ISSN: 0973-7510
E-ISSN: 2581-690X
Pest infestations present a significant challenge to agricultural productivity, and the excessive, indiscriminate utilization of chemical insecticides has led to issues like pest resistance, the emergence of new pest species, and serious environmental concerns. Consequently, adopting biopesticides is vital for sustainable pest management strategies. Entomopathogenic fungi (EPF) are efficient biological control agents that regulate pests through direct contact and penetration in the host’s cuticle, releasing toxins and proliferating by utilizing nutrients in the haemocoel while evading the host’s immune defences. Researchers have aimed to create effective fungal biocontrol formulations for managing plant diseases and pest infestation. One such formulation, “Biomix” was developed by VNMKV Parbhani. This microbial consortium contains beneficial microorganisms, including EPF and decomposer fungi such as Beauveria bassiana, Lecanicillium lecanii, Metarhizium anisopliae, and Aspergillus niger. These microbes collectively improve soil health, crop productivity through nutrient cycling, pathogen suppression and pest control. Given the significant role of “Biomix” it is essential to characterize these fungal isolates for integrated pest management. In this study, fungal isolates underwent morphological characterization, documenting growth patterns, colony color, texture, sporulation time, and hyphal structure. Molecular characterization involved extracting genomic DNA from mycelial mats, followed by PCR amplification of ITS rDNA region using ITS-1/ITS-4 primers. Based on the query coverage (QC), E-value, as well as sequence similarity, Beauveria bassiana isolate VNMKV 1 demonstrated the highest similarity (99.65%) to Beauveria bassiana strain 23D112, the Lecanicillium lecanii isolate VNMKV 1 showed 99.65% similarity to Lecanicillium lecanii isolate Vl-5, Aspergillus niger isolate VNMKV 1 exhibited the highest similarity (99.33%) with Aspergillus sp. isolate AUMS56 and Metarhizium anisopliae isolate VNMKV 1 displayed the highest similarity (99.82%) with Metarhizium anisopliae strain CN094G8.
Entomopathogenic Fungi, Organic Matter Decomposer Fungi, ITS-rDNA, Phylogenetic Analysis
Insect pests have posed a significant threat to agriculture since ancient times. Approximately 10,000 insect species attack crops, along with about 50,000 species of fungi, 1,800 species of weeds, and 15,000 species of nematodes globally accounting for an annual loss of about 6 to 50 billion $.1 While the application of chemical pesticides plays a crucial role in reducing crop yield losses, their use must be judicious and based on pest population levels surpassing established economic threshold limits. Synthetic pesticides are the most commonly used method for pest control, they have detrimental effects on soil health, water quality, and crop quality. They also lead to issues such as insecticide resistance, pest resurgence, secondary pest outbreaks, pesticide residues, and harm to non-target organisms. Biopesticides offer a promising alternative, potentially transforming chemocentric agriculture. They can promote a greener environment, protect farmers’ health, and foster an ecologically sustainable agricultural landscape without negatively impacting the economy.2
Entomopathogenic fungi (EPF) are among the most adaptable biological control agents and natural antagonists of arthropod pests, providing effective ecological regulation of various insect species in a sustainable manner which is characterized by a broad host range that frequently leads to natural epizootics.3 These insect-pathogenic fungi predominantly belong to the orders Moniliales (Deuteromycotina: Hyphomycetes, also known as Deuteromycetes) and Entomophthorales (Zygomycotina:Zygomycetes). While these entomopathogenic fungi have the capability to manage certain insect pests, their effectiveness is not universal, making it crucial to identify suitable targets for mycoinsecticide applications. A key advantage of these fungi is their mode of infection, which occurs through direct contact and subsequent penetration of the host.4 Entomopathogenic fungi upon infection, penetrate the cuticle, release toxins, and grow by exploiting nutrients in the haemocoel while evading the insect’s immune system.5 This diverse group includes over 100 genera and approximately 750 species, documented across various insect hosts, many of which show significant potential for pest management. Key fungal pathogens in this group include Beauveria bassiana, Metarhizium anisopliae, and Lecanicillium lecanii, which are capable of infecting various insects being a part of the orders Coleoptera, Hemiptera, Lepidoptera, as well as Isoptera.6
The white muscardine fungus, Beauveria bassiana, & the green muscardine fungus, Metarhizium anisopliae, are among the most prominent fungal entomopathogens used to manage sucking as well as chewing insect pests in agriculture. They play a crucial role in IPM (integrated pest management) approaches.7 Lecanicillium spp. Serve as a naturally occurring biological control agent and are considered highly effective mycoinsecticides for managing destructive cotton pests such as whiteflies and mealybugs. These species exhibit a broad host range, targeting insects, phytopathogenic fungi, and plant-parasitic nematodes.8
Aspergillus species are vital decomposers in agriculture, improving soil health through the cycling of vital nutrients like phosphorus, nitrogen, carbon, as well as sulfur as well as the degradation of organic debris. They improve soil fertility through the solubilization of nutrients and have biofertilizer potential. Additionally, some Aspergillus strains produce antifungal compounds that suppress soil-borne pathogens, reducing plant disease.9
Researchers have long focused on developing effective fungal biocontrol formulations to manage plant diseases. “Biomix” a bioagent consortia product developed at VNMKV, Parbhani, was designed for controlling citrus decline and combating rhizome rot and white grub in turmeric. It contains beneficial microorganisms, including entomopathogenic fungi and organic matter decomposer fungi, i.e. Beauveria bassiana, Verticillium lecanii, Metarhizium anisopliae, and Aspergillus niger, etc., which collectively enhance soil fertility, promote plant growth, and boost crop yield through nutrient solubilization, pathogen suppression, and organic matter decomposition.10 Accurate identification and characterization of entomopathogenic fungal strains are crucial for integrated pest management.11 Although traditional fungal identification relies on morphological traits such as conidia shape and size, it has limitations due to phenotypic variability.12,13 Advances in molecular techniques, particularly DNA analysis via PCR targeting ribosomal DNA and ITS regions, have improved fungal detection and phylogenetic analysis, highlighting the need to integrate morphological and molecular methods for precise identification of biotechnologically significant fungi.14-16 Considering the crucial role of ‘Biomix’ in enhancing development as well as proliferation of plants, the present study is designed to conduct comprehensive morphological and molecular characterization of the entomopathogenic and organic matter-decomposing fungi present in the VNMKV-produced ‘Biomix’ formulation and to evaluate the phylogenetic relationships among the bioagents using ITS rDNA sequence analysis which facilitated the accurate identification and classification of this fungi, providing insights into their evolutionary relationships and functional dynamics within the consortium, thereby optimizing the formulation for improved agricultural outcomes.
Collection of fungal isolates
The EPF and organic matter-decomposing fungal isolates involved in the VNMKV-produced ‘Biomix’ formulation were obtained from the Biomix Research and Production Unit, Department of Plant Pathology, Vasantrao Naik Marathwada Krishi Vidyapeeth, Parbhani. The fungal isolates included Beauveria bassiana, Lecanicillium lecanii, Aspergillus niger, and Metarhizium anisopliae.
