ISSN: 0973-7510
E-ISSN: 2581-690X
This study addresses the critical demand for sustainable energy by optimizing bioethanol production, focusing on maximizing yield from the ethanologenic bacterial strain Enterobacter mori SVIDE3 through a systematic one-factor-at-a-time (OFAT) approach. Key fermentation parameters, such as temperature, pH, nitrogen source, and incubation time, were individually examined to determine the conditions that enhance bioethanol production. The optimized model supports scaling bioethanol production, underscoring the efficiency of Enterobacter mori SVIDE3 and the power of statistical design in microbial bioethanol optimization. The OFAT approach revealed Enterobacter mori SVIDE3’s strong carbohydrate fermentation potential, achieving ethanol yields of 4.25 g/L from glucose at optimum conditions of 72 hrs incubation, 35 °C, yeast extract as a nitrogen source, and pH 9.0, and 2.69 g/L from xylose at optimum conditions of 72 hrs incubation, 35 °C, yeast extract, and pH 7.0. This research contributes valuable insights into improving fermentation processes and addresses key scalability and commercial feasibility challenges associated with Enterobacter mori SVIDE3 for sustainable bioethanol production.
Bioethanol, Renewable Energy, Fermentation, Ethanologenic Microbes, Enterobacter mori
The escalating expansion of the global economy has sparked apprehension regarding rising energy consumption and the consequent buildup of atmospheric greenhouse gases, exacerbating the profound impacts of climate change.1 In response, nations worldwide are actively directing investments to renewable energy sources, notably emphasizing the advancement of biofuels.2 Derived from biomass, particularly organic waste materials, biofuels offer a significant reduction in ecological footprint compared to traditional fossil fuels. Ethanol, a prominent biofuel, is distinguished for its environmental advantages, primarily due to its low pollution levels upon combustion. The pursuit of cost-effective ethanol production has spurred substantial research endeavors, with bioethanol emerging as a leading contender, anticipated to surpass 130 billion litres annually in global production, led by the United States and Brazil.3,4 Beyond its role as an energy source, ethanol assists as a precursor for various bio-based products, showcasing its versatility.5
Produced through microbial fermentation of carbohydrates extracted from diverse sources such as plants or algae, including corn, sugarcane, wheat, and lignocellulosic biomass (LCB), bioethanol holds promise as a renewable fuel, dominating the transportation biofuel sector. Despite corn starch and cane sugar traditionally serving as primary sources, innovations in ethanol production from lignocellulosic biomass have broadened possibilities to utilize abundant and cost-effective raw materials. However, extracting sugar monomers from lignocellulose demands extensive thermochemical or biochemical processing.6 The natural process of alcoholic fermentation, which involves decompose complex organic molecules into simpler ones, is harnessed to transform simple sugars into carbon dioxide, ethanol, and other by-products. Concerns have surfaced regarding the historical reliance on food crops for fermentation, prompting exploration into alternative biomass sources like lignocellulosic materials and algae. Technological advancements now facilitate ethanol production from a broader array of biomass resources, sorted into three generations based on feedstock utilization, reflecting ongoing advancements. Common processes across all generations include pretreatment, hydrolysis (except for sugar cane fermentation), and fermentation to convert sugars into ethanol. Certain feedstocks, like lignocellulosic materials and algal biomass, necessitate pretreatment to release fermentable sugars, classifying them as new-generation processes.4
The One-Factor-At-A-Time (OFAT) method simplifies the optimization by assessing individual variables while keeping others constant. Though time-consuming and limited in capturing interactions, it remains valuable, especially with resource constraints. While advanced methods like RSM and ANN offer deeper insights, OFAT’s ease of use ensures its continued relevance. Integrating it with modern techniques could enhance optimization.7 This study presents the first report of Enterobacter mori as an ethanol-producing bacterium. While its fermentative potential has not been previously explored, the Enterobacter genus is well known for bioethanol production. Enterobacter aerogenes is recognized for its efficiency in ethanol and biohydrogen synthesis, while Enterobacter hormaechei has been documented for bioethanol production.8,9
Lignocellulosic biomass fermentation involves pretreatment and hydrolysis to release fermentable sugars, but its resistance to chemical and enzymatic treatments presents challenges. Hydrolysates contain hexose, pentose, and inhibitory compounds like furfural and phenols, which hinder fermentation. Most industrial microbes efficiently utilize hexose (e.g., glucose) but struggle with pentose (e.g., xylose) due to missing pathways and glucose-induced repression. Enhancing substrate utilization requires engineering sugar transporters and isolating microbes capable of utilizing both sugars.10-12 This study aims to isolate ethanologenic bacteria that ferment both hexose and pentose for ethanol production while optimizing key physical and chemical parameters for improved bioethanol yield.
