ISSN: 0973-7510
E-ISSN: 2581-690X
Cosmetics are often contaminated by pathogenic bacteria, such as Staphylococcus spp. and Pseudomonas spp. During manufacturing, preservatives are added to cosmetic products to protect them from microbial contamination. In this study, the effects of four preservatives, phenoxyethanol (PE), benzyl alcohol (BA), propylparaben (PP), and methylparaben (MP), against seven microbial strains isolated from contaminated cosmetics were examined. The isolated bacterial strains included Bacillus cereus, Bacillus subtilis, Escherichia coli, and Staphylococcus aureus and the isolated fungal strains were Aspergillus flavus, Aspergillus fumigatus, and Aspergillus welwitschiae. Bacterial strains were identified using 16S rRNA sequencing, whereas the fungal strains were identified using 18S rRNA sequencing. The minimum inhibitory concentrations (MICs) were calculated to determine the sensitivity of the strains to the four preservatives (PE, BA, PP, and MP) at different concentrations (3.2, 1.6, 0.8, 0.4, 0.2, 0.1, and 0.05 mg/mL). The results showed that most of the bacterial strains were resistant to PP, and their growth was inhibited at high concentrations (0.8-3.2 mg/mL) of each preservative. B. subtilis was the most sensitive bacterial strain to preservatives, at low concentrations (0.1-0.8 mg /mL). The MICs of MP ranged from 0.05-0.2 mg/mL, which indicates the strength of their inhibitory effect on fungi at low concentrations. Growth of microorganisms inside cosmetic products is an issue that needs to be reconsidered by researchers and preservative systems should be evaluated. For safety and health purposes of customers usually use cosmetics, this study is prepared.
Cosmetics, Preservatives, MICs, Microbial Contamination
Cosmetics are personal care products aimed at enhancing beauty and hygiene, with skin care, makeup, perfumes, and hair care being the main categories of items.1
These products are frequently contaminated by pathogenic microorganisms, such as bacteria (Staphylococcus aureus and Pseudomonas aeruginosa), fungi ( Aspergillus and Penicillium), and yeast-like (Candida).2,3
Cosmetics are at risk of contamination due to imperfect manufacturing processes or user involvement.3 The packaging surface or inside of the product can become contaminated by microbes during filling, leading to infections in the user or product spoilage.4 Furthermore, cosmetics usually consist of natural ingredients, which are exposed to microbial contamination prior to processing.5
In tropical countries, humidity promotes the existence and growth of microorganisms, especially in highly contaminated nutrient-rich cosmetic products, where prompt growth and reproduction are common.6
One solution to reduce contamination is the addition of preservatives to cosmetics to protect them against microbial contamination.7 To ensure microbiological quality, cosmetic products should have an appropriate preservative system to prevent microbial contamination and ensure compatibility and stability.8,9
To ensure consumer safety, the preservation of cosmetics is governed by stringent global standards. The Food and Drug Administration (FDA), USA recommend that products should be free from risk when used under labeled conditions or as commonly used products.10
The European Union (EU) has made a list of approved preservatives, their dosage amounts, and specific product categories in which they can be used to ensure safety. The Annex V-List presents approved cosmetic preservatives (EU No. 1223/2009).1,11
Generally, preservatives consist of ingredients that can prevent bacterial growth by destroying cell membranes, denaturing proteins, or interfering with bacterial metabolism.12
Preservatives can be divided into two types: natural, such as essential oils or grapefruit seed extract; and synthetic, such as parabens or formaldehyde releases, including example, DMDM Hydantoin, isothiazolinones (e.g., MCT and MI), and phenolic types (e.g., phenoxyethanol and benzyl alcohol). Each has its own mechanism of action, spectrum, and consumer perceptions.13
Some preservatives, such as benzoic acid, phenoxyethanol, and sorbic acid are used in combination. Formaldehyde releasers, parabens, and Methylchloroisothiazolinone (MCI)/MI have been found to be more effective than sorbic acid, phenoxyethanol, and benzoic acid. In addition, some acidic preservatives require an acidic pH to be activated.14,15
Methylisothiazolinone (MIT) and MCI preservatives can prevent microorganism contamination at rates above 90%, which is much lower than the rates of P. aeruginosa and S. aureus.16
Thus, preservatives are crucial for upholding the microbiological safety of cosmetics, with the choice of preservatives being critical to effectively mitigate microbial threats.6,17,18
This study aimed to evaluate the effectiveness of four common preservatives, phenoxyethanol (PE), benzyl alcohol (BA), propylparaben (PP), and methylparaben (MP), against seven microbial strains isolated from contaminated cosmetics.