Subculturing of fungal culture on PDA
Conidia from the collected fungal isolates were inoculated onto PDA medium by using monosporic inoculation method & incubated at 27 ± 2 °C for a period of 5 to 7 days. All subculturing steps were performed aseptically in a laminar airflow cabinet (Microfilt MFI LAF-V). The fungal growth on the slants was monitored regularly, and microscopic examinations were conducted to assess the development.
Preparation of a fungal mycelial mat
To generate sufficient mycelium, a pure fungal culture was expanded by inoculating mycelial tissue disc of 2 mm diameter into 100 ml of PDB and incubating it at 27 ± 2 °C for seven days without disturbing the flasks. Subsequently, the mycelial mat from the broth was placed on Whatman filter paper and allowed to air dry for 20 to 24 minutes at room temperature. This mycelial mat was then utilized for the isolation of genomic DNA.
Morphological and microscopic examination for fungal identification
The fungal isolates were cultivated on PDA slants for morphological characterization. Colony morphology, including growth pattern, color, texture, sporulation period, and hyphal structure, was recorded. Microscopic identification of fungal structures was performed by preparing smears of the microcultures on clean glass slides, which were subsequently stained with lactophenol cotton blue for examination under light microscope. The slides were left at room temperature to air dry and were then observed under a light microscope at various magnifications (10X, 40X, and 100X).12,13
Molecular characterization of fungal isolates
Extraction of genomic DNA from fungal isolates
The DNA from the fungal isolates was extracted with specific modifications following the protocol by Prabhu et al. using the SDS method.17 Two hundred milligrams of mycelium from each isolate were ground with a mortar as well as pestle in 2 ml of 3% SDS buffer [0.5 M EDTA (pH 8), 1 M Tris-HCl (pH 8), 3% SDS]. The homogenate had been moved to a microcentrifuge tube with a capacity of 2 ml, and 1-2 µl of β-mercaptoethanol was added. The sample was incubated in a water bath at 65 °C for 30 minutes, with occasional inversions. Following incubation, the supernatant was moved to a fresh 1.5 ml tube & centrifuged for 10 minutes at 12,000 rpm. After a gentle mixing an equal volume of chloroform: isoamyl alcohol (24:1) added mixture and centrifuged for 10 minutes at 4 °C at 10,000 rpm. This procedure was repeated using isoamyl alcohol, phenol, and chloroform (25:24:1). After that, the top aqueous phase was combined with cold isopropanol, incubated for two hours at -20 °C, then centrifuged for ten minutes at 4 °C at 10,000 rpm. The supernatant was decanted and the pellet was washed with chilled 70 percent ethanol before being air-dried on blotting paper at normal room temperature. The pellet was dissolved in 200 µl of 1X TE buffer (1 mM EDTA, 10 mM Tris-HCl, pH 8.0) as well as allowed to sit at room temperature to facilitate DNA dissolution. After adding heat-treated RNase A of concentration 10 µg/ml, added to dissolved DNA and incubated for 30 minutes at 37 °C. All isolates’ DNA was extracted in the same way and kept for later use at -20 °C.
Agarose gel electrophoresis
The extracted DNA’s purity was evaluated using agarose gel electrophoresis. Ethidium bromide was added to a 0.8% agarose gel in 1X TAE (pH 8.0) buffer at a concentration of 0.5 µg/ml. The wells were loaded with 6 µl of the DNA preparations blended with 2 µl of 6X gel loading buffer. A 100 bp DNA ladder was used for reference. Electrophoresis was conducted at 100 V for 60 minutes at room temperature. Using a UV transilluminator, DNA bands were seen under UV light and then recorded.
PCR amplification targeting the ITS-rDNA region of fungal isolates
The ITS1-5.8S-ITS2 region of the rDNA had been amplified & sequenced utilizing the ITS-1 (TCCGTAGGTGAACCTGCGG) and ITS-4 (TCCTCCGCTTATTGATATGC) primers in order to molecularly identify the target fungal strains.14 2.5 µl of 10X Taq buffer, 1.5 µl of 25 mM MgCl2, 0.3 µl of 10 mM dNTPs, 1 µl of 10 pM forward primer, 1 µl of 10 pM reverse primer, 0.33 µl of 1U/ µl Taq DNA polymerase, 2 µl of DNA template, & 16.37 µl of nuclease-free water were used in the PCR reactions, which were carried out in a final volume of 25 µl. In a control tube, sterile deionized water was used in place of DNA as a negative control to guarantee the assay’s validity. Ten minutes of “1st denaturation at 95 °C was part of the amplification program. This was followed by 35 cycles of denaturation at 94 °C, annealing at 56 °C, extension at 72 °C, along with final extension at 72 °C for 10 minutes”.
On a 2 percent agarose gel with 4 µl of a 10 mg/ml ethidium bromide solution, 10 µl of PCR product from each tube was combined with 2 µl of 6X DNA loading dye and electrophoresed next to a 100 bp to 1000 bp marker in order to confirm the targeted PCR amplicons. The electrophoresis was carried out in 1X TAE buffer at 50V for 120 minutes. Estimating the amplified fragment sizes was made easier by the markers. Using a gel documentation system, the amplified PCR products were observed under UV light as single, compact bands of the anticipated size.
Purification and sequencing of the PCR product of the ITS-rDNA region
PCR products from all fungal isolates, amplified with ITS-1 and ITS-4 primers, yielded fragments of approximately 600 bp compared with 100 bp standard DNA ladder. These fragments were excised from a low-melting agarose gel (0.7%) utilizing a sharp, sterile scalpel blade while the gel was placed on a low-intensity (70%) UV transilluminator. The agarose gel pieces containing the fragments were collected in sterile, pre-weighed microcentrifuge tubes. The extracted PCR segments were purified by eluting the DNA from the agarose gel using a Qiagen gel extraction kit.18 At Eurofins Genomics India Private Limited in Bengaluru, India, the purified ITS-rDNA PCR products of the fungal isolates were sequenced using the Sanger dideoxy sequencing method with the forward and reverse primers (ITS-1 and ITS-4).
Alignment and phylogenetic analysis of fungi based on ITS rDNA sequences
Partial ITS rDNA gene sequences obtained from fungal isolates were compared with known sequences present at the GenBank database utilizing the BLAST algorithm accessible at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The sequences have been submitted to GenBank, and accession numbers have been acquired. The ITS rDNA gene sequences from the isolates were aligned and compared with known sequences in GenBank to identify the closest matches. MSA was performed using Clustal W.19 The Neighbor-Joining method in MEGA 11.0 software had been utilized to create a phylogenetic tree that shows the evolutionary interactions among the organisms. For accuracy, 1000 bootstrap replications were used.20-22 Based on the bootstrap values, the evolutionary relationships of the isolates with known fungal sequences were inferred using the ITS rDNA data.
