ISSN: 0973-7510
E-ISSN: 2581-690X
Hypervirulent Klebsiella pneumoniae (hvKp) is an emerging global pathogen associated with severe, invasive infections and increasing antimicrobial resistance. While hvKp poses significant clinical challenges, data on its prevalence and resistance patterns in India remain limited. A cross-sectional study was conducted between June and July 2021 at a tertiary care hospital in Puducherry. A total of 80 clinical isolates of K. pneumoniae were analyzed for hypermucoviscosity using the string test. Antimicrobial resistance patterns were assessed using the Kirby-Bauer disk diffusion method, and extended-spectrum beta-lactamase (ESBL) production was evaluated using phenotypic methods. Among the 80 isolates, 18 (22.5%) were identified as hvKp, while 62 (77.5%) were classified as classical K. pneumoniae (cKp). hvKp predominantly affected males (83.34%) and individuals aged 45-65 years (72.23%). Resistance rates were significantly higher in hvKp for gentamicin (50% vs. 24.19%, p = 0.038) and cefotaxime (44.45% vs. 27.42%). Furthermore, 55.56% of hvKp strains were ESBL producers, compared to 27.42% of cKp strains. This study highlights the clinical significance of hvKp, with a notable prevalence and a concerning resistance profile. Enhanced surveillance, routine phenotypic identification, and the development of tailored antibiotic protocols are crucial to addressing the challenges posed by hvKp in healthcare settings. These findings provide valuable insights for optimizing the management of hvKp infections in the Indian context.
Antimicrobial Resistance, Extended-spectrum Beta-Lactamases, Gram-Negative Bacterial Infections, Klebsiella pneumoniae, Multidrug-resistant Organisms
Klebsiella pneumoniae (K. pneumoniae) is a gram-negative bacterium commonly implicated in infections such as pneumonia, bloodstream infections, liver abscesses, and surgical site infections. A particularly concerning subtype, hypervirulent Klebsiella pneumoniae (hvKp), has emerged due to its ability to cause severe, life-threatening infections, even in healthy individuals and largely attributed to its hypermucoviscosity phenotype, detectable by a simple string test. Unlike classical strains (cKp), hvKp is strongly associated with metastatic infections such as hepatic abscesses, meningitis, and endophthalmitis, and carries high morbidity and mortality (3%-42%). First identified in the Asia-Pacific region, hvKp has since been reported globally and poses a growing public health concern due to its potential for severe, invasive infections. Unlike most gram-negative pathogens, hvKp exhibit a notable tendency to induce metastatic infections.1-3
A significant challenge associated with hvKp is its capacity to develop antimicrobial resistance, which complicates treatment strategies. This capability has resulted in its classification among the ESKAPE pathogens, namely Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, all of which are acknowledged globally for their potential for multidrug-resistance (MDR).4 The emergence of resistance mechanisms, such as extended-spectrum beta-lactamases (ESBLs) and carbapenemases, has rendered numerous commonly utilized antibiotics ineffective, thereby further complicating the clinical management of hvKp infections.5,6
The emergence of hvKp poses a notable challenge for clinicians, particularly in regions where its prevalence and resistance profiles are still being explored. The significance of early detection and accurate identification of hvKp cannot be overstated, as these factors play a crucial role in facilitating timely and effective treatment, particularly in cases involving metastatic infections or those localized to specific sites.7
To address these important considerations, a study was conducted at a tertiary care hospital in Puducherry, India. The primary objectives of this investigation were to assess the prevalence of hvKp in various clinical samples and to examine its antimicrobial resistance patterns. The insights gained from this study are anticipated to be valuable for both microbiologists and clinicians, ultimately contributing to a better understanding and more effective management of hvKp infections.
Study design and setting
This study employed a prospective, cross-sectional design to evaluate the prevalence and characteristics of K. pneumoniae isolates in a clinical setting. It was conducted at the Department of Microbiology, a tertiary care hospital. The study was designed to provide insights into the hypervirulent phenotype, antimicrobial resistance patterns, and ESBL production among K. pneumoniae isolates obtained from various clinical specimens.