Isolation of microorganisms
Microbial strains were isolated from different contaminated skin and body care products based on a method described in a previous study of Alshehrei.2 Thirty-two cosmetic products used for body and skin collected from different stores and outlets in Mecca region, SA. Samples are transported to the lab in sterilized condition. Serial dilutions (10-1, 10-2, 10-3, 10-4, and 10-5) are used to isolate microorganisms from cosmetic products, each dilution transferred to two different plates, one includes Tryptone Soy Agar (TSA) to grow bacteria, plates then are incubated for 48 hours at 37 °C, and the other one includes Sabouraud Dextrose Agar (SDA) to grow fungi, plates are incubated at 25 °C for 5-7 days. After isolation, microbial stains undergo molecular analysis; 16S rRNA in case of bacteria and 18S rRNA in case of fungi.
Identification of bacteria using 16S rRNA sequencing
Extraction of DNA
Bacterial isolates were grown in nutrient agar for 48 h at 37 °C, and then the bacterial culture was harvested using centrifugation at 5000 rpm for 10 min. One milliliter of a solution containing 5 × 108 bacterial cells was prepared, and DNA was extracted using the Qiagen Bacterial DNA extraction kit, as per the manufacturer’s protocol.
PCR amplification
The universal forward primer (5′-GAG TTT GAT CCT GGC TCAG-3′), and reverse primer (5′-GTT ACC TTG TTA CGA CTT-3′) were used to amplify DNA.19
Polymerase chain reaction (PCR) was performed in a thermal cycler with the following cycle conditions: The total volume of the reaction mixture was 25 µL; PCR was conducted for 35 cycles comprising the following steps: denaturation at 96 °C for 50 s, annealing at 50 °C for 45 s, and extension at 72 °C for 2 min. In addition to the 35 cycles, the PCR also included an initial denaturation step at 96 °C for 10 min and a final extension step at 72 °C for 10 min.
Identification of fungi using 18S rRNA sequencing
Extraction of DNA
Fungal species were detected using molecular internally transcribed spacer regions. Fungal isolates were grown in Sabouraud dextrose agar for 72 h at 30 °C. After harvesting, a 1 ml saline suspension containing 1-5 × 106 conidia were prepared. DNA was extracted from each sample using a DNeasy kit (Qiagen) to isolate genomic DNA from fungi (cultures and blood samples) according to the manufacturer’s instructions.