Chemicals
Luria Bertani growth medium and potassium dichromate were bought from HiMedia Laboratories Private Limited, India. Tri-n-butyl phosphate (TBP) was bought from Sisco Research Laboratories Pvt. Ltd., India. All media, and solutions were formulated with sterile distilled water.
Isolation and initial screening
Wild-type bacteria sourced from sugarcane juice was obtained from local market and allowed to incubate for one week at 30 °C to encourage natural bacterial growth. Subsequently, the sample underwent serial dilution up to a 10-6 dilution factor. From dilutions of 10-4 and 10-5, 100 µL of sample was spread onto Luria Bertani agar (LBA), composed of Tryptone (10 g/L), peptone (10 g/L), yeast extract (5 g/L), sodium chloride (10 g/L), agar (15 g/L), and sterile distilled water. The inoculated agar plates were then incubated at 30 °C for 72 hours. Distinct colonies displaying varying cultural characteristics were selected and sub-cultured onto separate LBA plates, followed by another 72-hour incubation period at 30 °C.13-15 The initial screening of ethanol production by the bacteria was determined using an iodoform test. This entailed mixing the cell-free fermentation broth with a 2 M Iodine solution, then adding a 2 M NaOH solution, while keeping a volumetric ratio of 1:3:4, respectively. The resulting mixture was subsequently placed in a hot water bath at 65 °C for a duration of 10 minutes.16,17
Secondary screening
Initial screening for sugar fermentation was performed using the Durham tube method. 0.1 mL of the bacterial inoculum was inoculated into a test tube containing 10 mL of MSM broth medium composed of NaCl (0.48 g/L), KH2PO4 (2.88 g/L), MgSO4 (0.12 g/L), CaCl2.2H2O (0.016 g/L), yeast extract (4.32 g/L), Na2HPO4 (5.76 g/L), and glucose (4.8 g/L) and Durham tube. The test tubes were then incubated at a temperature of 35 ± 2 °C for 48 hours to observe gas production.17,18
Carbohydrate fermentation test
Several carbohydrates such as glucose, sucrose, maltose, galactose, fructose, lactose, mannose, arabinose, and xylose were utilized in the carbohydrate fermentation test. This test was conducted with slight modifications. Specifically, a 0.1 mL aliquot of a 24-hour-old bacterial culture isolate was inoculated into 10 mL of MSM broth medium in a test tube, then incubated at 30 °C for 72 hours. Gas formation was monitored every 12 hours. The fermentation of each carbohydrate molecule by each bacterial isolate was assessed by observing gas formation in the Durham tube, with or without a change in medium color, after 72 hours of incubation.13
Ethanol tolerance study
The objective of this study was to examine the ethanol tolerance of SVIDE3. Triplicate cultures of the SVIDE3 bacterial strain were inoculated at varying concentrations (1-10% v/v) of ethanol into 10 mL of Luria Bertani broth (pH 7). Samples of 2 mL were withdrawn after 24 hours, and the absorption at 600 nm was measured for analysis.19
Bacterial identification
SVIDE3 was discovered via partial sequencing of the 16S rRNA gene. DNA extracted from the provided culture was isolated using the QIAGEN DNeasy UltraClean Microbial Kit. Following this, a segment of the 16S rRNA gene was amplified using universal primers 27F and 1492R. Subsequently, the PCR amplicon underwent purification to eliminate contaminants using the QIAGEN QIAquick PCR Purification Kit. The sequencing was performed using the BDT v3.1 Cycle sequencing kit on the Applied Biosystems™ MiniAmp™ Plus Thermal cycler and analyzed on the ABI 3730xl Genetic Analyzer. (Himedia, Mumbai, India). The obtained sequences underwent noise removal using the BioEdit Tool (BioEdit v7.0.5), and Basic Local Alignment Search Tool for nucleotides (BLASTn) was utilized in the National Centre for Biotechnology Information (NCBI) database to obtain homologous sequences. Then, multiple sequence alignment was conducted using CLUSTALW, employing homologous sequences sourced from the NCBI database. Finally, a phylogenetic tree was generated utilizing the neighbor-joining method in MEGA 11, aiming to ascertain the relationship between the test strain and other bacterial strains.17,20
Preparation of the inoculum
The bacterial inoculum was prepared by transferring one loopful of bacterial culture into an Erlenmeyer flask containing 100 mL of nutrient broth. The flask was subsequently incubated in an incubator at 37 °C for a duration of 18 hours. The resulting 18-hour bacterial suspension was used as the inoculum for further study.