Inclusion and exclusion criteria
Clinical samples that yielded positive cultures for K. pneumoniae were included in the analysis. Specimens were collected from diverse sources, including blood, pus, urine, and sputum, ensuring a comprehensive representation of infections caused by this pathogen. Samples that lacked adequate clinical data or those exhibiting co-infections with other bacterial species were excluded to maintain the specificity of the findings and minimize confounding factors.
Sample size
A total of 80 isolates of K. pneumoniae were collected and analyzed. This sample size was deemed sufficient to provide statistically meaningful insights into the prevalence of hypervirulent strains and resistance patterns within the study’s timeframe.
Isolation and identification of isolates
All clinical specimens were processed following standard microbiological techniques. For non-blood samples, direct Gram staining was performed to assess bacterial morphology and preliminary identification. Specimens were then streaked onto blood agar and MacConkey agar plates to promote the growth and differentiation of bacterial colonies. Identification of K. pneumoniae was carried out using established culture techniques and biochemical tests, including indole, citrate, and urease tests, as well as sugar fermentation assays. These methods ensured accurate identification of the isolates.
String test for hypervirulence
The hypermucoviscosity phenotype, indicative hvKp, was assessed using the string test. A bacterial colony was stretched using an inoculation loop, and the formation of a viscous string measuring more than 5 mm was considered a positive result. This test allowed for the differentiation of hypervirulent strains from cKp isolates, which do not exhibit the same level of Mucoviscosity.8
Antimicrobial susceptibility testing
The antimicrobial susceptibility profile of each isolate was determined using the Kirby-Bauer disk diffusion method. Antibiotic susceptibility was tested against a panel of commonly used antimicrobials, including amoxiclav (20/10 µg), cefotaxime (30 µg), ceftazidime (30 µg), piperacillin-tazobactam (100/10 µg), meropenem (10 µg), gentamicin (10 µg), amikacin (30 µg), and ciprofloxacin (5 µg). The testing procedure adhered to the Clinical and Laboratory Standards Institute (CLSI) guidelines, ensuring the reliability of results. Escherichia coli ATCC 25922 was used as a quality control strain to validate the accuracy of the susceptibility testing.
Detection of ESBL production
Extended-spectrum beta-lactamase (ESBL) production was detected using the phenotypic double-disk synergy test (DDST). This method involved testing ceftazidime and cefotaxime alone and in combination with clavulanate. ≥5 mm increase in zone diameter for either antimicrobial agent tested in combination with clavulanic acid vs zone of diameter of the agent when tested alone was considered indicative of ESBL production.
Statistical analysis
Data were compiled and managed using Microsoft Excel and analyzed with SPSS version 23.0. Descriptive statistics, such as frequencies and percentages, were used to summarize the characteristics of the isolates and their resistance patterns. Comparative analyses were conducted using the chi-square test or Fisher’s exact test, depending on the data distribution and sample size. A p-value of <0.05 was considered statistically significant, allowing for the identification of meaningful associations between variables. This statistical approach ensured a robust analysis of the study’s findings.
Distribution of hvKp and cKp strains
A total of 80 strains of K. pneumoniae were isolated from various clinical specimens, including urine, blood, pus, and sputum. Among these, 18 (22.5%) isolates tested positive for the string test, identifying them as hvKp. The remaining 62 (77.5%) isolates were classified as cKp. This distribution indicates that hvKp comprises a significant subset of K. pneumoniae infections, warranting closer clinical monitoring due to its hypervirulence.
Gender distribution of hvKp strains
Out of the 18 hvKp isolates, infections were more prevalent in males (15, 83.34%) compared to females (3, 16.67%) (Table 1). This male predominance could reflect underlying health or lifestyle factors, such as higher exposure to risk factors in men, or differences in immune responses between genders.