Internally transcribed spacer-PCR
A forward primer (5′-TCC GTA GGT GAA CCT GCG G-3’) and reverse primer (5′-TCC TCC GCT TAT TGA TAT GC-3’) were used to amplify ITS4 from the PCR reactions, consisting of 20 ng genomic DNA in a total volume of 25 L. PCR was performed in a thermal cycler. The reaction mixture contained 0.5 µM of both the forward and reverse primer, 50-100 ng of template DNA, 0.5 U of Taq DNA polymerase, 200 µM of dNTPs, 10 mM Tris, 50 mM KCl, and 1.5 mM MgCl2. The total volume of the reaction mixture was 25 µL. PCR was conducted for 35 cycles, with each cycle consisting of the following steps: denaturation at 96 °C for 50 s, annealing at 50 °C for 45 s, and extension at 72 °C for 2 min. In addition to the 35 cycles, the PCR process also included an initial denaturation step at 96 °C for 10 min and a final extension step at 72 °C for 10 min.20
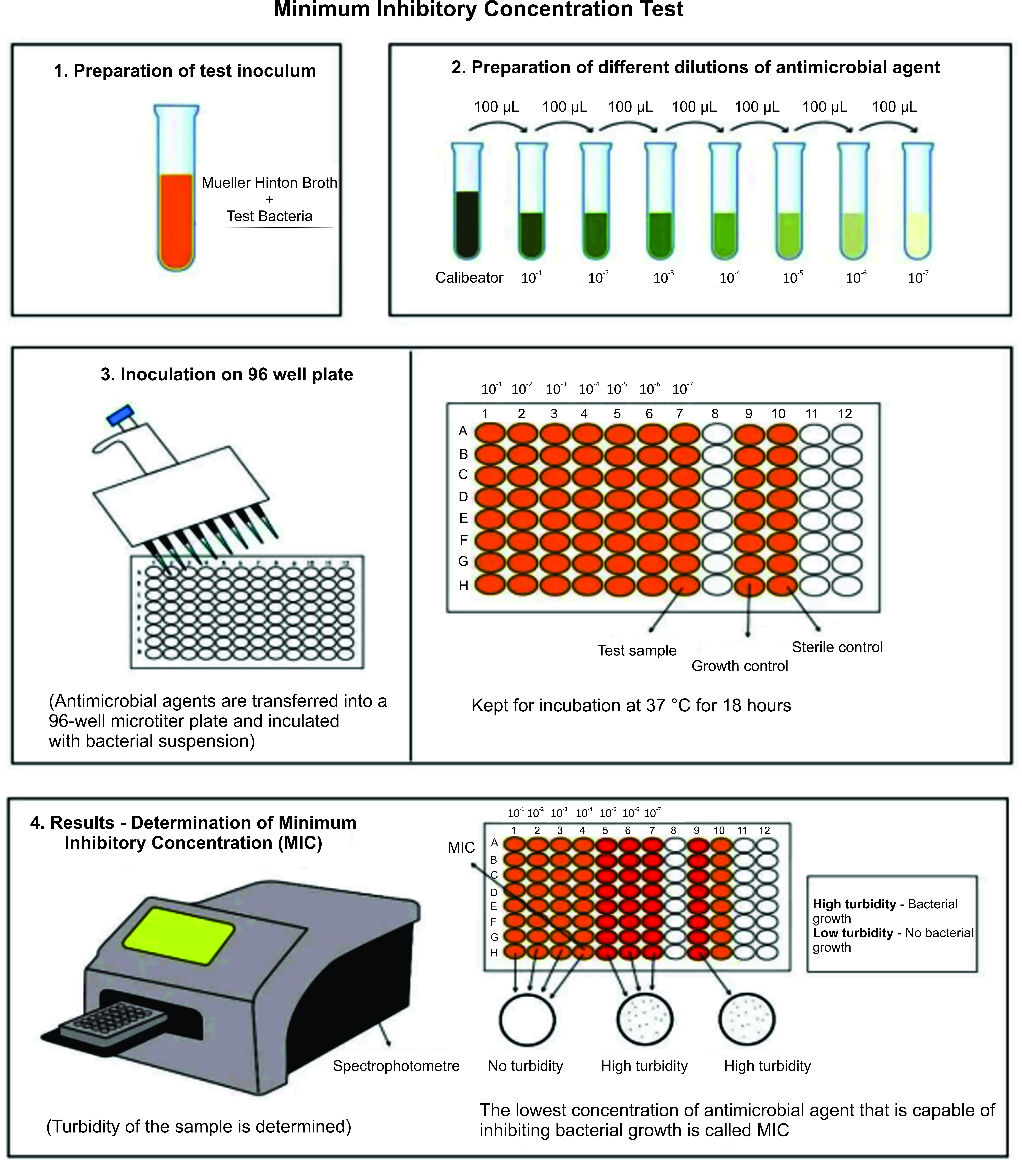
Minimum inhibitory concentrations (MICs)
The effects of four preservatives that are used broadly in cosmetics preserving, PE, BA, PP, and MP, on seven strains isolated from contaminated cosmetics were examined.