Morphological characterization and microscopic examination of fungal isolates
Morphological characteristics such as growth pattern, colour, texture, sporulation time, and hyphal structure were documented. After 4-6 days of incubation, the isolates displayed typical colony development, with growth patterns ranging from dense to radial. Colony colours included white, black, and deep green, and textures varied from cottony to powdery. Sporulation occurred within 3-5 days. The cultural features of the isolates are detailed in Figure 1 and Table 1.
Table (1):
Morphological and microscopic characterization of fungal isolates
Fungal isolate |
Growth pattern |
Colour |
Texture |
No. of days for sporulation |
Hyphae |
|---|---|---|---|---|---|
Beauveria bassiana isolate VNMKV1 |
Radial and Dense |
White |
Velvety, Smooth, Cottony |
5 |
Present, septate hyphae |
Lecanicillium lecanii isolate VNMKV1 |
Radial |
White |
Flat cottony, Velvety |
6 |
Present, septate hyphae with thin wall |
Aspergillus niger isolate VNMKV1 |
Radial |
Black |
Powdery |
3-5 |
Present, septate hyphae |
Metarhizium anisopliae isolate VNMKV1 |
Radial |
Deep green |
Cottony and Powdery |
4 |
Present, septate hyphae |
Figure 1. Fungal mats grown on potato dextrose broth media
A. Beauveria bassiana; B. Lecanicillium lecanii; C. Aspergillus niger; D. Metarhizium anisopliae showing fully grown fungal mat on potato dextrose broth media; E. Beauveria bassiana showing cottony white appearance; F. Lecanicillium lecanii showing cottony white appearance; G. Aspergillus niger showing inner black and white colour outside; H. Metarhizium anisopliae showing deep green powdery mat
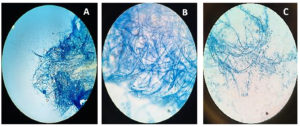
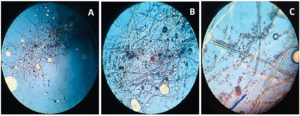
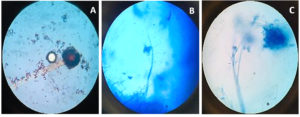
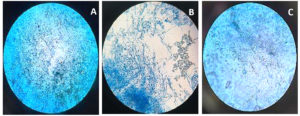
Beauveria bassiana, an entomopathogenic filamentous fungus, formed white colonies with a velvety and smooth cottony texture during conidia production. It displayed both radial and dense growth patterns on PDB, with sporulation occurring 5 days post-inoculation. Microscopically, the hyphae were fine, smooth, and septate, with distinct septations. The conidiophores were short and exhibited a zigzag arrangement for conidia formation (Figure 2). Aspergillus niger displayed a radial growth pattern, forming black colonies due to dense conidial production. The colony texture was powdery, and sporulation began 3 to 5 days after inoculation. Microscopically, septate hyphae were evident, with hyaline conidiophores ending in a globose vesicle, generating chains of conidia, resulting in the black powdery appearance (Figure 3). Lecanicillium lecanii showed radial growth, producing white colonies with a cottony to powdery texture. Septate hyphae were observed under 100X magnification, with sporulation occurring after 6 days (Figure 4). Metarhizium anisopliae exhibited a radial growth pattern, with colonies initially white that gradually turned green as sporulation advanced. The colonies had a powdery texture, indicating significant conidial production. Sporulation was observed 4 days post-inoculation. Microscopic analysis showed hyaline, branched, and septate hyphae, with conidiophores forming chains of conidia, contributing to the powdery consistency and greenish appearance of the mature colonies (Figure 5).
Figure 2. Microscopic observation of fungi Beauveria bassiana
A. Beauveria bassiana structure at 10X; B. Beauveria bassiana structure at 40X; C. Beauveria bassiana structure at 100X
Figure 3. Microscopic observation of fungi Aspergillus niger
A. Aspergillus niger structure at 4X; B. Aspergillus niger structure at 10X; C. Aspergillus niger structure at 40X
Figure 4. Microscopic observation of fungi Lecanicillium lecanii
A. Lecanicillium lecanii structure at 10X; B. Lecanicillium lecanii structure at 40X; C. Lecanicillium lecanii structure at 100X
Figure 5. Microscopic observation of fungi Metarhizium anisopliae
A. Metarhizium anisopliae structure at 10X; B. Metarhizium anisopliae structure at 40X; C. Metarhizium anisopliae structure at 100X
The results aligned with previous findings reported by Castillo et al., who documented similar morphological traits.23 Comparable characteristics were also noted by Krull et al. during their investigation into the morphology of Aspergillus niger.24 Humber observed analogous features in the morphological assessments of Beauveria bassiana and Metarhizium anisopliae isolates.13 These consistencies across studies support the reliability of the observed morphological patterns and validate the identification methods employed.
Molecular characterization of fungal isolates
Isolation of DNA from fungal isolates
In a comparative study by Serna-Dominguez et al., various DNA extraction techniques, including SDS, were evaluated.25 Their findings indicated that the SDS method was highly effective in producing high-quality DNA from EPF like B. bassiana & M. anisopliae. Similarly, Chauhan et al. examined several extraction approaches and found that SDS-based protocols were particularly suitable for isolating DNA from filamentous fungi, emphasizing the significance of reliable extraction for successful PCR amplification.26
PCR Screening of the ITS rDNA gene in fungal isolates
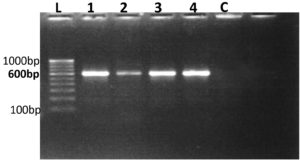
The ITS (internal transcribed spacer) region is commonly employed for fungal identification due to its distinctive features that enable precise species-level resolution. This segment of ribosomal DNA is well-suited for differentiating closely related fungal species. Serving as a reliable DNA barcode, the ITS region facilitates the accurate identification of fungi, including cryptic species that exhibit morphological similarity.27 The PCR performed with the universal primers ITS-1 & ITS-4 produced a monomorphic band of approximately 550 base pairs (bp) across all fungal isolates, confirming their identification (Figure 6).
