Environmental and biotic stresses are increasing globally due to anthropogenic activities. Omics approach including metagenomics, metatranscriptomics and interactome network analysis provide an insight into a comprehensive understanding of the plant’s response to abiotic stress as heat-cold, drought and salinity. Understanding the structure and function of plant-associated microbial communities, their taxonomic composition, functional potential, dynamics of plant soil processes along with plant-soil interactions, is essential for strategizing sustainable agricultural strategies and advancing plant improvement tools, like CRISPR-Cas technologies. Transcriptome profiling using techniques, such as digital gene expression (DGE), RNA sequencing (RNA-seq), or SAGE (serial paired oligo-nucleic acid sequencing) have been done in crops like Angelica sinensis, Zea mays and other major cereal crops like wheat revealing information regarding the key regulators which play a positive role in controlling the abiotic stress responses. Chromatography techniques like gas chromatography-MS (GC-MS) and LC-MS/MS are widely used in metabolomics research due to their vast coverage of large metabolites in crops like mangosteen (Garcinia mangostana Linn.). In this article, we explain with examples, the network of transcriptional factors, plant immune hormones crosstalk and the signalling molecules involved in improved plant tolerance to abiotic stresses. We outline the instances where ‘omics’ research has pushed the boundaries of information about plant metabolites, plant gene expression pattern, soil and endophytic plant community composition, with a comprehensive view of recent advances in omics-driven research on plant gene expression, metabolites, and plant-soil-microbe interactions.
Metagenomics, Plant Abiotic Stresses, Gene Regulation, Plant Omics, Proteomics, Transcriptomics
The production of sustainable agriculture in the twenty-first century faces a grave threat from climate change. Numerous studies have shown that biotic stressors (30%) and abiotic stressors (50%) are responsible for losses in global agricultural production, which is the main threat to future food security worldwide.1 Abiotic stressors primarily consist of heat stress, cold stress, salinity, moisture, light intensity, drought, and lack of nutrition.2 As projected by the IPCC, atmospheric CO2 concentrations are also expected to rise to 605-755 ppm by 2070, with global temperatures increasing by 1.5 °C between 2015-2050 and up to 3.0 °C by 2100. Simulation studies assess the combined impact of elevated CO2 and temperature levels suggesting how they negatively affect the yields of key crops such as rice (13%), maize (13%), jowar (21%), bajra (39%), wheat (25%) and barley (46%) during the kharif season when the temperature was increased by 3 °C.3
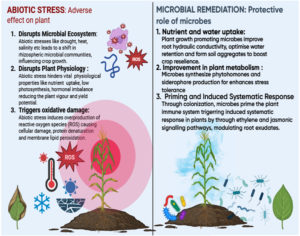
Microbial interactions are crucial for survival of soil ecosystem since climate change has altered plant chemoattractant profiles and the carbon-to-nitrogen (C/N) ratio, thereby influencing plant-microbe interactions. Awareness about the associations and roles of various microbial communities requires an awareness of these interactions in soil microbial communities and biotic-abiotic variables. Among the several microbial interactions, plant-microbe interaction is the most important for maintaining the equilibrium in an ecosystem.4 Arbuscular mycorrhizal fungi (AMFs) mitigate abiotic stress through multiple mechanisms, including enhanced mineral uptake, improved water absorption, ionic balance, phytohormone modulation, increased photosynthetic efficiency, and elevated antioxidant enzyme production. Elevated soil Naz and Cl– levels under stress can hinder the transport of essential nutrients like Ca2+, P, K+, and Mg2+.5 Plants create a multitude of inorganic and organic substances that contribute to the establishment of a nutrient-enriched environment that is advantageous for the substantial colonization of a diverse range of microorganisms.6 These interactions will help us decipher the role of microbes in enhancing plant health with reduced chemical inputs. Osmoprotectants like proline, serine, GABA (g-aminobutyric acid), myo-inositol, D-pinitol, sucrose, trehalose, etc., are secreted by plants to help maintain cellular balance during stress conditions. Another aspect of plant microbe interaction is knowing how plant pathogens are responsible for causing diseases the plant. It is widely reported that effector proteins manipulate plant defense mechanisms and metabolism to the pathogen’s benefit, there is a significant knowledge gap about the specific roles played by the numerous effectors.7 All of this demonstrates that the diversity of microbial communities and their interactions with the plants are responsible for maintaining the ecological stability and for providing several benefits for the plant growth, disease resistance, and stress tolerance.8 Microbes enhance plant defenses, promote health, and improve adaptability to environmental stress. During drought, they activate molecular pathways that boost water uptake, stimulate root growth, and increase stress-related hormones and enzymes. They also produce osmoprotectants, which help maintain cell turgor and water balance. Additionally, some microbes modify root structure, enhancing nutrient and water absorption, thereby increasing the plant’s resilience to drought conditions.9 Certain soil bacteria produce polyextremophilic enzymes such as cellulases, xylanases, proteases, amylases, lipases, and gelatinases, which aid in plant resilience. Under drought conditions, the root microbiomes of crops like rice (Oryza sativa) and sorghum (Sorghum bicolor) are enriched with Actinobacteria, which may enhance stress tolerance even under hypoxic conditions. Additionally, many bacteria produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase to regulate ethylene levels, reducing stress-induced growth inhibition.10 Additionally, it has been reported that Pseudomonas cedrina, Brevundimonas terrae, and Arthrobacter nicotianae promote plant health under low-temperature stress.11
Deciphering the impact of the environment on plant-microbe interactions is crucial to understand the symbiotic associations, inhibiting phytopathogens, and developing effective biocontrol agents for maximum crop productivity. Better understanding of plant-microbe interactions can be attained by integrating multi-omics approaches, which combine experimental and computational methods to identify the unique microbial role in the system and various other mechanism that are involved in abiotic stress tolerance Comprehensive information about plant soil processes is essential for developing newer varieties and to modify cultivation practices for boosting crop productivity, increased stress tolerance and phytopathogen control and also for optimizing nutrient use efficiency.