Preparation of the bacterial inoculum
Bacterial strains were isolated from various contaminated skin and body care cosmetics, based on a previous study.2 The identified strains were B. cereus, B. subtilis, S. aureus, and E. coli. For each bacterium, nutrient broth medium was inoculated with a single colony and incubated at 37 °C with shaking. Next, suspensions were adjusted to the same OD60 for quantification purposes. MIC tests were performed using bacterial strains, maintained in nutrient broth medium, and inoculated with 105 CFU/mL of each bacterium. Serial dilutions were carried out in test tubes, which were incubated for 24 h at 37 °C in a rotary shaker at 180 rpm.
Determination of minimum inhibitory concentrations (MICs)
A serial dilution method was used to determine the MICs of the four preservatives (PE, BA, PP, and MP) at different concentrations (3.2, 1.6, 0.8, 0.4, 0.2, 0.1, and 0.05 mg/mL). Medium without preservatives was used as the control. The MICs of the preservatives were calculated in triplicate.
MICs experiments were performed in round-bottomed 96-well plates (Figure 1). First, 100 µL of each preservative agent was aliquoted into the 96-well microplates (1 to 10 wells). Growth and control groups were included. Next, 100 µL of RPMI1640 broth medium and 100 µL of fungal/bacterial inoculum were added to the preservative agents. Additionally, a negative control was prepared using 25 µL of preservative agent and 75 µL of RPMI1640 broth medium. Culture plates were heated at 37 °C for 24 hrs.
The MICs were determined at 24 and 48 hrs as the concentrations of the preservatives that achieved 100% growth inhibition.
Preparation of fungal inoculum
Fungal strains were isolated from different contaminated skin and body care products based on a method described in a previous study.3 These strains were identified on a molecular level at the Laboratory of Microbiology, Mr. Alqura University to be A. flavus, A. fumigatus, and A. welwitschiae.
Determination of minimum fungicidal concentration (MFC)
After 48 hrs of incubation, 10 L of the contents were removed from the wells that showed no growth and spread on Sabouraud dextrose agar. MFCs were determined after incubation for 72 hrs.
Statistics
Data were analyzed by Excel program. Standard deviation for each treatment was calculated. Error bars were visualized as the value of STDEV on the charts.
Molecular Identification
Bacterial strains were identified using 16S rRNA, whereas fungal strains were identified using 18S rRNA. The sequences of the isolates were aligned with those in the NCBI database. The matching ratios of sequencing are shown in Table, and Figure 2-8 shows the phylogenetic trees of each strain.
Table:
Results of NCBI database analysis showing the match ratios of the isolated strains
No. |
Microorganism |
Percentage of Identification |
|---|---|---|
1 |
Bacillus cereus |
94.56% |
2 |
Bacillus subtilis |
95.45% |
3 |
Escherichia coli |
97.9% |
4 |
Staphylococcus aureus |
98.58% |
5 |
Aspergillus flavus |
100% |
6 |
Aspergillus fumigatus |
99.42% |
7 |
Aspergillus welwitschiae |
100% |
Figure 2. Phylogenetic tree of 16S ribosomal RNA gene sequences of B. cereus isolated from cosmetics
Figure 3. Phylogenetic tree of 16S ribosomal RNA gene sequences of B. subtilis isolated from cosmetics
Figure 5. Phylogenetic tree of 16S ribosomal RNA gene sequences of S. aureus isolated from cosmetics
Figure 6. Phylogenetic tree of 18S ribosomal RNA gene sequences of A. flavus isolated from cosmetics
Figure 7. Phylogenetic tree of 18S ribosomal RNA gene sequences of A. fumigatus isolated from cosmetics
Figure 8. Phylogenetic tree of 18S ribosomal RNA gene sequences of A. welwitschiae isolated from cosmetics
MICs determination
The MIC is defined as the lowest concentration of an antibacterial agent that completely inhibits the growth of microbes after 24 hours.21 The MFC is defined as the lowest drug concentration that kills 95% of an inoculum.22
Determining the MIC is a good method for measuring the sensitivity of bacteria to preservatives, and the MIC value is inversely proportional to the sensitivity of the bacteria.