Figure 6. PCR Amplification of fungal isolates using ITS rDNA sequences
L-100 bp to 1000 bp ladder, 1. Beauveria bassiana 2. Lecanicillium lecanii 3. Aspergillus niger 4. Metarhizium anisopliae C – Control
Previous studies have demonstrated that the ITS-1 and ITS-4 primers consistently generated a monomorphic band of around 600 bp across diverse fungal samples, including pathogenic fungi.28,29 Similarly, ITS region amplification via PCR of rDNA in Beauveria bassiana and Metarhizium anisopliae yielded a single amplicon of approximately 545 bp in all isolates. This was comparable to fragment sizes of around 500 bp reported for ITS-rDNA regions amplified with ITS1-ITS4 primers in eight isolates of B. bassiana & M. anisopliae, as well as a 560 bp fragment in other isolates of Beauveria bassiana.30,31
Purification and sequencing of PCR product
PCR products, approximately 550 bp in size, corresponding to the ITS rDNA region amplified using ITS1 and ITS4 primers, were purified with the Qiagen Gel Extraction Kit. The purified PCR amplicons underwent further processing, and sequencing was performed on the ~550 bp products derived from fungal isolates, with technical assistance from Eurofins Genomics India Private Limited, Bengaluru, India. The Sanger dideoxy sequencing method was utilized, and the consensus ITS rDNA sequences were stored in FASTA format. Similarly, Mahbouba et al. employed the Qiagen Gel Extraction Kit (France) for the purification of amplified fragments.32 In addition, Ramteke et al. conducted the genetic characterization of Lecanicillium sp. strains, where they obtained PCR amplicon products ranging from 600 to 800 bp from the ITS1/ITS4 rDNA regions.33
BLAST analysis of fungal isolates
The ITS rDNA gene sequences for the four fungal isolates, obtained in FASTA format, have been compared with the ones found in the NCBI database using the BLASTn search tool (Table 2). Comparative sequence analysis based on BLASTn parameters from NCBI indicated that one isolate belonged to the genus Akanthomyces and another to the genus Metarhizium, while the remaining isolates were identified as Beauveria and Aspergillus, respectively.
Table (2):
The BLAST analysis of 4 fungal isolates based on ITS rRNA gene sequencing showing related species with similarity and accession numbers
Fungal Isolate and Accession Number |
Related species |
Query cover (QC %) |
E-value |
Similarity (%) |
Accession number |
|---|---|---|---|---|---|
Beauveria bassiana isolate VNMKV 1 PQ279517.1 |
Beauveria bassiana strain 23D112_091_104 SSU rRNA gene, partial sequence; ITS 1, 5.8S rRNA gene, and ITS 2, complete sequence; and large subunit rRNA gene, partial sequence |
100 |
0.0 |
99.65 |
OQ891309.1 |
Lecanicillium lecanii isolate VNMKV 1 PQ270490.1 |
Lecanicillium lecanii isolate Vl-5 18S rRNA gene, partial sequence; ITS 1, 5.8S rRNA gene, & ITS 2, complete sequence; & 28S rRNA gene, partial sequence |
100 |
0.0 |
99.65 |
JN206647.1 |
Aspergillus niger isolate VNMKV 1 PQ272547.1 |
Aspergillus sp. isolate AUMS56 SSU-rRNA gene, partial sequence; internal transcribed spacer 1, 5.8S rRNA gene, & ITS 2, complete sequence; & large subunit rRNA gene, partial sequence |
99 |
0.0 |
99.33 |
MT802125.1 |
Metarhizium anisopliae isolate VNMKV 1 PQ270494.1 |
Metarhizium anisopliae strain CN094G8 SSU rRNA gene, partial sequence; ITS 1, 5.8S rRNA gene, & ITS 2, complete sequence; & large subunit rRNA gene, partial sequence |
100 |
0.0 |
99.82 |
ON006868.1 |
Based on the query coverage (QC), E-value, as well as sequence similarity, Beauveria bassiana isolate VNMKV 1 demonstrated the highest similarity (99.65%) to Beauveria bassiana strain 23D112, which included the partial SSU rRNA, ITS1, 5.8SrRNA gene, complete ITS2, & partial large subunit rRNA gene, under accession number OQ891309.34 The association between Akanthomyces lecanii and Lecanicillium lecanii is rooted in their taxonomic history and their common role as entomopathogenic fungi, with phylogenetic studies reclassifying Lecanicillium lecanii as Akanthomyces lecanii.35 In this investigation, the Lecanicillium lecanii isolate VNMKV 1 showed 99.65% similarity to Lecanicillium lecanii isolate Vl-5, comprising the partial 18S rRNA gene, complete ITS 2, 5.8S rRNA gene, ITS 1, as well as partial 28S rRNA gene, with accession number JN206647.
Furthermore, Aspergillus niger isolate VNMKV 1 exhibited the highest similarity (99.33%) with Aspergillus sp. isolate AUMS56, which includes the partial SSU rRNA gene, ITS 1, 5.8S rRNA gene, complete ITS 2, and partial large subunit rRNA gene, with accession number MT802125.1, a query cover of 99%, and an E-value of 0.0. The Metarhizium anisopliae isolate VNMKV 1 displayed the highest similarity (99.82%) with Metarhizium anisopliae strain CN094G8, comprising the partial SSU rRNA gene, 5.8S rRNA gene, ITS 1, complete ITS 2, as well as partial large subunit rRNA gene, with the accession number ON006868.
The sequences of these fungal isolates were submitted to the NCBI database, and accession numbers were assigned as follows: Beauveria bassiana isolate VNMKV 1 (PQ279517.1), Lecanicillium lecanii isolate VNMKV 1 (PQ270490.1), Aspergillus niger isolate VNMKV 1 (PQ272547.1), and Metarhizium anisopliae isolate VNMKV 1 (PQ270494.1), as shown in Table 2.
Similarly, the findings of Al Qadi et al. were consistent with the identification of fungal isolates (Beauveria bassiana, Lecanicillium lecanii, Aspergillus niger, Metarhizium anisopliae) based on ITS rDNA gene sequences and NCBI BLASTn analysis.36
Phylogenetic analysis using fungal isolates’ ITS rDNA sequences
Ribosomal DNA (rDNA) comprises several components, including the 18S, 5.8S, and 28S rDNA genes, along with the ETS (External Transcribed Spacer) as well as Internal Transcribed Spacer regions (ITS1 & ITS2).37 Both conserved gene regions and variable spacer regions serve as molecular markers for resolving phylogenetic relationships.38 While conserved regions such as 18S and 28S rDNA are typically used for higher-level phylogeny, the more variable ITS1 & ITS2 regions are favored for species-level identification due to their length variability.39,40 In this study, the ITS region was utilized for species-level identification of fungal isolates due to its high polymorphism and non-coding nature, which supports accurate species discrimination.41 The ITS region is recognized as the standard DNA barcode for fungi, providing precise identification at species and subspecies levels due to its hypervariable sequences.42,43
Phylogenetic analysis was carried out using ITS1-5.8S-ITS2 rDNA sequences from four fungal isolates: Beauveria bassiana VNMKV 1 (PQ279517.1), Lecanicillium lecanii VNMKV 1 (PQ270490.1), Aspergillus niger VNMKV 1 (PQ272547.1), and Metarhizium anisopliae VNMKV 1 (PQ270494.1) along with related sequences retrieved from GenBank (NCBI). Bootstrap analysis, conducted with 1,000 replicates, indicated the percentage of replicate trees in which taxa grouped together, shown below the branches.44 The tree was scaled to represent evolutionary distances calculated via the Maximum Composite Likelihood method, expressed as base substitutions per site.45
Phylogenetic analysis of Beauveria bassiana isolate VNMKV 1 based on ITS rDNA gene sequence
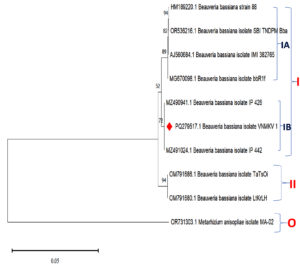
The phylogenetic analysis of Beauveria bassiana isolate VNMKV 1 (PQ279517.1) was conducted utilizing partial sequences from the small and large subunit rRNA genes, the complete 5.8S ribosomal RNA gene, and ITS regions 1 & 2. The analysis included eight closely related sequences identified via NCBI BLASTn, consisting of B. bassiana strain 88 (HM189220.1), isolate SBI TNDPM Bba (OR536216.1), isolate IMI 382785 (AJ560684.1), isolate bbR1f (MG670098.1), isolate IP 426 (MZ490941.1), isolate IP 442 (MZ491024.1), isolate TaTsOi (OM791686.1), and isolate LtKrLH (OM791680.1). The Metarhizium anisopliae isolate MA-02 (OR731303.1) was utilized as an outgroup to anchor the tree and facilitate the assessment of genetic relationships among the B. bassiana strains (Figure 7).