Plant response to stresses
The plant immune system is triggered by microbial signals of two different kinds and is comprised of an intricate network of transcriptional factors, hormone crosstalk, and signaling molecules. Broadly conserved microbe/pathogen-associated molecular patterns (PAMPs) such as chitin and flagellin are included in the first category. PAMP-triggered immunity (PTI), which is thought to be the main element of basal defense against all microorganisms, is due to recognition of PAMPs. However, PTI is frequently inhibited by developed pathogens, primarily through virulence “effector” proteins that the pathogens introduce into the plant cell. Plants have developed the ability to detect individual effector proteins, which are a type of microbial signal that counteracts pathogen virulence. This recognition is made possible by nucleotide-binding leucine-rich repeat (NLR) immune receptors, which trigger a more potent type of immunity known as effector-triggered immunity (ETI). When plant immune receptors (R proteins) detect pathogen effectors, they trigger ETI. This often involves a signaling cascade that leads to localized cell death (hypersensitive response) and systemic resistance. Calcium signaling plays a key role in this process-recognition of effectors causes a rapid and sustained rise in cytoplasmic calcium, which activates defense-related genes, strengthens cell walls, and initiates programmed cell death to stop the spread of infection.12 PTI and ETI have distinct ways of perceiving signals, but they have several downstream reactions in common. Elevated abiotic stress brought on by climate change and global warming further worsens the problem of plant response against stress. It is well established that the microbial community that colonizes the rhizosphere and the surrounding soil influences plant’s ability to withstand abiotic stresses as presented in the Figure 1. In continuation to this, effect of microbes in plant abiotic stress response has been briefly described in Table 1. Plants defense mechanism, mainly hormonal, is strengthened by biological and chemical priming. Priming, also known as acclimatization, is a complicated process that involves preconditioning the plant’s abiotic and immunological systems to increase the speed, potency and effectiveness of its responses to stress.13 Multi-omics studies have demonstrated the potential to greatly advance our understanding of rhizospheric science by enabling the characterization of beneficial microorganisms associated with plants and their roles.14
Table (1):
Effect of microbes in plant abiotic stress response
Abiotic Stress |
Effect on Plant & Microbiome |
Microbial Role |
Ref. |
|---|---|---|---|
Salt stress |
Alters microbial community in rhizosphere and phyllosphere; affects plant genotype |
Pseudomonas spp. modulate phytohormones, improve water and ion uptake, and soil structure |
15 |
Low temp. |
Affects plant metabolism and survival |
Microbe-plant interactions enhance starch and carbohydrate metabolism for cold tolerance |
16 |
High temp. |
Induces oxidative stress in plants |
Some microbes trigger production of ROS to help plants endure heat |
17 |
Plant omics
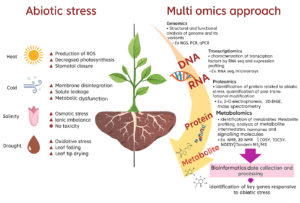
Use of Omics tool is a modern approach that offers technological advancement and multidisciplinary approach to enhance our understanding of all genomic and transcriptomic events that occur inside the plants.18 It helps the plant during abiotic stress by recognizing key genes responsible as presented in Figure 2. Genomic, transcriptomic and metabolomic techniques provide data for over-expression/under-expression of gene of interest.17 Reference genetic databases, transcriptomic, metabolomic databases of plant response under varied situations include KBase, MOCAT2, Metawrap, DRAM/DRAM-v. Bioinformatic programmes included in metagenome assembled genome (MAG) refinement programme use tools like FastQC, MultiQC, Meta-IDBA, Maxbin2, Check M and Meta Gene Mark for sequence reads.19 Arabidopsis, used as a starting point for developing models that aid in examining different aspects of the complex interaction between bacteria and plants.20 The effective application of different markers for early-generation population monitoring, such as in marker-assisted crossbreeding programmed has been made possible by these resources in crops. The paradigm of crop