Optimization of the various factors
Bioethanol production was optimized using the classical one-factor-at-a-time (OFAT) approach to maximize yield from an isolated bacterial strain. To prevent interference from inhibitors generated during pretreatment, commercially available glucose and xylose were used as substrates to evaluate the strain’s ethanol production potential. Key process parameters were systematically optimized, including incubation time (24-120 h), pH (5, 7, and 9), nitrogen sources at 0.05% concentration (beef extract, yeast extract, ammonium sulfate, urea, and peptone), and temperature (25 °C, 35 °C, and 45 °C).
Dichromate assay for bioethanol determination
Sample collection
After every incubation period, samples were obtained from triplicate flasks. Then centrifuged at 10,000 rpm for 10 minutes. Following centrifugation, the supernatants were meticulously stored and utilized for bioethanol determination.
Determination of Bioethanol by dichromate method
Solvents, specifically Tri-n-butyl phosphate (TBP) with a density of 0.9727 and a water solubility of 0.028% (w/v), were utilized for extracting ethanol in an aqueous solution. In a 2.0 ml microtube, 1 ml of TBP was mixed with 1 ml of either an ethanol standard solution or a sample, and the resulting mixture underwent vigorous vortexing for 60 seconds using a vortex mixer. Following phase separation, 750 µL of the upper solvent phase was transferred to a new microtube. Subsequently, 750 µL of dichromate reagent was introduced, and the mixture was vortexed vigorously for 10 minutes. After another phase separation, the lower phase developed a blue-green color. One hundred microlitres of the oxidation products (from the lower phase) were diluted with 900 µL of deionized water. The optical density at 595 nm (A595) of the tested sample was assessed using a spectrophotometer. The ethanol concentration in the sample was estimated from the ethanol standard curve, depicting the relationship between absorption at 595 nm and ethanol concentrations. However, the solvent extraction and dichromate assay may face sensitivity issues due to interference from culture components, affecting color development. While reliable for ethanol concentrations up to 8% (v/v), higher concentrations may cause inaccuracies due to saturation effects or incomplete ethanol extraction.21,22
Statistical evaluation
Each experiment was performed in triplicate flasks, and the data provided in the tables are presented as the average ± standard error (S.E). The calculations and graphs are made with the help of OriginPro software version 2024b and Microsoft Excel version 2016.
Rising CO2‚ levels and fossil fuel depletion drive the search for alternative energy sources, with lignocellulosic ethanol emerging as an eco-friendly option. However, challenges like pretreatment and efficient sugar extraction limit commercialization.23 Biomass pretreatment releases fermentable sugars from complex lignocellulosic structures but often results in toxic byproducts, which necessitates engineering microbial hosts to co-utilize sugars and tolerate toxins.24,25 Ethanol production, typically from glucose via glycolysis, involves pyruvate fermentation catalyzed by Alcohol Dehydrogenase (ADH) and Pyruvate Decarboxylase (PDC), studied extensively in Saccharomyces cerevisiae and Escherichia coli. The glycolysis pathways Entner-Doudoroff (ED) and Embden-Meyerhof-Parnas (EMP) show varied Adenosine triphosphate (ATP) yields, influencing biomass production.24 Xylose metabolism occurs via isomerase, oxidoreductase, and oxidative pathways, each feeding into the pentose phosphate pathway (PPP) differently, depending on the organism type.26,27 Most industrial microbes utilize hexoses like glucose efficiently but struggle with pentoses such as xylose due to lack of suitable pathways. Engineering efforts focus on pentose-preferential sugar transporters and isolating microbes that co-utilize glucose and xylose, vital for biofuels.10 This study isolates ethanologenic bacteria from spoiled fruits and juices using iodoform and carbohydrate fermentation tests, aligning with similar findings on ethanologenic microbial diversity in spoiled juices.13,14,28