Figure 7. Phylogenetic analysis based on ITS-5.8S-ITS4 rDNA sequences of Beauveria bassiana isolate VNMKV 1 and other related sequences deposited in GeneBank of NCBI. Phylogenetic tree constructed based on the neighbor-joining method using MEGA 11 software. Bootstrap value >50% are showed and Metarhizium anisopliae isolate MA-02 was displayed as out-group taxa
The resulting phylogenetic tree divided the isolates into two primary clades. Clade I, supported by a bootstrap value of 52, encompassed B. bassiana strain 88, SBI TNDPM Bba, IMI 382785, bbR1f, IP 426, IP 442, and VNMKV 1, and was further subdivided into subclades IA and IB. Subclade IA showed strong support (89 bootstrap value) for the grouping of strain 88, SBI TNDPM Bba, IMI 382785, and bbR1f. A branch within this subclade, with a bootstrap value of 94, indicated a highly confident close evolutionary relationship between isolates TaTsOi and LtKrLH, suggesting that they likely share a recent common ancestor and exhibit minimal genetic divergence. Additionally, a bootstrap value of 82 in subclade IA indicated a moderately robust relationship between bbR1f and the cluster containing SBI TNDPM Bba, IMI 382785, and strain 88, indicating a close yet distinct evolutionary connection.
In subclade IB, the isolates IP 426, IP 442, and VNMKV 1 formed a cluster with a bootstrap value of 72, indicating a moderately high confidence in their close evolutionary relationship. Separately, in Clade II, a highly supported grouping (bootstrap value of 94) between isolates TaTsOi and LtKrLH suggested a strong phylogenetic closeness, likely due to a recent shared ancestor.
The phylogenetic tree showed that isolate VNMKV 1 was closely related to IP 426 and IP 442, suggesting a close evolutionary affinity among these isolates. In contrast, other isolates, such as strain 88, IMI 382785, and SBI TNDPM Bba, were more distantly related, occupying separate branches with distinct bootstrap support values. This pattern suggested that these isolates may share a more ancient common ancestor compared to the cluster containing VNMKV 1. The cluster including strain 88 and IMI 382785 appeared to have diverged earlier than the group containing VNMKV 1. As an outgroup, M. anisopliae MA-02 was positioned on a separate branch, exhibiting no significant bootstrap support with any B. bassiana isolates, thus affirming its distant relationship.
Phylogenetic analysis of Lecanicillium lecanii isolate VNMKV 1 based on ITS rDNA gene sequence
The phylogenetic relationships of Akanthomyces lecanii isolate VNMKV 1 (PQ270490.1) were deduced through an analysis of genetic sequences, including the SSU rRNA gene (partial), ITS regions 1 & 2, the full-length 5.8S rRNA gene, and the large subunit rRNA gene (partial). The classification recognized Lecanicillium and Akanthomyces as distinct genera, despite their close affiliation as entomopathogenic fungi, which infect insects. The genus Akanthomyces encompasses species previously categorized within Lecanicillium, such as Akanthomyces lecanii, whose reclassification was prompted by recent molecular data.46
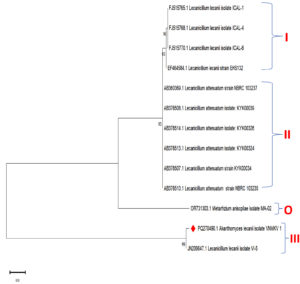
A phylogenetic tree was constructed to depict relationships among the eleven most closely related strains identified through NCBI BLASTn analysis. The sequences included Lecanicillium lecanii isolate ICAL-1 (FJ515765.1), isolate ICAL-4 (FJ515768.1), isolate ICAL-6 (FJ515770.1), strain EHS132 (EF464584.1), isolate VI-5 (JN206647.1), and Lecanicillium attenuatum strains such as NBRC 103237 (AB360369.1), KYK00039 (AB378508.1), KYK00326 (AB378514.1), KYK00324 (AB378513.1), KYK00034 (AB378507.1), and NBRC 103235 (AB378510.1). Metarhizium anisopliae isolate MA-02 (OR731303.1) served as an outgroup for rooting the tree (Figure 8).
Figure 8. Phylogenetic analysis based on ITS-5.8S-ITS4 rDNA sequences of Lecanicillium lecanii isolate VNMKV 1 and other related sequences deposited in GeneBank of NCBI. Phylogenetic tree constructed based on the neighbor-joining method using MEGA 11 software. Bootstrap value >50% are showed and Metarhizium anisopliae isolate MA-02 was displayed as out-group taxa
The phylogenetic analysis revealed three main clades. Clade I comprised L. lecanii isolates, specifically ICAL-1, ICAL-4, ICAL-6, and strain EHS132. Clade II encompassed L. attenuatum isolates, including strains NBRC 103237, KYK00039, KYK00326, KYK00324, KYK00034, and NBRC 103235. Clade III included Akanthomyces lecanii isolate VNMKV 1 and L. lecanii isolate VI-5, which formed a distinct cluster. In Clade I, the grouping of L. lecanii isolates ICAL-1, ICAL-4, and ICAL-6 exhibited high bootstrap support (values of 90 and 93), indicating a close evolutionary relationship and suggesting a shared recent ancestor. L. lecanii strain EHS132 was also closely associated with the other members of this clade. Clade II comprised a well-supported cluster (bootstrap value of 95) of L. attenuatum isolates, indicating that these isolates form a distinct lineage with a more recent common ancestor within L. attenuatum. Clade III was characterized by a close relationship between Akanthomyces lecanii isolate VNMKV 1 and Lecanicillium lecanii isolate VI-5, with a bootstrap value of 99, indicating strong support for their shared evolutionary history. Despite being categorized under different genera, their positioning indicated a close phylogenetic relationship. The outgroup, M. anisopliae MA-02, formed a separate branch on the tree, highlighting its distant genetic relationship with the Lecanicillium and Akanthomyces isolates, consistent with its divergent evolutionary history.