breeding has been greatly altered by use of genomic selection and CRISP-Cas systems like CRISPR-Cas9, CRISPRi (CRISPR interference) and CRISPR-Cpf1.21 When combined, the abundance of omics resources and breeding techniques enabled by omics would improve genetic gain in crop development and hasten the release of crop varieties that satisfy growing productivity and quality standards in crops like wheat, maize, soybean and tomato.22 In a study, Paraburkholderia phytofirmans PsJN displayed a distinct gene expression response when exposed to osmotic stress within its host plant since plants frequently experience shifts in their cellular redox state in response to environmental changes and developmental cues.23 The effective application of different markers for early-generation population monitoring, such as in marker-assisted crossbreeding programs has been made possible by these that enhance productivity, maximize nutrient resource efficiency are being developed.24
Genomics
DNA and RNA are the nucleic acids that carry genetic information collectively known as a genome. Advances in molecular biology methods have sped up the process of characterizing the genome, analyzing gene expression, and sequencing high-throughput genomes. It’s through the high throughput next-generation sequencing (NGS) technology, that a genome can be decoded. The process involves isolating genomic DNA, amplifying it through Polymerase Chain Reaction (PCR), sequencing the DNA and evaluating the integrity of the sequence. Large-scale genomic studies have been made possible by the sequencing, assembly and structural and functional annotation of DNA, which also explains the relationships between genomic products on organism as well as cellular levels RNA interference (RNAi) approaches have been employed to reveal the functions of different genes in plant species’ responses to abiotic stress.
The overexpression of the Tubby-like protein gene GmTLP8 in soybeans increased their resistance to salt stress and drought, whereas its silencing reduced their tolerance.25 Many genes that regulate a plant’s response to drought have been found through studies on gene expression. The majority of the genes that are regulated by drought also respond to light stress (KNAT3, KNAT4, SEN1, DIN9, DIN10, and ACP4) and/or circadian rhythm (e.g., CCA1, WNK1, and FSD1), according to the results, suggesting that drought may have an impact on a plant’s ability to respond to light and/or circadian cycles. Accumulation of abscisic acid (ABA) during drought conditions triggers a series of events for closing the stomata to stop transpiration of water. Consequently, stomatal movement assisted by ABA is a desirable target in the quest to increase agricultural drought resistance. Under drought stress, abscisic acid blocks the action of plasma membrane proton (H+)-ATPases, which causes stomatal closure. Tissue-specific Cas9 cassette has been successfully used to modify the gene encoding OPEN STOMATA 2 (OST2), formerly known as AHA1, in Arabidopsis. This has improved ABA-induced stomatal closure and reduced transpirational water loss when compared to the wild-type and T-DNA insertion mutant. MicroRNAs or miRNAs are viable targets for genetic engineering aimed at enhancing crop resistance to abiotic stress. Transgenic plants with modified miRNA expression have shown increased sensitivity or tolerance to various abiotic stressors, contingent on the target genes.26 Similarly, drought tolerance is demonstrated by the null mutant, or so-called miR169a gene knockout, which is a negative factor of drought tolerance regulated by the ABA-dependent pathway generated using the dual-sgRNA CRISPR/Cas9 system. It suggests functional knockout of miRNA using CRISPR/Cas system as a viable strategy for miRNA-based crop breeding. Molecular markers are useful for marking genomic features, including tolerance to abiotic stress, quantitative analysis, and disease resistance. It has opened up new possibilities for the genetic enhancement of features resistant to pressures like salt, drought, and other environmental factors. Thus far, a number of molecular markers have been reported to assist in the identification of polymorphism in plants. These markers consist of sequence-tagged sites (STS), restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), single-nucleotide polymorphism (SNP) and simple sequence repeats (SSR).