Table (1):
Results of hvKp Based on Gender
Gender |
Total hvKp sample (n = 18) Number and Percentage (%) |
|---|---|
Male |
15 (83.34%) |
Female |
3 (16.67%) |
Age distribution of hvKp strains
The majority of hvKp isolates (13, 72.23%) were found in patients aged 45-65 years. Infections were less common in the 25-45 years age group (2, 11.12%) and above 65 years (3, 16.67%), with no cases reported in individuals below 25 years (Supplementary Table 1). This age distribution highlights that individuals in the middle-aged to older population are at higher risk, potentially due to comorbid conditions or weakened immune systems.
Clinical sample distribution of hvKp and cKp
Among hvKp isolates, most were recovered from pus (13, 72.23%), followed by blood (2, 11.12%), sputum (2, 11.12%), and urine (1, 5.56%). For cKp, the majority were isolated from pus (41, 66.13%), followed by urine (15, 24.19%) and sputum (6, 9.67%) (Table 2). The higher prevalence of hvKp in pus samples suggests its association with severe soft tissue infections, whereas cKp showed a broader distribution across different sample types.
Table (2):
Results of hvKp and cKp Based on Clinical Samples
Clinical Sample |
hvKp (n = 18) |
Number and Percentage (%) |
cKp (n = 62) |
Number and Percentage (%) |
|---|---|---|---|---|
Blood |
2 |
11.12% |
0 |
0 |
Urine |
1 |
5.56% |
15 |
24.19% |
Sputum |
2 |
11.12% |
6 |
9.67% |
Pus |
13 |
72.23% |
41 |
66.13% |
Antimicrobial resistance patterns
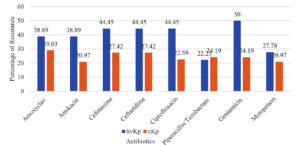
The hvKp isolates exhibited higher resistance to gentamicin (9, 50%) compared to cKp isolates (15, 24.19%), with this difference being statistically significant (p = 0.038). Resistance to other antibiotics, including amoxiclav, amikacin, ceftazidime, cefotaxime, ciprofloxacin, meropenem, and piperacillin-tazobactam, was also higher in hvKp isolates but not statistically significant (Supplementary Table 2, Figure 1). These findings suggest that hvKp strains may possess enhanced resistance mechanisms, raising concerns about treatment efficacy.
ESBL production in hvKp and cKp
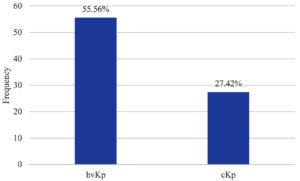
The hvKp isolates showed significantly higher rates of ESBL production (10, 55.56%) compared to cKp isolates (17, 27.42%), with a statistically significant difference (p = 0.01) (Supplementary Table 3, Figure 2). This increased ESBL production in hvKp underscores its potential for multidrug-resistance, making it a formidable pathogen in clinical settings.
The escalating global incidence of hvKp infections over recent decades presents a significant public health challenge. Our study, conducted within a one-month period from June to July 2021 at our institution, involved 80 patients with culture-confirmed K. pneumoniae, among which 18 (22.5%) were identified as hvKp. This rate of hypervirulence is noteworthy when juxtaposed against the backdrop of global reports, which suggest considerable geographical variability in hvKp prevalence.2
Comparative studies in Asia have demonstrated higher prevalence rates, such as 33% in China,9 38% in Taiwan,10 and an alarming 42.4% in Korea.11 Conversely, lower prevalence has been reported in the West, with Spain and Alberta, Canada documenting rates of 5.4% and 8.2%, respectively.2,7 This variation may reflect differences in genetic predispositions of strains circulating in different regions, healthcare practices, or even diagnostic criteria for identifying hvKp, which is predominantly characterized by its hypermucoviscosity, detectable through a simple string test on agar plates.