Phylogenetic analysis of Aspergillus niger isolates VNMKV 1 based on ITS rDNA gene sequence
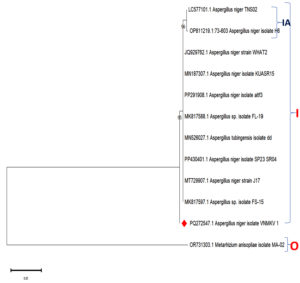
The phylogenetic relationships of Aspergillus niger isolate VNMKV 1 (PQ272547.1) were elucidated through the analysis of genetic sequences, including a partial sequence of the SSU rRNA gene, complete ITS regions 1 & 2, the full 5.8S rRNA gene, and a partial sequence of the large subunit rRNA gene. A phylogenetic tree had been constructed to explore evolutionary connections among the 10 closest strains identified through NCBI BLASTn analysis. The sequences used were A. niger isolate TNS02 (LC577101.1), A. niger isolate H6 (OP811219.1), A. niger strain WHAT2 (JQ929762.1), A. niger isolate KUASR15 (MN187307.1), A. niger isolate altf3 (PP291908.1), Aspergillus sp. isolate FL-19 (MK817588.1), A. tubigensis isolate dd (MN526027.1), A. niger isolate SP23 SR04 (PP430401.1), A. niger strain J17 (MT729907.1), and Aspergillus sp. isolate FS-15 (MK817597.1). The tree also included the target isolate, A. niger VNMKV 1, while Metarhizium anisopliae isolate MA-02 (OR731303.1) was utilized as an outgroup for comparison (Figure 9).
Figure 9. Phylogenetic analysis based on ITS-5.8S-ITS4 rDNA sequences of Aspergillus niger isolate VNMKV 1 and other related sequences deposited in GeneBank of NCBI. Phylogenetic tree constructed based on the neighbor-joining method using MEGA 11 software. Bootstrap value >50% are showed and Metarhizium anisopliae isolate MA-02 was displayed as out-group taxa
The phylogenetic analysis revealed a prominent cluster, designated Clade I, with a high bootstrap value of 95, indicating strong support. This clade encompassed multiple A. niger isolates, including TNS02, H6, WHAT2, KUASR15, altf3, and FL-19, as well as Aspergillus sp. isolate FL-19, A. tubigensis isolate dd, A. niger SP23 SR04, strain J17, and Aspergillus sp. isolate FS-15. Notably, A. niger isolate VNMKV 1 formed a distinct branch separate from Clade I, suggesting that it diverged earlier from the other A. niger strains. Within Clade I, a subclade (IA) emerged, consisting of A. niger isolates TNS02 and H6, supported by a high bootstrap value of 96, indicating robust evolutionary affinity.
In subclade IA, A. niger isolates TNS02, H6, and strain WHAT2 formed a tightly grouped cluster, supported by a 96% bootstrap value, which strongly suggests a shared recent ancestor among these strains. Clade I also included other A. niger isolates such as KUASR15, altf3, and FL-19, which were closely related & formed a subgroup with a bootstrap value of 95, indicating a well-supported evolutionary relationship. Although A. tubigensis isolate dd belonged to a different species, it was integrated within the larger A. niger cluster, indicating a closer phylogenetic relationship with these isolates despite its divergence.
Other A. niger isolates, including SP23 SR04, strain J17, and isolate FS-15, were found to be more distantly related within Clade I but still belonged to the same overarching A. niger grouping. The distinct branch of A. niger isolates VNMKV 1 indicated that while it was related to the other isolates, it may have diverged earlier, suggesting some level of genetic differentiation. The placement of Metarhizium anisopliae isolate MA-02 as an outgroup further emphasized its distant evolutionary relationship from the Aspergillus species, forming a separate branch as anticipated.
Phylogenetic analysis of Metarhizium anisopliae isolate VNMKV 1 based on ITS rDNA gene sequence
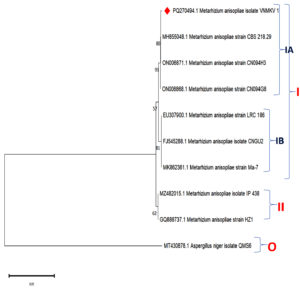
The evolutionary relationships of Metarhizium anisopliae isolate VNMKV 1 (PQ270494) were examined using partial sequences of several genetic markers, including the SSU rRNA gene, complete ITS regions 1 & 2, the 5.8S rRNA gene, and the large subunit rRNA gene. A phylogenetic tree was constructed to evaluate the evolutionary connections among eight closely related M. anisopliae strains identified through NCBI BLASTn analysis. The sequences used were from M. anisopliae strain CBS 218.29 (MH855048.1), strain CN094H3 (ON006871.1), strain CN094G8 (ON006868.1), strain LRC 186 (EU307900.1), isolate CNGU2 (FJ545288.1), strain Ma-7 (MK862361.1), and isolate IP 438 (MZ482015.1), in addition to isolate VNMKV 1. Aspergillus niger isolate QMS6 (MT430878.1) was included as an outgroup to provide a reference point for the tree’s root (Figure 10).
Figure 10. Phylogenetic analysis based on ITS-5.8S-ITS4 rDNA sequences of Metarhizium anisopliae isolate VNMKV 1 and other related sequences deposited in GeneBank of NCBI. Phylogenetic tree constructed based on the neighbor-joining method using MEGA 11 software. Bootstrap value >50% are showed and Aspergillus niger isolate QMS6 was displayed as out-group taxa
The phylogenetic tree displayed two primary clades. Clade I encompassed M. anisopliae strains CBS 218.29, CN094H3, CN094G8, LRC 186, isolate CNGU2, strain Ma-7, and isolate VNMKV 1, while Clade II comprised M. anisopliae isolates IP 438 and strain HZ1, with the latter two grouped together at a bootstrap value of 62, indicating moderate support. Within Clade I, two subclades were distinguished: IA and IB, with a bootstrap support of 57 separating them. Subclade IA, well-supported by a bootstrap value of 91, contained isolate VNMKV 1, strain CN094H3, strain CN094G8, and strain CBS 218.29, which were further clustered with an internal bootstrap value of 80. In contrast, Subclade IB included M. anisopliae strains LRC 186, CNGU2, and Ma-7, grouped at a bootstrap value of 81.
Clade I, representing a large cluster of M. anisopliae isolates, indicated a close evolutionary relationship among these entomopathogenic fungi, which are commonly used for insect pest management. The strong bootstrap values within Clade I suggest recent evolutionary divergence among the strains. Subclade IA demonstrated a high degree of genetic affinity, especially among isolate VNMKV 1 and strains CN094H3, CN094G8, and CBS 218.29, indicating a shared recent ancestor. While isolate VNMKV 1 formed a distinct branch within subclade IA, it maintained a close association with strain CBS 218.29, suggesting an early divergence in the evolutionary history of these strains.
