protein variability server (http://imed.med. ucm.es/PVS/) to analyse the variability of proteins from Indian and Malaysian sources. The results of the analysis have shown that these proteins remained highly conserved over a period of 2.5 decades, without any significant variations. Hence, the utilization of these proteins for developing a cocktail-based multiepitope vaccine would expect to be highly effective and could subsequently progress for appropriate regulatory studies, both in vitro and in vivo.
In this study, a total of ten T-cell epitopes and linear B cells for each protein, i.e. F, G & W, based on the percentile rank were predicted, and were found are stimulating antibody-mediated immunity as well as cellular immunity. The quality attributes of each epitope were analysed using commercially available immunoinformatic tools such as VaxiJen, Allertop, and Toxinpred. Based on potential factors of vaccine candidates such as toxicity prediction, allergenicity prediction, antigenicity score, and immunogenicity prediction, the epitopes were assessed (Table 5-12). Furthermore, the epitopes were subjected to linking and constructing CAPPV candidates (Figure 3). The 2D and 3D structures of these constructed vaccines were predicted using PSIPRED and Alphafold, respectively (Figures 4 and 5).
Table 12 shows the physiochemical characteristics of those constructed vaccines-PNiV Vac-I and PNiV Vac-M, such as molecular weight, amino acid number and composition, hydropathicity index (GRAVY), aliphatic index, instability index, extinction coefficient, theoretical PI, and quality attributes such as toxicity, allergenicity and antigenicity.
The molecular weight of the peptides impacts the immune response in different ways. The size of the protein can influence its ability to be processed and presented to immune cells, and the size and shape of the protein influence its ability to interact with immune cells and signalling molecules. The molecular weight of proteins can also have an impact on stability and solubility, and the molecular weights of proteins have a great impact on immune function through a variety of cellular mechanisms.46 The molecular weights of our vaccine constructs were 28982.80 D and 27201.24 D for PNiV Vac-I and PNiV Vac-M, respectively.
As it has been shown that the amino acid number and composition are influential factors of immune function and the risk of infectious and inflammatory diseases.47 The present study determined that the amino acid residues of PNiV Vac-I and PNiV Vac-M were determined to be 279 and 245, respectively.
Further, it has been shown that PNiV Vac-I has hydropathicity index of -0.292 and that PNiV Vac-M has a hydropathicity index of -0.426, as the hydropathicity index (GRAVY) influences various aspects of immune functions, including antigen presentation, antibody recognition, and immune cell trafficking. Proteins that possess a GRAVY score above 0 exhibit hydrophobic characteristics, while those with a GRAVY score below 0 display hydrophilic properties. Here, both vaccine constructs were found maintaining their hydrophilic nature.
Considering the significance of aliphatic index in the thermal stability of proteins, our study, determined the aliphatic index of PNiV Vac-I and PNiV Vac-M to be 67.53 and 77.51, respectively, which indicates their thermal stability.
The instability index is a protein measurement used to ascertain its stability. Any protein with an instability index less than 40 was considered stable and those with an instability index greater than 40 were considered unstable.48 From this study, it was found that the instability indices of PNiV Vac-I and PNiV Vac-M were 26.41 and 35.24, respectively, which indicates that both are stable.
The extinction coefficient is a measure of the amount of light absorbed by a sample at a particular wavelength and depends on the composition and structure of the sample.49 Our study revealed that the extinction coefficients of PNiV Vac-I and PNiV Vac-M were 25900 and 27055, respectively.
The pI of a protein indicates the pH at which the protein has an overall neutral charge in a particular solution, and it can influence the interplay between the immune system and the vaccine construct in different ways. The pI has an impact on solubility, stability, antigenicity and the ability to elicit an immune response, bioavailability and distribution throughout the body.50 According to this research, the pI of PNiV Vac-I was determined to be 9.30 and that of PNiV Vac-M was 10.08.
Analysis of the physiochemical parameters revealed that both vaccine constructs satisfied all the necessary requirements for a compound to be considered effective as a vaccine.
Molecular docking studies of PNiV Vac-M & PNiV Vac-I were conducted with TLR 4 to confirm the binding potential, and the results revealed that the interactions were thermodynamically stable. Further dynamic simulations were performed to assess the physical movements of atoms or molecules. The RMSD confirmed that PNiV Vac-I is more stable than PNiV Vac-M.