Transcriptomics
The term “transcriptome” refers to the complete set of transcripts in an organism’s cell. Study of transcriptome is known as transcriptomics. It primarily aids in the identification of RNA or gene transcripts linked to a plant’s phenotypic expression under various environmental circumstances. A group of genes known as regulatory genes and functional genes are elicited by different stressors in plants. These stressors induce the initiation of different proteins that initiate signalling pathways for providing tolerance against stress. The transcription factors (TFs) encoded by the regulatory group of genes control many genes that respond to stress in a collective and distinct manner. To conduct a transcriptome study, a range of techniques, such as digital gene expression (DGE), RNA sequencing (RNA-seq), or SAGE (serial paired oligo-nucleic acid sequencing) are used in crops like Angelica sinensis, a medicinal plant, known for its pharmacological and anti-inflammatory properties. The genes annotated through DGE technique gave information on the biosynthesis of secondary metabolites, signal transduction, and transcriptome analysis in regard to the early flowering of the plant.27 Metagenomes are characterized using True-Seq (soil and rhizosphere) or Nextera XT Low-Input methods (root endosphere). NGS technologies, which include the 454 and MiSeq platforms, facilitate the identification of complex microbial networks and the connections between the roles of the corresponding niche and the microbial community.28 Researchers use well-established databases like PlantExp, PPRD, Genevestigator, ePlant, and PlantGenIE, which offer a variety of transcriptomic data sets for many crop species, to access bulk transcriptome data for crop plants.29 Recent transcriptomic studies have provided valuable insights into how plants respond to environmental and nutrient-related stress at the molecular level. In the case of drought stress, certain histidine kinases like AHK1/ATHK1 have been identified as positive regulators of abscisic acid (ABA)-dependent stress responses, helping the plant cope with water scarcity. Conversely, cytokinin-related receptors such as AHK2, AHK3, and CRE1 appear to negatively influence the drought response, suggesting a fine balance between different hormonal pathways. Similarly, under iron deficiency, plants like tomato and Arabidopsis activate specific genetic mechanisms to manage nutrient uptake. The FER gene in tomato and its counterpart FIT in Arabidopsis play key roles in iron absorption. Additionally, several transcription factors from the bHLH family-including AtbHLH38, AtbHLH39, AtbHLH100, and AtbHLH101-are significantly upregulated in Arabidopsis under iron stress, indicating a strong transcriptional response to nutrient limitations. These findings help deepen our understanding of how plants adapt to stress and offer potential genetic targets for improving crop resilience. Transcriptome profiling of Bacillus mycoides EC18 revealed upregulation of genes related to amino acid metabolism, regulatory proteins, and signaling pathways, suggesting their role in ecological adaptation of the endophyte.30 In rice, inoculation with Trichoderma asperellum SL2 triggered significant upregulation of genes involved in photosynthesis, hormone signaling and nutrient uptake. Specifically, genes like RBCS and its isoforms promoted Rubisco biosynthesis has been corrected to specifically, the nuclear-encoded RBCS multigene family, which produces the small subunits of Rubisco, promoted Rubisco biosynthesis while CYP38 and CYP20-2 were linked to improved stress tolerance. The expression of OsGAE1 and MOC1 pointed to enhanced growth regulation through gibberellin pathways and tiller development, respectively. A comprehensive tabulated form of transcriptomic approaches to reveal gene functions is presented in Table 2.
Table (2):
Transcriptomic approaches to reveal gene functions and implications in crop stress responses
Crop |
Stress |
Method |
Key Findings |
Implications |
Ref. |
|---|---|---|---|---|---|
Wheat (Triticum aestivum) |
Nitrogen Use Efficiency (NUE) |
RNA-Seq (Comparative transcriptomics) |
Gene expression shifts regulating NUE identified through transcriptome profiling. |
Supports development of wheat lines with improved nitrogen efficiency. |
31 |
Maize (Zea mays) |
Abiotic stress (heat, cold, salinity, drought) |
RNA-Seq (B73 seedling leaves) |
5,330 DEGs identified; stress response linked to hormone signaling and TFs. |
Aids breeding of stress-resilient maize cultivars. |
32 |
Arabidopsis (Arabidopsis thaliana) |
Drought stress, ABA signaling |
Transcriptome profiling |
AHK1/ATHK1 (positive ABA response); AHK2, AHK3, CRE1 (negative regulators) identified. |
Reveals hormone cross-talk in drought adaptation. |
33 |
Tomato (Solanum lycopersicum) & Arabidopsis (Arabidopsis thaliana) |
Iron (Fe) deficiency stress |
Transcriptome analysis |
FER in tomato; FIT and bHLH TFs upregulated in Arabidopsis. |
Key regulatory genes identified for improving iron uptake under Fe stress. |
34 |
Rice (Oryza sativa) |
Microbial stimulation, photosynthesis, stress tolerance |
RNA-Seq (with T. asperellum SL2 inoculation) |
Upregulation of genes for Rubisco synthesis (RBCS, OsRBCS1, OsRBCS2), stress tolerance (CYP38, CYP20-2), growth (MOC1), phosphorus uptake (OsPHR2). |
Suggests enhanced photosynthetic efficiency, nutrient acquisition, and growth from microbial interaction. |
35 |
Proteomics