Isolation and identification of ethanologenic bacteria
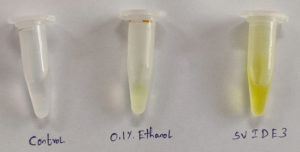
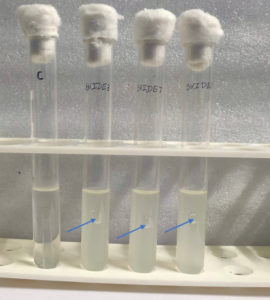
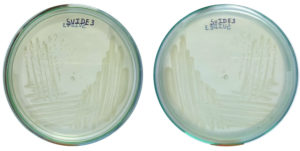
Spoiled sugarcane juice samples were subjected to serial dilution and subsequently plated onto agar plates containing Luria Bertani (LB) medium. This process led to the isolation of 17 bacterial strains. The strains were then transferred to new LB agar plates for additional analysis. Initially, ethanol production screening was conducted using the iodoform test with potassium iodide and sodium hydroxide. Out of the 17 strains, only three (SVIDE3, SVIDE7, SVIDE11) exhibited a positive result, as indicated by the yellow color formation after boiling for 10 minutes at 65 °C (Figure 1), suggesting their ability to produce ethanol. Following this, three bacterial strains were screened for gas production in Durham’s tubes within a glucose-containing medium as a secondary assessment. Gas or carbon dioxide production is a byproduct of ethanol production. The SVIDE3, SVIDE7 and SVIDE11 bacterial strain showed gas production in Durham’s tube (Figure 2), consistent with previous studies.16,17 In the carbohydrate fermentation test, ethanol producing bacteria vary in their fermentation and assimilation of carbohydrate molecules. A broader carbohydrate fermentation of ethanologenic microbe is crucial for the effective conversion of various sugars in the biomass to ethanol.29 Results of this study showed that pre-selected isolate SVIDE3 (Figure 3A) ferment glucose, xylose, sucrose, Arabinose, and fructose to different extents after 48 hrs of incubation. SVIDE7 (Figure 3B) and SVIDE11 (Figure 3C) showed gas production in glucose, sucrose and fructose containing test tubes.
Figure 1. Iodoform test confirming bioethanol production by SVIDE3 bacterial culture, indicated by distinct yellow precipitate, a characteristic reaction of ethanol with iodine in a basic medium
Figure 2. Secondary screening of bioethanol production using the Durham tube method, where gas formation in SVIDE3, SVIDE7 and SVIDE11 (marked by arrows) indicates microbial fermentation and ethanol production
Figure 3. Carbohydrate fermentation test showing gas formation in Durham’s tube, confirming carbohydrate utilization by bacterial isolates (A) SVIDE3, (B) SVIDE7, and (C) SVIDE11 using different sugars (A – Arabinose, F – Fructose, G – Glucose, S – Sucrose, X – Xylose)
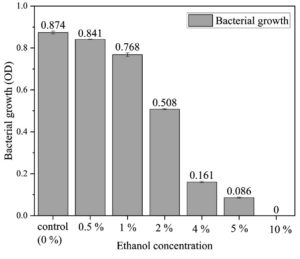
Ethanol tolerance is crucial for ethanologenic microbes, as high ethanol levels disrupt cellular integrity by affecting both hydrophobic proteins in membranes and hydrophilic proteins in the cytoplasm, impacting stability and function.30,31 Kluyveromyces marxianus exhibits moderate tolerance at 5-7% v/v ethanol, beneficial for bioethanol production,32 and the SVIDE3 (Figure 4) strain also shows growth up to 5% ethanol in Luria Bertani medium before inhibition occurs (Figure 5). Common strains like Zymomonas mobilis and Saccharomyces cerevisiae are widely used for bioethanol due to high ethanol yield, rapid sugar uptake, glucose tolerance, ethanol resistance, and Generally Regarded as Safe (GRAS) status.33,34 Compared to these, SVIDE3 shows similar ethanol tolerance, marking it as a viable candidate for optimized bioethanol production.