Similar efforts on the development of NiV vaccine candidates have been documented in the literature. Several investigations have focused on the design, development and production of NiV vaccines.51 According to the studies of Benjamin et al., a subunit vaccine comprising a soluble glycoprotein (Sg) from Hendra virus (HeV), a related henipavirus that causes diseases in humans and equines, the HeV Sg vaccine is presently employed as an animal vaccine to protect horses against Hendra Virus (Equivac HeV, Zoetis) in Australia and is considered as a vaccine candidate for humans against Nipah virus that is still not approved by regulators.
Various approaches utilize viral vectors such as canarypox which codes for NiV G or NiV F,25 vaccinia viruses, which code for NiV G or NiV F,24 recombinant AAV, which codes Niv G,29 recombinant rhabdoviruses (VSV and rabies), which express NiV G or NiV F,52 and recombinant measles virus vectors, which express NiV G.30 These approaches have been demonstrated to protect Pigs, Syrian hamsters, Ferrets, and/or AGMs from NiV infection (Table 3).
Recently, an attempt on developing DNA vaccine (DNA-G) and a recombinant chimpanzee adenovirus vaccine (AdC68-G), expressing NiV-G and induce long-term immunity against NiV infection was undertaken.53
All of these earlier studies were found focusing on a single protein/epitope of the protein only, despites the single protein-based vaccines may suffer from limited effectiveness, and susceptibility to antigenic variation and have some drawbacks that can compromise their ability to provide broad and durable protection. In our study, we focused on a cocktail approach involving highly antigenic and immunogenic NiV proteins through an immunoinformatic method, which revealed a more comprehensive and resilient immune response and reduced the time, manpower and cost associated with the design and development of newer vaccine candidates.
The vaccine candidate, which was designed using immunoinformatic methods, underwent comprehensive characterization, suggesting its strong immunogenicity, lack of toxicity and allergenicity, and stability under various suitable environmental conditions. The availability of computational tools promisingly facilitates hassle-free designing of an epitope-based peptide vaccine, that is potentially antigenic and highly immunogenic in nature.
Nipah virus infection is an emerging serious public health problem worldwide. Considering the high mortality rates and the nonavailability of a safe and effective vaccine against NiV, swift actions should be prioritized for its design and development exploiting modern innovative tools. The present study has demonstrated the application of immunoinformatic tools and successfully predicted certain promising immunogenic cocktail antigen-presenting peptides (CAPP) as promising vaccine candidates, which could be further tested for its immunogenic potential using appropriate in vivo methods.
ACKNOWLEDGMENTS
The authors would like to acknowledge The Director, Pasteur institute of India, Coonoor, for research support; Dr. Paramasivan Rajiah, Scientist E, ICMR, Vector Control Research Center, Madurai, Tamil Nadu, India, for mentorship; Ms. Reshma and Mr. S. Raju, Research scholars, Pasteur institute of India, Coonoor, for assistance with editing; Dr. Vijayakumar, IISC, Bangalore, for guidance on bioinformatics; Mr. Muhammed Noufel and Mr. Mathanagobal for their support during the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NKP and SJ conceptualized the study. SJ performed Research Co-ordination and supervision. NKP, SJ and SBP applied methodology. NKP and SJ performed computational work. NKP, SJ and RP performed data analysis. NKP wrote the manuscript. SKC, AAP, MM, KD, PB, SS, SJ and SBP reviewed the manuscript. RP, SJ, SKC, AAP, MM, KD, PB, SS and SBP edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not Applicable.
- Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R S B Biol Sci. 2001;356(1411):983-989.
Crossref - Mohapatra P, Khatib MN, Shabil M, et al. Addressing the Nipah virus threat: a call for global vigilance and coordinated action. Clinical Infection in Practice. 2024;24:10390
- Singh RK, Dhama K, Chakraborty S, et al. Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies-a comprehensive review. Veterinary Quarterly. 2019;39(1):26-55.
Crossref - Magdum M, Chowdhury MAT, Khandaker M, et al. Nipah virus unveiled: A review article. Adv Biosci Biotechnol. 2024;15(3):161-173
- Adiga R. Emergence of nipah virus: a review. J Biomed Pharm Res. 2019;8(1):48-54.