Proteomics is the study of the entire protein sequence that is present in a cell, species or an organ in a specific moment/experimental set-up.36 A gene expresses itself differently in response to varied environments, including biotic and abiotic stresses. Consequently, cells synthesize different sets of proteins. Therefore, some proteins act as distinct biomarkers during abiotic stress conditions.37 Shotgun proteomics helped identify drought-responsive genes in Oryza sativa present in the vegetative tissues and leaf proteins. Similar study was conducted using comparative proteomics technique, to understand the complexity of protein abundance in chickpea roots during drought stress.38 Proteomics relates proteins with stressful environmental conditions, together with their activity patterns, post-translational modifications (PTMs) and their interactions with each other.39 Different proteins’ up- and down-regulation primarily impacts photosynthesis by preserving protein production, energy metabolism, and detoxification in salty environments. Quantitative proteome investigation of Malus halliana exposed to salt-alkali mixed stress revealed 179 differentially expressed salt-responsive proteins using proteomics.40
Proteomics investigations include mass spectrometry-based protein identification and protein electrophoresis. Two-dimensional electrophoresis (2-DE) and Difference In-Gel Electrophoresis (DIGE) are two examples of conventional gel-based protein electrophoresis techniques.41 The drawbacks of 2-DE have been widely addressed by the non-gel approach known as Multi-dimensional Protein Identification Technology (MudPIT), which allows for both qualitative and quantitative proteomic investigations. Techniques including liquid chromatography-Mass Spectrometry (LC-MS/MS), Ion Trap-MS (IT-MS), and matrix-assisted laser desorption/ionization-MS (MALDI-MS) are employed in the mass spectrometry (MS) method.42 The development of label-free MS-based quantification and fluorophore-tagged protein immune-precipitation techniques has led to a greater degree of accuracy in the identification of regulatory protein complexes and low-abundance signaling.43
Metabolomics
Metabolites carry out vital functions in a spatiotemporal way and are present in the quantitative, qualitative and dynamic study of all endogenous, low molecular weight molecules (less than 1000-1500 daltons) within the cells, tissues or organs. There are roughly 0.2-1.0 million different macromolecules in the kingdom of plants. The amounts of these molecules differ throughout species. The classifications, physiochemical characteristics, chemical structures and polarity levels of these compounds differ from one another.44 Metabolomics offers an advantage over proteomics, transcriptomics and genomes. The by-product of gene and protein activity that determines the impact on physiological activity and other aspects of living phenotype is called a metabolite.45 Metabolites are divided into two categories-primary and secondary metabolites. Secondary metabolites are crucial for defense responses in the face of a variety of abiotic challenges. Primary metabolites are vital for plant growth and have a broad role in physiological activities.46 Secondary metabolites are particular to certain species and conditions whereas primary metabolites, such sugars, amino acids, and TCA (Krebs) cycle intermediates (citric acid, α-ketoglutarate), are directly involved in the regular growth and development of plants. As a result, the overall metabolite profile of a particular plant species reveals the degree of integration of several regulatory systems, including gene expression and gene-protein interaction. Metabolomics also reveal how plants adjust their metabolite profiles by revealing temporal and spatial insights over time and across tissues in response to stress as presented in Table 3. This knowledge supports breeding efforts by identifying stress-protective compounds like proline, glycine betaine, and trehalose, which help plants manage drought and salinity by maintaining cell stability and osmotic balance. In oxidative stress, metabolomics highlights the increased production of antioxidants such as ascorbate, glutathione, and flavonoids that protect cells by neutralizing harmful reactive oxygen species.47 Advances like imaging mass spectrometry (MSI) have improved spatial metabolomics by providing sensitive and detailed metabolite coverage. Additionally, microtechnologies such as lab-on-a-chip and microfluidics, when combined with ESI or MALDI-MS, enhance throughput, sensitivity, and data reliability, making metabolomics more efficient and precise.48 Plants display a range of reactions in response to unfavorable environmental conditions, many of which are linked to specific stress tolerance and metabolic changes. Work on metabolite alterations brought on by stress is receiving more focus in the twenty-first century.49 When a metabolic pathway is triggered, bioactive substances such as antioxidants, signalling molecules, biosynthesis intermediates for cellular structures and storage compounds are synthesized. The controller triggers the synthesis of other substances or mediators that can feedback activate or deactivate certain metabolic processes.50 The organism’s metabolite profile is kept in balance by changes in a variety of metabolic pathways found in plant cells and organs. The metabolomic profile of an organism is now visualized by the combination of various detection methods and analytical separation techniques. Numerous metabolite analyses and investigations have been very useful for scientists due to advancements in chromatographic, nuclear magnetic resonance (NMR) and MS techniques.51 The techniques of gas chromatography-MS (GC-MS) and LC-MS/MS are widely used in metabolomics research due to their vast coverage of large metabolites and unmatched sensitivity in crops 52 like mangosteen (Garcinia mangostana Linn.) fruit during ripening.53 More precise descriptions of metabolite interactions in a particular plant species are possible due to the development of new analytical techniques such capillary electrophoresis (CE), Fourier transform infrared spectroscopy (FTIR), GC, and LC coupled to MS, NMR, and FTIR.54