Subclade IB, which included strains LRC 186, CNGU2, and Ma-7, showed significant genetic similarity, supported by a bootstrap value of 81. This clustering suggested that these strains evolved from a common ancestral lineage, albeit slightly more distantly related to subclade IA. The inclusion of these isolates in Clade I highlighted their overall evolutionary proximity to other members of the group. Clade II featured the more genetically distinct M. anisopliae isolates IP 438 and HZ1, separated from Clade I by a moderate bootstrap value of 62. The close relationship between isolate IP 438 and strain HZ1 suggested a more recent common ancestry compared to the strains in Clade I. Nevertheless, these isolates were still part of the broader M. anisopliae group. The outgroup, Aspergillus niger isolate QMS6, was positioned on a separate branch, serving as a reference for rooting the phylogenetic tree. Its placement outside the Metarhizium clusters underscored its distant evolutionary relationship with the entomopathogenic fungi, confirming its divergence from the group.
Perez-Gonzalez et al. conducted a comparable study involving Beauveria and Metarhizium species, utilizing internal transcribed spacer (ITS) sequences for species identification and submitting the sequence data to GenBank to obtain accession numbers.47 Gebremariam et al. validated the identity of fungal isolates through phylogenetic analysis of ITS-rDNA sequences, identifying seven isolates as Beauveria bassiana and five as Metarhizium anisopliae.48 Similarly, ITS-rDNA amplification and sequencing have been widely accepted as reliable methods for distinguishing fungal species, particularly for taxa like B. bassiana & M. anisopliae.49
Fungal isolates displayed diverse colony textures, ranging from cottony to powdery, with growth patterns from dense to radial. Colony colours included white (B. bassiana, L. lecanii), black (A. niger), and dark green (M. anisopliae) with sporulation times of 3 to 6 days and septate hyphae observed in all isolates.
Phylogenetic analysis based on ITS1-5.8S-ITS4 rDNA sequences for four fungal isolates revealed that clade of B. bassiana isolate IP 426 (MZ490941.1), B. bassiana isolate IP 442 (MZ491024.1), and B. bassiana isolate VNMKV 1 (PQ279517.1) was formed with a moderate to high bootstrap value of 72, suggesting a close evolutionary relationship. Also, L. lecanii isolate VNMKV 1 (PQ270490.1) and L. lecanii isolate VI-5 (JN206647.1), which formed a well-supported clade with a high bootstrap value of 99, A. niger isolate VNMKV 1 included ten closely related strains, with a well-supported Clade I having a bootstrap value of 95. M. anisopliae isolate VNMKV 1 (PQ270494.1), strain CBS 218.29 (MH855048.1), strain CN094H3 (ON006871.1), and strain CN094G8 (ON006868.1). formed a clade at the strong bootstrap value of 91, suggesting a high degree of confidence in the evolutionary relationships within this clade, indicating that these strains shared a recent common ancestor.
ACKNOWLEDGMENTS
The authors would like to thank Rajiv Gandhi Science and Technology Commission (RGSTC), Mumbai, for financial support to carry out this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
The research work is funded by Rajiv Gandhi Science and Technology Commission (RGSTC), Mumbai, India, vide grant number CAET/RGSTC/144/2024 dated 15.05.2024.
DATA AVAILABILITY
All datasets generated or analysed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Koul OKO. Microbial biopesticides: opportunities and challenges. Cabi Reviews. 2011;6(056):1-26.
Crossref - Ayilara MS, Adeleke BS, Akinola SA, et al. Biopesticides as a promising alternative to synthetic pesticides: A case for microbial pesticides, phytopesticides, and nanobiopesticides. Front Microbiol. 2023;16(14):1040901.
Crossref - Litwin A, Nowak M, Rozalska S. Entomopathogenic fungi: unconventional applications. Rev Environ Sci Biotechnol. 2020;19(1):23-42.
Crossref - Erler F, Ozgur AA. Potential of two entomopathogenic fungi, Beauveria bassiana and Metarhizium anisopliae (Coleoptera: Scarabaeidae), as biological control agents against the June beetle. J Insect Sci. 2015;15(1):44.
Crossref - Shin TY, Lee MR, Park SE, Lee SJ, Kim WJ, Kim JS. Pathogenesis-related genes of entomopathogenic fungi. Arch Insect Biochem Physiol. 2020;105(4):e21747.
Crossref - Nadeau MP, Dunphy GB, Boisvert JL. Development of Erynia conica (Zygomycetes: Entomophthorales) on the Cuticle of the Adult Black Flies Simulium rostratum and Simulium decorum (Diptera: Simuliidae). J Invertebr Pathol. 1996;68(1):50-58.
Crossref - Malekan N, Hatami B, Ebadi R, Akhavan A, Radjabi R. Evaluation of entomopathogenic fungi Beauveria bassiana and Lecanicillium muscarium on different nymphal stages of greenhouse whitefly Trialeurodes vaporariorum in greenhouse conditions. Biharean Biologist. 2015;9(2):108-112.
- Reddy SGE. Lecanicillium spp. for the management of aphids, whiteflies, thrips, scales and mealy bugs: Review. In Ramon E, Rebolledo R (eds.), Arthropods – Are They Beneficial for Mankind? Published by Intech Open, London (U.K.). 2020.
Crossref - Nayak S, Samanta S, Mukherjee AK. Beneficial role of Aspergillus sp. in agricultural soil and environment. In: Nayak SK, Mishra BB R (eds.), Frontiers in Soil and Environmental Microbiology 2020;3:17-36.
Crossref - Borase GS, Kulkarni MV, Kharge AP. Knowledge of farmers regarding Biomix. The Pharma Innovation Journal. 2022;11(3);303-306.
- Wang L, Keyhani NO, Xia Y, Xie J. The potential and limitations of entomopathogenic fungi as biocontrol agents for insect pest management. Entomologia Generalis. 2024; 44(4):797.
Crossref - Gautam AK, Verma RK, Avasthi S, et al. Current insight into traditional and modern methods in fungal diversity estimates. Journal of Fungi. 2022; 8(3):226.
Crossref - Humber RA. Identification of entomopathogenic fungi. In: Lacey LA (eds.), Manual of Techniques in Invertebrate Pathology. Elsevier 2012;2:151-187.
Crossref - Bich GA, Castrillo ML, Kramer FL, Villalba LL, Zapata PD. Morphological and molecular identification of entomopathogenic fungi from agricultural and forestry crops. Floresta e Ambiente. 2021; 28(2):e20180086.
Crossref - Driver F, Milner RJ, Trueman JWH. A taxonomic revision of Metarhizium based on a phylogenetic analysis of rDNA sequence data. Mycol Res. 2000;104(2):134-150.
Crossref - Alviti Kankanamalage HP, Yang JY, et al. Entomopathogenic fungi: insights into recent understanding. World Journal of Microbiology and Biotechnology. 2025;41(6):179.