Crossref - A Ternhag, Penttinen P. [Nipah virus—another product from the Asian “virus factory”]. Lakartidningen.2005;102(14):1046-7
- Bellini WJ, Harcourt BH, Bowden N, Rota PA. Nipah virus: An emergent paramyxovirus causing severe encephalitis in humans. J Neurovirol. 2005;11(5):481-487.
Crossref - Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. Endocytosis of the Nipah Virus Glycoproteins. J Virol. 2005;79(6):3865-3872.
Crossref - Ortega V, Zamora JLR, Monreal IA, et al. Novel Roles of the Nipah Virus Attachment Glycoprotein and Its Mobility in Early and Late Membrane Fusion Steps. mBio. 2022;13(3):e0322221.
Crossref - Loomis RJ, Stewart-Jones GBE, Tsybovsky Y, et al. Structure-Based Design of Nipah Virus Vaccines: A Generalizable Approach to Paramyxovirus Immunogen Development. Front Immunol. 2020;11.
Crossref - Talukdar P, Dutta D, Ghosh E, Bose I, Bhattacharjee S. Molecular Pathogenesis of Nipah Virus. Appl Biochem Biotechnol. 2023;195(4):2451-2462.
Crossref - Raju S, Sahoo D, Bhari VK. In silico design of multi-epitope vaccine against Nipah virus using immunoinformatics approach. J Pure Appl Microbiol. 2021;15(1):212-231.
Crossref - Pariyapurath NK, Jagannathan S, Mathanmohun M, et al. Targeted Immunization Strategies and Designing Vaccine against Indian Nipah Virus Strain (NiV B) and Malaysian Variant (NiV M). Int J Pharm Investig. 2024;14(4):1201-1207.
Crossref - Yoneda M, Guillaume V, Sato H, et al. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One. 2010;5(9):1-8.
Crossref - Liew YJM, Ibrahim PAS, Ong HM, et al. The Immunobiology of Nipah Virus. Microorganisms. 2022;10(6):1162.
Crossref - Wolf MC, Freiberg AN, Zhang T, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107(7):3157-3162.
Crossref - Dang HV, Cross RW, Borisevich V, et al. Broadly neutralizing antibody cocktails targeting Nipah virus and Hendra virus fusion glycoproteins. Nat Struct Mol Biol. 2021;28(5):426-434.
Crossref - Bossart KN, Zhu Z, Middleton D, et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute Nipah virus infection. PLoS Pathog. 2009;5(10):1000642.
Crossref - Georges-Courbot MC, Contamin H, Faure C, et al. Poly(I)-Poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob Agents Chemother. 2006;50(5):1768-1772.
Crossref - Mathieu C, Porotto M, Figueira TN, Horvat B, Moscona A. Fusion inhibitory lipopeptides engineered for prophylaxis of nipah virus in primates. J Infect Dis. 2018;218(2):218-227.
Crossref - Lo MK, Feldmann F, Gary JM, et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci Transl Med. 2019;11(494):aau9242.
Crossref - Dawes BE, Kalveram B, Ikegami T, et al. Favipiravir (T-705) protects against Nipah virus infection in the hamster model. Sci Rep. 2018;8(1):7604.
Crossref - Gomez Roman R, Tornieporth N, Cherian NG, et al. Medical countermeasures against henipaviruses: a review and public health perspective. Lancet Infect Dis. 2022;22(1):e13-e27.
Crossref - Guillaume V, Contamin H, Loth P, et al. Nipah Virus: Vaccination and Passive Protection Studies in a Hamster Model. J Virol. 2004;78(2):834-840.
Crossref - Weingartl HM, Berhane Y, Caswell JL, et al. Recombinant Nipah Virus Vaccines Protect Pigs against Challenge. J Virol. 2006;80(16):7929-7938.
Crossref - Mire CE, Versteeg KM, Cross RW, et al. Single Injection Recombinant Vesicular Stomatitis Virus Vaccines Protect Ferrets against Lethal Nipah Virus Disease. Virol J. 2013;10-353.
Crossref - Lo MK, Bird BH, Chattopadhyay A, et al. Single-dose replication-defective VSV-based Nipah virus vaccines provide protection from lethal challenge in Syrian hamsters. Antiviral Res. 2014;101(1):26-29.
Crossref - Woolsey C, Borisevich V, Fears AC, et al. Recombinant vesicular stomatitis virus-vectored vaccine induces long-lasting immunity against Nipah virus disease. J Clin Invest. 2023;133(3):JCI164946.