Table (3):
Metabolomic approaches to understand crop responses and tolerance mechanisms under stress
Crop |
Stress |
Metabolomics technique |
Key Metabolomics Insight |
Ref. |
|---|---|---|---|---|
Rice (Oryza sativa) |
Drought stress |
GC-MS |
Metabolomic profiling has revealed drought-induced shifts in metabolite levels, helping to understand stress adaptation. |
55 |
Wheat (Triticum aestivum) |
Drought stress |
UPLC-MS |
Changes in specific metabolite pathways under drought conditions suggest metabolic strategies for stress resilience. |
56 |
Maize (Zea mays) |
Drought stress |
GC-TOF-MS |
Identified metabolite markers and altered pathways linked to tolerance, aiding stress-resilient breeding efforts. |
57 |
Barley (Hordeum vulgare) |
Salinity stress |
LC-MS |
Salinity stress led to changes in root phytohormones and metabolite composition, shedding light on salt tolerance mechanisms. |
58 |
Rice (Oryza sativa) & Maize (Zea mays) |
mQTL mapping for trait discovery |
LC-MS |
Metabolome QTL analysis linked specific metabolite concentrations to genetic loci, offering candidate biomarkers for breeding. |
59 |
Multiomics approaches enable the construction of regulatory networks that depict the interactions among genes, proteins, metabolites, and epigenetic factors involved in stress response pathways. Understanding these regulatory networks helps decipher the complex signaling cascades and regulatory mechanisms governing stress adaptation in plants. The various omics techniques currently used are coupled to one another, making it possible to identify integrated cellular processes that result in a plant’s tolerance levels and stress responses. Using multiple omics approaches together offers a powerful way to understand how plants respond to stress at every level-from genes to metabolites. Instead of looking at just one type of data, integrating these layers helps reveal how different parts of the plant work together to adapt. Multi-omics data collected via different omics pipelines must be integrated to properly comprehend and draw conclusions about the primary cell response cascades that differ between tolerant and sensitive plants in specific abiotic stress situations. This approach helps identify important regulatory networks, biomarkers and candidate genes that can be targeted for breeding efforts and for enabling precision agriculture strategies. Research has demonstrated the potential benefits of combining multiple omics techniques to discover putative candidate genes and associated pathways. The likelihood of successfully creating crop types resistant to stress is increased by this strategy. The large data acquired from the multi-omics layers along with sophisticated bioinformatics and computational tools can potentially be utilized for precision breeding and predictive modelling. These accomplishments help to ensure food security in the face of shifting environmental conditions by advancing the creation of crop types that can withstand stress and sustainable agricultural techniques.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Kumar A, Verma JP. Does plant-microbe interaction confer stress tolerance in plants: a review? Microbiol Res. 2018;207:41-52.
Crossref - Ray A, Kundu S, Mohapatra SS, et al. An Insight into the Role of Phenolics in Abiotic Stress Tolerance in Plants: Current Perspective for Sustainable Environment. J Pure Appl Microbiol. 2024;18(1):64-79.
Crossref - Yadav MK, Singh RS, Singh KK, et al. Assessment of climate change impact on productivity of different cereal crops in Varanasi, India. J Agrometeorol. 2015;17(2):179-184.
Crossref - Nadarajah K, Abdul Rahman NSN. Plant-microbe interaction: Aboveground to belowground, from the good to the bad. Intl J Mol Sci. 2021;22(19):10388.
Crossref - Ahmed N, Li J, Li Y, et al. Symbiotic synergy: How Arbuscular Mycorrhizal Fungi enhance nutrient uptake, stress tolerance, and soil health through molecular mechanisms and hormonal regulation. IMA Fungus. 2025;16;144989.
Crossref - Sharma M, Sudheer S, Usmani Z, Rani R, Gupta P. Deciphering the omics of plant-microbe interaction: Perspectives and new insights. Curr Genomics. 2020;21(5):343-362.
Crossref - Zhang J, Coaker G, Zhou JM, Dong X. Plant immune mechanisms: from reductionistic to holistic points of view. Mol Plant. 2020;13(10)1358-378.
Crossref - Schirawski J, Perlin MH. Plant-microbe interaction 2017-the good, the bad and the diverse. Int J Mol Sci. 2018;19(5):1374.
Crossref - Muhammad M, Wahab A, Waheed A, et al. Navigating climate change: Exploring the dynamics between plant-soil microbiomes and their impact on plant growth and productivity. Globl Chang Biol. 2025;31(2);70057.
Crossref - Mukhopadhyay S, Maiti SK. Trace metal accumulation and natural mycorrhizal colonisation in an afforested coalmine overburden dump: a case study from India. Int J Min Reclam Environ, 2011;25(2): 187-207.