The antibacterial activities of four preservatives against bacteria are studied. Bacterial strains were isolated from contaminated cosmetics. In this experiment, the MICs of four common preservatives were determined in vitro for seven microbial strains using a serial dilution method.
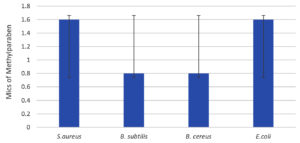
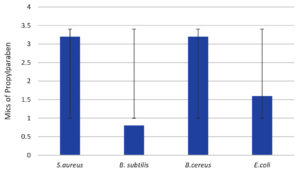
In Figures 9-12, the tested bacterial strains showed different sensitivity to the preservatives. B. subtilis was the most sensitive bacterial strain against all the preservatives, even under low concentrations (0.1-0.8). While S.aureus is the most resistant strain, growth inhibition observed at high concentrations (1.6-3.2) against all preservatives.
Figure 9. Minimal inhibitory concentrations of preservative phenoxy ethanol (PE) against various bacterial strains. The standard deviations are indicated by the bars S. aureus, B. subtilis, B. cereus and E. coli
Figure 10. Minimal inhibitory concentrations of preservative benzyl alcohol (BA) against various bacterial strains. The standard deviations are indicated by the bars S. aureus, B. subtilis, B. cereus and E. coli
Figure 11. Minimal inhibitory concentrations of preservatives Methylparaben against various bacterial strains. The standard deviations are indicated by the bars S. aureus, B. subtilis, B. cereus and E. coli
Figure 12. Minimal inhibitory concentrations of the preservative propylparaben (PP) against various bacterial strains. The standard deviations are indicated by the bars S. aureus, B. subtilis, B. cereus and E. coli
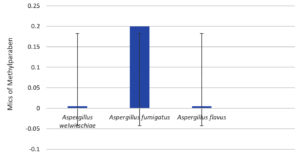
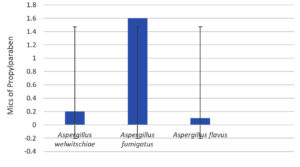
Regarding the fungal strains, Figures 13-16 shows the MICs of the preservatives;
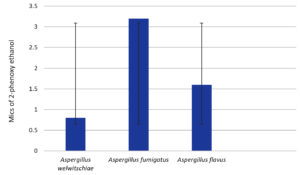
A. welwitschiae is the most sensitive fungal strain in low concentrations for PE and PB at 0.05 mg/ml, and for MB, BA at 0.8 and 1.6 mg/ml, respectively.
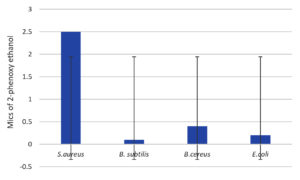
Figure 13. Minimal inhibitory concentrations of the preservative 2-phenoxyethanol (PE) against fungal strains. The standard deviations are indicated by the bars A. welwitschia, A. fumigatus and A. flavus
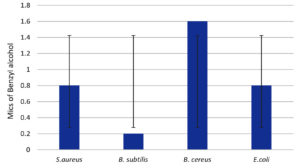
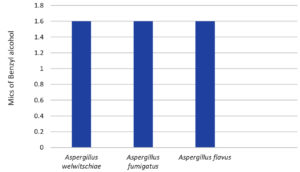
Figure 14. Minimal inhibitory concentrations of the preservative benzyl alcohol (BA) on various fungal strains. (Values are equal at 1.6 mg/ml)
Figure 15. Minimal inhibitory concentrations of the preservative methylparaben (MP) against various fungal strains. The standard deviations are indicated by the bars A. welwitschia, A. fumigatus and A. flavus
Figure 16. Minimal inhibitory concentrations of the preservative propylparaben (PP) against various fungal strains. The standard deviations are indicated by the bars A. welwitschia, A. fumigatus and A. flavus
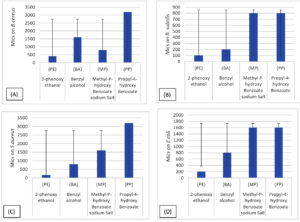
Figure 17 shows that PE had the strongest inhibitory effect on each bacterial strain at low concentrations (0.05-0.4 mg/mL), while MP had less effect on bacterial strains than BA.