Figure 5. Influence of ethanol concentration on SVIDE3 growth, assessing tolerance and adaptability under different ethanol levels
SVIDE3’s high ethanol tolerance makes it a strong candidate for industrial bioethanol production by reducing product inhibition. Ethanol fermentation involves glycolysis, where pyruvate is converted by Alcohol Dehydrogenase (ADH) and Pyruvate Decarboxylase (PDC). The Entner-Doudoroff (ED) and Embden-Meyerhof-Parnas (EMP) pathways produce varying ATP levels, influencing biomass, while xylose metabolism integrates into the pentose phosphate pathway (PPP) through different enzymatic routes.24,26,27
Notably, bacterial isolate SVIDE3 exhibited remarkable carbohydrate fermentation capacity, leading to its selection for further study in ethanol production. To classify strain SVIDE3 phylogenetically, the 16S rRNA gene sequence (1476 bp) was analyzed. Comparison showed high similarity to the Enterobacter genus, specifically Enterobacter mori YIM Hb-3 (99.93% similarity) and Leclercia adecarboxylata CIP 82.92 (98.44% similarity), with slightly lower homology to Enterobacter cloacae subsp. dissolvens strain ATCC 23373 (98.04% similarity). The neighbor-joining phylogenetic tree (Figure 6) grouped strain SVIDE3 with members of the Enterobacter genus, confirming its classification as Enterobacter mori. The sequence was deposited in the NCBI GenBank database and assigned accession number PP549909. Enterobacter, a genus of Gram-negative bacteria, has been found in various habitats and is associated with environments and hosts such as water, plants, humans, and animals. These rod-shaped, non-spore-forming bacteria are widely distributed in nature. Some species act as plant growth-promoting bacteria in agriculture, while others are frequently isolated from spoiled fruits, spoiled fruit juices, brewery wastewater.35 The fermentative potential of Enterobacter is widely acknowledged. For instance, Enterobacter aerogenes is recognized for its proficiency in ethanol and biohydrogen synthesis.9,36
Figure 6. Phylogenetic tree of SVIDE3, illustrating its evolutionary relationship with closely related bacterial species
Optimization of bioethanol production
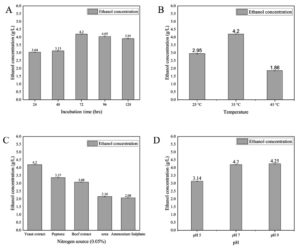
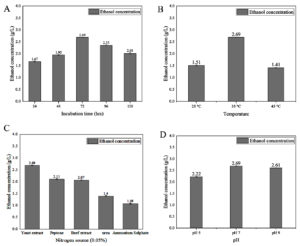
The optimization of physical and chemical factors, including temperature, pH, inoculum volume, nitrogen source, and incubation time, is essential for efficient bioethanol production, as slight variations in these conditions can significantly impact bacterial growth and product yields. This study utilized commercially available pentose (C5) and hexose (C6) sugars, such as xylose and glucose, to investigate bioethanol production by Enterobacter mori SVIDE3 under different environmental conditions, applying a one-factor-at-a-time (OFAT) approach to assess the effects on bioethanol yield.
Table:
Optimization through OFAT-Bioethanol production
| Category | S. No. | Incubation time (hrs) | Temp. | Nitrogen source (0.05 g) | pH | Ethanol Concen. (g/L) | |
|---|---|---|---|---|---|---|---|
| Glucose (1%) | Xylose (1%) | ||||||
| OFAT -Incubation time | 1. | 24 | 35 °C | Yeast extract | 7.0 | 3.04 ± 0.01 | 1.67 ± 0.02 |
| 2. | 48 | 35 °C | Yeast extract | 7.0 | 3.13 ± 0.02 | 1.95 ± 0.02 | |
| 3. | 72 | 35 °C | Yeast extract | 7.0 | 4.20 ± 0.01 | 2.69 ± 0.01 | |
| 4. | 96 | 35 °C | Yeast extract | 7.0 | 4.05 ± 0.04 | 2.35 ± 0.03 | |
| 5. | 120 | 35 °C | Yeast extract | 7.0 | 3.91 ± 0.04 | 2.01 ± 0.04 | |
| OFAT – Temperature | 6. | 72 | 25 °C | Yeast extract | 7.0 | 2.95 ± 0.03 | 1.51 ± 0.02 |
| 7. | 72 | 35 °C | Yeast extract | 7.0 | 4.20 ± 0.01 | 2.69 ± 0.01 | |
| 8. | 72 | 45 °C | Yeast extract | 7.0 | 1.86 ± 0.02 | 1.41 ± 0.02 | |
| OFAT -Nitrogen source | 9. | 72 | 35 °C | Yeast extract | 7.0 | 4.20 ± 0.01 | 2.69 ± 0.01 |
| 10. | 72 | 35 °C | Beef extract | 7.0 | 3.08 ± 0.01 | 2.07 ± 0.02 | |