Crossref - Gonzalez-Mendoza VM, de la Torre M, Rocha J. Plant-Growth Promoting Endophytic Bacteria and Their Role for Maize Acclimatation to Abiotic Stress. In: Oliveira, M, Fernandes-Silva, A. (Eds). Abiotic Stress in Plants – Adaptations to Climate Change. : IntechOpen; 2023:259.
Crossref - Sang T, Zhang Z, Liu G, Wang P. Navigating the landscape of plant proteomics. J Integr Plant Biol. 2025;67(3);740-761.
Crossref - Lohani N, Jain D, Singh MB, Bhalla PL. Engineering multiple abiotic stress tolerance in canola, Brassica napus. Front Plant Sci. 2020;11:3.
Crossref - Mishra AK, Sudalaimuthuasari N, Hazzouri KM, Saeed EE, Shah I, Amiri KMA. Tapping into plant-microbiome interactions through the lens of multi-omics techniques. Cells. 2022;11(20):3254.
Crossref - Li H, La S, Zhang X, Gao L, Tian Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. The ISME J. 2021;15(10):2865-82.
Crossref - Miliute I, Buzaite O, Baniulis D, Stanys V. Bacterial endophytes in agricultural crops and their role in stress tolerance: a review 2015;465-78.
Crossref - Shekhawat K, Saad MM, Sheikh A, et al. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021;22(3):51049.
Crossref - Kumar P, Choudhary M, Halder T, et al. Salinity stress tolerance and omics approaches: Revisiting the progress and achievements in major cereal crops. Heredity. 2020;128(6):497-518.
Crossref - Jha UC, Bohra A, Nayyar H. Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 2020;139(1):1-27.
Crossref - Tas N, de Jong AEE, Li Y, Trubl G, Xue Y and Dove NC. Metagenomic tools in microbial ecology research. Curr Opin Biotechnol. 2021;67:184-191.
Crossref - Biswas S, Zhang D and Shi J. CRISPR/Cas systems: opportunities and challenges for crop breeding. Plant Cell Rep. 2021;40(6): 979-98.
Crossref - Hasan N, Choudhary S, Naaz N, Sharma N, Laskar RA. Recent advancements in molecular marker-assisted selection and applications in plant breeding programs. J Genet Eng Biotechnol. 2021;19(1):1-26.
Crossref - Pandey AK, Rubiales D, Wang Y, et al. Omics resources and omics-enabled approaches for achieving high productivity and improved quality in pea (Pisum sativum L.). Theor Appl Genet. 2021;134(3):755-776.
Crossref - Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBio. 2015;6(5):e00621.
Crossref - Kumar U, Raj S, Sreenikethanam A, et al. Multi-omics approaches in plant-microbe interactions hold enormous promise for sustainable agriculture. Agronomy. 2023;13(7):1804.
Crossref - Xu HR, Liu Y, Yu TF, et al. Comprehensive profiling of tubby-like proteins in soybean and roles of the GmTLP8 gene in abiotic stress responses. Front Plant Sci. 2022;13:844545.
Crossref - Zhao D, Xia X, Su J, Wei M, Wu Y, Tao J. Overexpression of herbaceous peony HSP70 confers high temperature tolerance. BMC Genomics. 2019;20(1):70.
Crossref - Rane NR, Tapase S, Kanojia A, et al. Molecular insights into plant-microbe interactions for sustainable remediation of contaminated environment. Bioresource Technol. 2022;344(Pt B):126246.
Crossref - Chao H, Zhang S, Hu Y, et al. Integrating omics databases for enhanced crop breeding. J Integ Bioinfo. 2024;20(4):20230012.
Crossref - Yi Y, de Jong A, Frenzel E, Kuipers OP. Comparative transcriptomics of Bacillus mycoides strains in response to potato-root exudates reveals different genetic adaptation of endophytic and soil isolates. Front. microbiol. 2017;8:1487.
Crossref - Kaur S, Shamshad M, Jindal S et al. RNA-seq-based transcriptomics study to investigate the genes governing nitrogen use efficiency in Indian wheat cultivars. Front Genet. 2022;13;853910.
Crossref - Li P, Cao W, Fang H, et al. Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front Plant Sci. 2017;8:290.
Crossref - Javaid MH, Khan AR, Salam A et al. Exploring the adaptive responses of plants to abiotic stresses using transcriptome data. Agriculture. 2022;12(2):211.
Crossref - Kim SA, LaCroix IS, Gerber SA, Guerinot ML. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc Natl Acad Sci USA. 2019;116(50):24933-24942.
Crossref - Doni F, Fathurrahman F, Mispan MS, Suhaimi NSM, Yusoff WMW, Uphoff N. Transcriptomic profiling of rice seedlings inoculated with the symbiotic fungus Trichoderma asperellum SL2. J Plant Growth Regul. 2019;38:1507-1515.