Figure 17. Effects of the preservatives phenoxyethanol (PE), benzyl alcohol (BA), propylparaben (PP), and methylparaben (MP) on the bacterial strains. The standard deviations are indicated by the bars : (A) B. subtilis, (B) B. cereus, (C) S. aureus, and (D) E. coli
Orus et al.8 proposed that benzoic acid has antibacterial activity, as it decreased the intracellular pH of microorganisms, thereby inhibiting their growth.
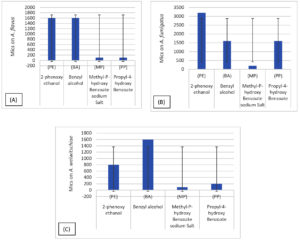
A. fumigatus showed good growth at high concentrations (3.2, 1.6, and 0.2) for PE, BA PB, and MB respectively. All fungal strains recorded (1.6 mg/mL) for BA. In Figure 18, MICs of MP ranged from 0.05-0.2 mg/mL are effective against all fungal strains in low concentrations, which indicates its strong preservative against fungi. MP and PP are derivatives of parabens. Many studies have demonstrated that parabens exert excellent bactericidal and fungicidal properties by preventing the cell replication of several microorganisms.7,23,24
Figure 18. Effects of preservatives phenoxyethanol (PE), benzyl alcohol (BA), propylparaben (PP), and methylparaben (MP) on the fungal strain. The standard deviations are indicated by the bars: (A) A. flavus, (B) A. fumigatus, and (C) A. welwitschiae
Growth of microorganisms inside cosmetics is an issue that needs to be reconsidered by researchers and preservative systems should be evaluated. Preservative Efficacy Testing (PET) and challenge tests are important tests that are usually used to check the quality of preservative system by inhibiting growth of microorganisms.25-27
In this study, seven microbial strains isolated from contaminated personal care products were studied. The MICs were calculated to determine the sensitivity of these strains to the four preservatives (PE, BA, PP, and MP) at different concentrations. B. subtilis is the most sensitive bacterial strain to preservatives at low concentrations (0.8-01). The MICs of MP ranged from 0.05-0.2 mg/mL, which indicates the strength of their inhibitory effect on fungi at low concentrations. The study showed variation in the strength of preservatives and variation of the interaction of microbes against preservatives. The choice of the best preservatives against bacteria or fungi requires a lot of studies for enhancing cosmetic products quality against microbial contamination.
ACKNOWLEDGMENTS
None.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Cosmetics Europe: The Personal Care Association. https://cosmeticseurope.eu/
- Alshehrei FM. Microbiological Quality Assessment of Skin and Body care Cosmetics by using Challenge test. Saudi J Biol Sci. 2024;31(4):103965.
Crossref - Lundov MH, Moesby L, Zachariae C, Johansen JH. Contamination versus preservation of cosmetics: a review on legislation, usage, infections, and contact allergy. Contact Dermatitis. 2009;60(2):70-78.
Crossref - Jildeh ZB, Wagner PH, Schoning MJ. Sterilization of Objects, Products, and Packaging Surfaces and Their Characterization in Different Fields of Industry: The Status in 2020. Phys Status Solidi A Appl Mater Sci. 2021;218(13):2000732.
Crossref - Stoffels KM. Modern and Safe antimicrobial stabilization of cosmetic products. Biocides and Preservation. 2012;7(1):18-22.
- Hugbo PG, Onyekweli AO, Lgwe L. Microbial contamination and preservative capacity of some brands of cosmetic creams. Trop J Pharm Res. 2003;2(2):229-234.
Crossref - Hill C. Preservation of cosmetics and toiletries. In: Rossmoore, H.W. (eds) Handbook of Biocide and Preservative Use. Springer, Dordrecht. 1995:39-115.