Hypermucoviscosity, a quintessential feature of hvKp, plays a pivotal role in the pathogen’s ability to cause severe infections.12 This characteristic, which enhances the bacterium’s ability to evade host immune responses, is easily identifiable by the positive result of the string test – a critical tool not only for diagnosis but also for epidemiological studies.13,14 The hypermucoviscous phenotype is associated with increased virulence, enabling the bacteria to cause disseminated infections even in young and healthy individuals, a pattern uncommon with cKp strains.13
In our study, the majority of infections were observed in males (83.3%), a demographic trend consistent with previous studies. This gender predisposition could be attributed to behavioral and physiological differences that may affect exposure or susceptibility to hvKp.15 Additionally, the predilection for infection in individuals in their fourth to sixth decades of life could suggest a role of occupational exposure or slight immunological decline with age, though this warrants further investigation.16
Our findings also underscore the growing concern of antimicrobial resistance among hvKp strains. In our study, hvKp isolates exhibited greater resistance compared to cKp isolates.17 This resistance can significantly complicate treatment strategies, as hvKp not only poses a challenge due to its inherent virulence but also due to its ability to withstand standard antibiotic regimens.12,18
Previous research has indicated that K1 subtype K. pneumoniae, often isolated from liver abscesses, is less resistant to antimicrobials than other strains.19 However, more recent studies suggest an increasing association between hvKp and antimicrobial resistance, which could be due to genetic factors that confer both virulence and resistance mechanisms.20-22
The mechanisms underlying the observed resistance patterns in hvKp are multifaceted. Our study indicated a higher prevalence of ESBL-producing strains among hvKp compared to cKp, suggesting that hvKp may acquire resistance through several pathways.19,23 These include the acquisition of conjugative plasmids and integrative conjugative elements (ICEs) that harbor resistance genes, or through mutations in chromosomal genes. Such mechanisms highlight the adaptive capabilities of hvKp, enabling it to thrive in antimicrobial-rich environments, which could be exacerbated by hospital settings or widespread antibiotic use.24-26
The clinical implications of our findings are profound. The increased resistance of hvKp strains necessitates heightened vigilance and proactive management strategies in clinical settings.27 This includes the rapid identification of hvKp through reliable diagnostic methods like the string test and tailored antibiotic regimens to combat the robust resistance profiles observed.28,29 From a public health perspective, our study highlights the need for global surveillance programs to monitor the prevalence and resistance patterns of hvKp, to better understand and mitigate its spread.30-32
Our study is not without limitations. The sample size, while adequate for preliminary observations, is relatively small and geographically confined, which may limit the generalizability of our results. Additionally, the study period of one month provides only a snapshot of hvKp prevalence and behavior. Future studies should aim to include larger, more diverse populations and extend over longer periods to capture seasonal and annual trends in hvKp dynamics. Further research should also explore the molecular mechanisms of hvKp virulence and resistance in greater detail, to develop targeted interventions.
The findings from our study confirm the concerning trend of increased antimicrobial resistance among hvKp strains, which poses a significant threat to effective clinical management. The emerging resistance of hvKp highlight an urgent need for enhanced diagnostic capabilities and treatment strategies that can address the unique challenges presented by this virulent pathogen. As the global health community continues to grapple with hvKp, we must improve our understanding of its epidemiology, mechanisms of resistance, and potential strategies for mitigation. This will require collaborative efforts across research, clinical, and public health domains to safeguard against this emerging threat.
Additional file: Additional Table S1-S3.
ACKNOWLEDGMENTS
The authors are thankful to Vinayaka Missions Research Foundation (Deemed to be University) for providing the necessary facilities for this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SM conceptualized the study and performed Investigation. KK performed supervision and project administration. SM and ACS applied methodology. RL and ACS performed formal analysis. BID performed visualization and data validation. RL performed data curation. SM and ACS wrote the original draft. KK reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This research was supported by the Indian Council of Medical Research (ICMR) under the Short-Term Studentship (STS) program vide grant number 2020-02687.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Human Ethical Commitee, Aarupadai Veedu Medical College and Hospital, Puducherry, vide ethical approval number AV/IEC/2020/066.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology, and risk factors. Ann Clin Microbiol Antimicrob. 2019;18(1):4.
Crossref - Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107-118.