Crossref - Conde D, Kirst M. Decoding exceptional plant traits by comparative single-cell genomics. Trends Plant Sci. 2020;27(11):1095-1098.
Crossref - Li J, Sohail H, Nawaz MA, Liu C, Yang P. Physiological and proteomic analyses reveals that brassinosteroids application improves the chilling stress tolerance of pepper seedlings. Plant Growth Regul. 2022;96(2):315-329.
Crossref - Gupta S, Mishra SK, Misra S, et al. Revealing the complexity of protein abundance in chickpea root under drought-stress using a comparative proteomics approach. Plant Physiol Biochem. 2020;151:88-102.
Crossref - Kausar R, Wang X, Komatsu S. Crop proteomics under abiotic stress: from data to insights. Plants. 2022;11(21):2877.
Crossref - Zhu Y, Jia X, Wu Y, et al. Quantitative proteomic analysis of Malus halliana exposed to salt-alkali mixed stress reveals alterations in energy metabolism and stress regulation. Plant Growth Regul. 2020;90(2):205-222.
Crossref - Yan S, Bhawal R, Yin Z, Thannhauser TW, Zhang S. Recent advances in proteomics and metabolomics in plants. Mol Hortic. 2022;2(1):17.
Crossref - Lohani N, Singh MB, Bhalla PL. Biological parts for engineering abiotic stress tolerance in plants. BioDesign Research. 2022;2022:9819314.
Crossref - Roychowdhury R, Das SP, Gupta A, et al. Multi-omics pipeline and omics-integration approach to decipher plant’s abiotic stress tolerance responses, Genes. 2023;14(6):1281.
Crossref - Alseekh S, de Souza LP, Benina M, Fernie AR. The style and substance of plant flavonoid decoration; towards defining both structure and function. Phytochemis. 2020;174:112347.
Crossref - Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20(6):353-367.
Crossref - Verslues PE, Bailey-Serres J, Brodersen C, et al. Burning questions for a warming and changing world: 15 unknowns in plant abiotic stress. The Plant Cell. 2023;35(1):67-108.
Crossref - Saleem MH, Noreen S, Ishaq I et al. Omics technologies: unraveling abiotic stress tolerance mechanisms for sustainable crop improvement. J Plant Growth Regul. 2025:1-23.
Crossref - Mashabela MD, Masamba P, Kappo AP. Applications of metabolomics for the elucidation of abiotic stress tolerance in plants: a special focus on osmotic stress and heavy metal toxicity. Plants. 2023;12(2);269.
Crossref - Patel J, Khandwal D, Choudhary B, et al. Differential physio-biochemical and metabolic responses of peanut (Arachis hypogaea L.) under multiple abiotic stress conditions. Intl J Mol Sci. 2022;23(2):660.
Crossref - Huchzermeyer B, Menghani E, Khardia P, Shilu A. Metabolic pathway of natural antioxidants, antioxidant enzymes and ROS providence. Antioxidants. 2022;11(4):761.
Crossref - Ghatak A, Chaturvedi P, Weckwerth W. Metabolomics Metabolomics in Plant Stress Physiology. In: Varshney R, Pandey M, Chitikineni A. (eds) Plant Genetics and Molecular Biology. Advances in Biochemical Engineering/Biotechnology, vol 164. Springer, Cham. 2018;164:187-236.
Crossref - Samtani H, Sharma A, Khurana P. Overexpression of HVA1 enhances drought and heat stress tolerance in Triticum aestivum doubled haploid plants. Cells. 2022;11(5):912.
Crossref - Mamat SF, Azizan KA, Baharum SN, Noor NM, Aizat WM. GC-MS and LC-MS analyses reveal the distribution of primary and secondary metabolites in mangosteen (Garcinia mangostana Linn.) fruit during ripening. Sci Horti. 2019;262:109004.
Crossref - Liu R, Bao ZX, Zhao PJ, Li GH. Advances in the study of metabolomics and metabolites in some species interactions. Molecules. 2021;26(11):3311.
Crossref - Casartelli A, Riewe D, Hubberten HM, Altmann T, Hoefgen R, Heuer S. Exploring traditional aus-type rice for metabolites conferring drought tolerance. Rice. 2018;11;1-16.
Crossref - Zhao YY, Wu SP, Liu S, Zhang Y, Lin RC. Ultra-performance liquid chromatography-mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem Biol Interact. 2014;220:181-192.
Crossref - Witt S, Galicia L, Lisec J, et al. Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol Plant. 2012;5(2):401-17.
Crossref - Cao D, Lutz A, Hill CB, Callahan DL, Roessner U. A quantitative profiling method of phytohormones and other metabolites applied to barley roots subjected to salinity stress. Front Plant Sci. 2017;7:2070.
Crossref - Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2012;403(6):1523-1548.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.