Crossref - Orus P, Perez LG, Leranoz S, Berlanga M. Increasing antibiotic resistance in preservative-tolerant bacterial strains isolated from cosmetic products. Int Microbiol. 2015;18(1):51-59.
Crossref - Nuzhath F. Bacteriological evaluation and comparison of unused brand and non-branded cosmetic products available in Jazan region of Saudi Arabia. J Microb World. 2014;16(1-2):57-67.
- Kattstrom D, Beronius A, Ruden C, Agerstrand M. Stricter regulation applies to antimicrobial substances when used as biocides compared to cosmetics under current EU legislation. Emerging Contaminants. 2022;8:229-242.
Crossref - European Chemical Agency. Cosmetic Products Regulation, Annex V – Allowed Preservatives. Regulation (EU) 2024/996. https://echa.europa.eu/es/cosmetics-preservatives
- Deyner SP. Mechanisms of Action of Antibacterial Biocides. Int Biodeter Biodegr. 1995;36(3-4):221-245.
Crossref - Aerts O, Goossens A. Contact Allergy to Preservatives. In: Johansen, J.D., Mahler, V., Lepoittevin, JP., Frosch, P.J. (eds) Contact Dermatitis. Springer, Cham. 2021:835-876.
Crossref - Nowak-Lange M, Niedzialkowska K, Lisowska K. Cosmetic preservatives: hazardous micropollutants in need of greater attention? Int J Mol Sci. 2022;23(22):14495.
Crossref - Steinberg DC. Preservatives for Cosmetics, 2nd edition, Allured Publishing, Illinois. 2006.
- Silva V, Silva C, Soares P, Garrido EM, Borges F, Garrido J. Isothiazolinones biocides: chemistry, biological, and toxicity profiles. Molecules. 2020;25(4):991.
Crossref - Neza E, Centini M. Microbiologically Contaminated and Over-Preserved Cosmetic Products According Rapex 2008–2014. Cosmetics. 2016;3(3):1-11.
Crossref - Noureddine H, Fernandes IP, Heleno SA, et al. Cosmetics Preservation: A Review on Present Strategies. Molecules. 2018;23(7).1-41.
Crossref - Aktar N, Karim MM, Khan SN, Rahman M, Begum A, Hoq MM. In silico Studies of Parasporin Proteins: Structural and Functional Insights and Proposed Cancer Cell Killing Mechanism for Parasporin 5 and 6. Microbial Bioactives. 2019;2(1):E082-E090.
Crossref - Shishir A, Roy A, Islam N, Rahman A, Khan SN, Hoq MM. Abundance and diversity of Bacillus thuringiensis in Bangladesh and their cry genes profile. Front Environ Sci. 2014;2:20.
Crossref - Lagier JC, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28(1):208-236.
Crossref - Shafi MS. Determination of antimicrobial MIC by paper diffusion method. J Clin Path. 1975;28(12):989-992.
Crossref - Sandel T. Microbiological challenges to the pharmaceuticals and healthcare. Pharmaceutical Microbiology. 2016:281-294.
Crossref - Saverina EA, Frolov NA, Kamanina OA, Arlyapov VA, Vereshchagin AN, Ananikov VP. From antibacterial to antibiofilm targeting: an emerging paradigm shift in the development of quaternary ammonium compounds (QACs). ACS Infect Dis. 2023;9(3):394-422.
Crossref - Hodges N, Hanlon C. Microbial preservative encase testing. In: Handbook of Microbiological Quality Control. Pharmaceuticals and Medical Devices (R. M. Baird, N. A. Hodges and S. P. Denyer, eds). Taylor & Francis, London. 2000:168-189. chapter_10
Crossref - Uzdrowska K, Gorska-Ponikowska M. Preservatives in cosmetics technology. Aesthetic Cosmetology and Medicine. 2023;12(2):73-78.
Crossref - Dini I, Laneri S. The new challenge of green cosmetics: Natural food ingredients for cosmetic formulations. Molecules. 2021;26(13):3921.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.