Crossref - Ploquin A, Szecsi J, Mathieu C, et al. Protection against henipavirus infection by use of recombinant adeno-associated virus-vector vaccines. J Infect Dis. 2013;207(3):469-478.
Crossref - Yoneda M, Georges-Courbot MC, Ikeda F, et al. Recombinant Measles Virus Vaccine Expressing the Nipah Virus Glycoprotein Protects against Lethal Nipah Virus Challenge. PLoS One. 2013;8(3):0058414.
Crossref - Mungall BA, Middleton D, Crameri G, et al. Feline Model of Acute Nipah Virus Infection and Protection with a Soluble Glycoprotein-Based Subunit Vaccine. J Virol. 2006;80(24):12293-12302.
Crossref - McEachern JA, Bingham J, Crameri G, et al. A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine. 2008;26(31):3842-3852.
Crossref - Pallister J, Middleton D, Wang LF, et al. A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine. 2011;29(34):5623-5630.
Crossref - Bossart KN, Rockx B, Feldmann F, et al. A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci Transl Med. 2012;4(146):3004241.
Crossref - Droppa-Almeida D, Franceschi E, Padilha FF. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinform Biol Insights. 2018;12.
Crossref - Sunita, Sajid A, Singh Y, Shukla P. Computational tools for modern vaccine development. Hum Vaccin Immunother. 2020;16(3):723-735.
Crossref - Rawat SS, Keshri AK, Kaur R, Prasad A. Immunoinformatics Approaches for Vaccine Design: A Fast and Secure Strategy for Successful Vaccine Development. Vaccines. 2023;11(2):11020221.
Crossref - Martinelli DD. In silico vaccine design: A tutorial in immunoinformatics. Healthcare Analytics. 2022;2:100044.
Crossref - Dubey KK, Luke GA, Knox C, et al. Vaccine and Antibody Production in Plants: Developments and Computational Tools. Brief Funct Genomics. 2018;17(5):295-307.
Crossref - Fadaka AO, Sibuyi NRS, Martin DR, et al. Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus. Sci Rep. 2021;11(1):19707.
Crossref - Shawan MMAK, Sharma AR, Halder SK, et al. Advances in Computational and Bioinformatics Tools and Databases for Designing and Developing a Multi-Epitope-Based Peptide Vaccine. Int J Pept Res Ther. 2023;29(4):60.
Crossref - Sarika PB, Selvaraj J, Sumitha J, Shivanandappa KC, Sivakumar S. Immunodominant Protein of Kyasanur Forest Disease Virus: A retrospective study. Res J Biotechnol. 2024;19(9):132-142.
Crossref - Hoof I, Peters B, Sidney J, et al. NetMHCpan, a method for MHC class i binding prediction beyond humans. Immunogenetics. 2009;61(1):1-13.
Crossref - Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics. 2015;67(11-12):641-650.
Crossref - Liao H, Yeh W, Chiang D, Jernigan RL, Lustig B. Protein sequence entropy is closely related to packing density and hydrophobicity. Protein Eng Des Sel. 2005;18(2):59-64.
Crossref - Bachmann MF, Jennings GT. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10(11):787-796.
Crossref - Kelly B, Pearce EL. Amino Assets: How Amino Acids Support Immunity. Cell Metab. 2020;32(2):154-175.
Crossref - Gamage DG, Gunaratne A, Periyannan GR, Russel TG. Applicability of instability index for in vitro protein stability prediction. Protein Pept Lett. 2019;26(5):339-347.
Crossref - Batabyal D, Lord H, Ahlstrom B, Vikstrom M. Determination of the experimental extinction coefficient of therapeutic proteins using the Edelhoch method. Biologicals. 2021;71:42-47.
Crossref - Heffron J, Mayer BK. Virus Isoelectric Point Estimation: Theories and Methods. Appl Environ Microbiol. 2021;87(3):1-17.
Crossref - Fulginiti VA, Eller JJ, Downe AW, Kempe CH. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202(12):1075-80.
Crossref - Keshwara R, Shiels T, Postnikova E, et al. Rabies-based vaccine induces potent immune responses against Nipah virus. NPJ Vaccines. 2019;4:15.
Crossref - Lu M, Yao Y, Zhang X, et al. Both chimpanzee adenovirus-vectored and DNA vaccines induced long-term immunity against Nipah virus infection. NPJ Vaccines. 2023;8(1):170.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.