Crossref - Juan CH, Fang SY, Chou CH, Tsai TY, Lin YT. Clinical characteristics of patients with pneumonia caused by Klebsiella pneumoniae in Taiwan and prevalence of antimicrobial-resistant and hypervirulent strains: a retrospective study. Antimicrob Resist Infect Control. 2020;9(1):1-8.
Crossref - Raj S, Sharma T, Pradhan D, et al. Comparative analysis of clinical and genomic characteristics of hypervirulent Klebsiella pneumoniae from hospital and community settings: experience from a tertiary healthcare center in India. Microbiology Spectrum. 2022;10(5):e00376-22
- Rupp ME, Fey PD. Extended spectrum b-lactamase (ESBL)-producing Enterobacteriaceae: considerations for diagnosis, prevention, and drug treatment. Drugs. 2003;63(4)353-365.
Crossref - Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther. 2019;17(2):71-73.
Crossref - Chen Y, Chen Y. Clinical challenges with hypervirulent Klebsiella pneumoniae (hvKP) in China. J Transl Intern Med. 2021;9(2):71-75.
Crossref - Hagiya H, Watanabe N, Maki M, Murase T, Otsuka F. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype Klebsiella pneumoniae. Acute Med Surg. 2014;1(4):245-246.
Crossref - Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115-6120.
Crossref - Liao CH, Huang YT, Hsueh PR. Multicenter surveillance of capsular serotypes, virulence genes, and antimicrobial susceptibilities of Klebsiella pneumoniae causing bacteremia in Taiwan, 2017-2019. Front Microbiol. 2022;13:783523.
Crossref - Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483.
Crossref - Peirano G, Pitout JDD, Laupland KB, Meatherall B, Gregson DB. Population-based surveillance for hypermucoviscosity Klebsiella pneumoniae causing community-acquired bacteremia in Calgary, Alberta. Can J Infect Dis Med Microbiol. 2013;24(3):e61.
Crossref - Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225-232.
Crossref - Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182.
Crossref - Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351-1358.
Crossref - Jung SW, Chae HJ, Park YJ, et al. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect. 2013;141(2):334-340.
Crossref - Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect. 2016;22(2):154-160.
Crossref - Beig M, Aghamohammad S, Majidzadeh N, et al. Antibiotic resistance rates in hypervirulent Klebsiella pneumoniae strains: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2024;38:376-388.
Crossref - Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031-41.
Crossref - Ikeda M, Mizoguchi M, Oshida Y, et al. Clinical and microbiological characteristics and occurrence of Klebsiella pneumoniae infection in Japan. Int J Gen Med. 2018;293-299.
Crossref - Lin YT, Jeng YY, Chen TL, Fung CP. Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001-2008. BMC Infect Dis. 2010;10:307.
Crossref - Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881-887.
Crossref - Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284-293.
Crossref - Cheng NC, Yu YC, Tai HC, et al. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis. 2012;55(7):930-939.
Crossref - Su SC, Siu LK, Ma L, et al. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum b-lactamase. Antimicrob Agents Chemother. 2008;52(2):804-805.
Crossref - Lobo AS, Moosabba MS. Antibiogram and hypermucoviscosity pattern among Klebsiella pneumoniae isolates from respiratory samples: A tertiary care hospital study in South India. IP Int J Compr Adv Pharmacol. 2019;4(4):134-138.
Crossref - Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62(7):10-128.
Crossref - Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1-12.
Crossref - Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709-11.
Crossref - Turton JF, Payne Z, Coward A, et al. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol. 2018;67(1):118-28.
Crossref - Yap AUJ, Tan BWY, Tay LC, Chang KM, Loy TK, Mok BYY. Effect of mouthrinses on microhardness and wear of composite and compomer restoratives. Oper Dent. 2003;28(6):740-746.
- Yang Y, Liu JH, Hu XX, et al. Clinical and microbiological characteristics of hypervirulent Klebsiella pneumoniae (hvKP) in a hospital from North China. J Infect Dev Ctries. 2020;14(6):606-613.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.