Crossref - Prabhu AS, Filippi MC, Araujo LG, Faria JC. Genetic and phenotypic characterization of isolates of Pyricularia grisea from the rice cultivars Epagri 108 and 109 in the State of Tocantins. Fitopatol Bras. 2002;27(6):566-573.
Crossref - Sambrook J, Russell DW. Detection of DNA in agarose gels. Cold Spring Harb Protoc. 2006;2006(1):4022.
Crossref - Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-4680.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229-1235.
Crossref - Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022-3027.
Crossref - Castillo MG, Rivera IA, Padilla AB, Lara F, Victoriano CN, Herrera RR. Isolation and identification of novel entomopathogenic fungal strains of the Beauveria and Metarhizium generous. BioTechnol Indian J. 2012;6:386-395.
- Krull R, Cordes C, Horn H, et al. Morphology of Filamentous Fungi: Linking Cellular Biology to Process Engineering Using Aspergillus niger. In: Wittmann, C., Krull, R. (eds) Biosystems Engineering II. Advances in Biochemical Engineering/Biotechnology, vol 121. Springer, Berlin, Heidelberg. 2010;121:1-21.
Crossref - Serna-Dominguez MG, Andrade-Michel GY, Arredondo-Bernal HC, Gallou A. Two efficient methods for isolation of high-quality genomic DNA from entomopathogenic fungi. J Microbiol Methods. 2018;148:55-63.
Crossref - Chauhan BM, Kunjadiya KM, Parekh SN. Comparison of Extraction Techniques for Isolation Fungal DNA. Int J Res Appl Sci Eng Technol. 2022;10(4):1413-1419.
Crossref - Harnelly E, Kusuma HI, Thomy Z, Samingan S. Internal Transcribed Spacer (ITS) gene as an accurate DNA barcode for identification of macroscopic fungus in Aceh. Biodiversitas Journal of Biological Diversity. 2022;23(5).
Crossref - Sousa GS, De Oliveira RS, Souza AB, et al. Development of PCR-Multiplex Assays for Identification of the Herpotrichiellaceae Family and Agents Causing Chromoblastomycosis. J Fungi. 2024;10(8):548.
Crossref - Salami SA, Babafemi EM, Ossai FE. Application of Electrical Resistivity Sounding Method for Groundwater Exploration in Ugboshi-Afe, Akoko-Edo, Southwestern Nigeria. J Appl Sci Environ Manag. 2024;28(5):1345-1353.
Crossref - Gebremariam A, Chekol Y, Assefa F. Phenotypic, molecular, and virulence characterization of entomopathogenic fungi, Beauveria bassiana (Balsam) Vuillemin, and Metarhizium anisopliae (Metschn.) Sorokin from soil samples of Ethiopia for the development of mycoinsecticide. Heliyon. 2021;7(5).
Crossref - Belay YC, Meressa BH, Alemu T, Hallmann J. Molecular detection of the entomopathogenic fungus Beauveria bassiana isolates from soils of coffee growing areas in Ethiopia using rDNA-ITS. J Appl Biosc. 2017;119:11943-53.
- Mahbouba B, Le Roux C, Nadir B, Nadia Y, Abdelhamid D. Phenotypic and molecular characterization of plant growth promoting Rhizobacteria isolated from the rhizosphere of wheat (Triticum durum Desf.) in Algeria. Afr J Microbiol Res. 2013;7(23):2893-2904.
Crossref - Ramteke BD, Thakur KD, Bramhankar SB, Gawande SP, Isokar SS, Ramteke D. Molecular characterization and Phylogenetic analysis of Lecanicillium spp. The Pharma Innovation Journal. 2022;11(8):1029-1032.
- Rizal LM, Hereward JP, Brookes DR, Furlong MJ, Walter GH. Hidden diversity within Beauveria and Metarhizium-comparing morphology, barcoding, multilocus phylogenies and whole-genome sequences. Fungal Ecology. 2024;67:101304.
Crossref - Manfrino R, Gutierrez A, Diez del Valle F, et al. First Description of Akanthomyces uredinophilus comb. nov. from Hemipteran Insects in America. Diversity. 2022;14(12):1118.
Crossref - Al Qadi I, Farrah I, Iraki N. Determination of Genetic variation among Beauveria bassiana and Metarhizium anisopliae Isolates from Palestine and other Geographic regions using ITS and B-tubulin Sequences, RFLP and ISSR Data. Rflp and Issr Data. Bethlehem University Journal. 2023.
Crossref - Hillis DM, Moritz C, Porter CA, Baker RJ. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science. 1991;251(4991):308-310.
Crossref - Wheeler WC, Bang R, Schuh RT. Cladistic relationships among higher groups of Heteroptera: congruence between morphological and molecular data sets. Insect Syst Evol. 1993;24(2):121-137.
Crossref - Schuh RT, Weirauch C, Wheeler WC. Phylogenetic relationships within the Cimicomorpha (Hemiptera: Heteroptera):a total evidence analysis. Syst Entomol. 2009;34(1):15-48.
Crossref - Marcilla A, Bargues MD, Ramsey JM, et al. The ITS-2 of the nuclear rDNA as a molecular marker for populations, species, and phylogenetic relationships in Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mol Phylogenet Evol. 2001;18(1):136-142.
Crossref - Haque Z, Iqbal MS, Ahmad A, Khan MS, Prakash J. Molecular characterization of Trichoderma spp. isolates by internal transcribed spacer (ITS) region sequencing technique and ITS use as a biocontrol agent. Open Biotechnol J. 2020;14(1):14-70.
Crossref - Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241-6246.
Crossref - Toju H, Tanabe AS, Yamamoto S, Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PloS one. 2012;7(7):40863.
Crossref - Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030-11035.
Crossref - Isma’il S, Deba FA, Ladan MA, et al. In vitro pathogenicity of Akanthomyces lecanii and Metarhizium anisopliae against the aphid Aphis craccivora. InAnales de Biologia Servicio de Publicaciones de la Universidad de Murcia. 2023;45:97-106.
Crossref - Perez-Gonzalez VH, Guzman-Franco AW, Alatorre-Rosas R, et al. Specific diversity of the entomopathogenic fungi Beauveria and Metarhizium in Mexican agricultural soils. J Invertebr Pathol. 2014;119:54-61.
Crossref - Gebremariam A, Chekol Y, Assefa F. Phenotypic, molecular, and virulence characterization of entomopathogenic fungi, Beauveria bassiana (Balsam) Vuillemin, and Metarhizium anisopliae (Metschn.) Sorokin from soil samples of Ethiopia for the development of mycoinsecticide. Heliyon. 2021;7(5):e07091.
Crossref - Garcia JL, Sotelo P, Monroy DM,et al. Identification and characterization of a Beauveria bassiana (Bals.) Vuill. isolate having a high potential for the control of the Diatraea sp. sugarcane stem borer. Biotecnologia Aplicada. 2018;35(1):1201-1207.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.