| 11. | 72 | 35 °C | Peptone | 7.0 | 3.37 ± 0.02 | 2.11 ± 0.02 | |
| 12. | 72 | 35 °C | Ammonium Sulphate | 7.0 | 2.08 ± 0.01 | 1.08 ± 0.02 | |
| 13. | 72 | 35 °C | Urea | 7.0 | 2.16 ± 0.03 | 1.40 ± 0.02 | |
| OFAT – pH | 14. | 72 | 35 °C | Yeast extract | 5.0 | 3.14 ± 0.02 | 2.22 ± 0.02 |
| 15. | 72 | 35 °C | Yeast extract | 7.0 | 4.20 ± 0.01 | 2.69 ± 0.01 | |
| 16. | 72 | 35 °C | Yeast extract | 9.0 | 4.25 ± 0.02 | 2.61 ± 0.01 | |
Values express mean + standard error, number of replicate (n = 3)
Ethanol production (Figure 7A, B, C, D and Table) reached its peak at 4.25 ± 0.01 g/L on the third day of incubation when glucose was utilized as the primary carbon source. The most favorable conditions for ethanol production with glucose involved yeast extract as the nitrogen source, maintaining a pH range between 7.0 and 9.0, and incubation at a temperature of 35 °C. Regarding ethanol production with xylose (as depicted in Figure 8A, B, C, D and outlined in Table), the highest yield of 2.69 ± 0.01 g/L was achieved on the third day of incubation. Yeast extract was found to be the optimal nitrogen source, while maintaining a pH between 7.0 and 9.0 was conducive to xylose-based ethanol production.
Figure 7. Effect of different factors on Bioethanol production using glucose (A) incubation time, (B) temperature, (C) nitrogen source, and (D) pH
Figure 8. Impact of different parameters on bioethanol production using xylose, analyzing (A) incubation time, (B) temperature, (C) nitrogen source, and (D) pH
Effect of temperature
The optimal temperature for achieving higher bioethanol production by E. mori SVIDE3 is 35 °C for both glucose and xylose substrate (Figure 7B and 8B). It contrasts with optimal temperature for bioethanol production by other bacterial strains like Z. mobilis ATCC 10988,37 Z. mobilis CP438 and Z. mobilis MTCC 2427.39 Periyasamy and colleagues documented the production of bioethanol from sugar molasses employing S. cerevisiae, achieving a maximum bioethanol yield of 53% at a temperature of 35 °C.40 Ado and colleagues described the process of ethanol production from cassava starch, employing a co-culture of Aspergillus niger and S. cerevisiae in a simultaneous saccharification, achieving peak ethanol production at 35 °C.41 Banerjee et al., examined the impact of temperature on bioethanol production from rice husk, noting the highest production at 30 °C.42
Effect of pH
The optimum pH for bioethanol production in our study was found to be 7 and 9 for both glucose and xylose substrates. This aligns with the findings of Sarkar et al., who reported peak bioethanol production at pH 8.0. Similarly, our optimized pH is consistent with the results of Pinilla et al. and Sharma et al., who achieved higher bioethanol yields of 93.5 g/L and 14.7 g/L, respectively, at pH 7.0.43,44 In contrast, Mohseni et al. observed an effective pH range of 6-8 for both microbial growth and ethanol production, differing somewhat from our findings.10
Effect of nitrogen source
Our study revealed that bioethanol production was enhanced by adding yeast extract. Organic nitrogen sources, such as yeast extract, are known to support microbial growth during fermentation.45 Research shows that supplementing crude glycerol and glucose fermentations with yeast extract can increase ethanol yields and biomass growth. The right nitrogen source can also reduce by-product formation, further boosting ethanol production. Nikel et al. demonstrated that yeast extract significantly enhanced bioethanol production in E. coli.46
Effect of Incubation time
Fermentation time denotes the period during which the fermentation process reaches its peak efficiency, converting fermentable sugars into alcohol as required. Prolonged incubation periods can lead to the production of high concentrations of bioethanol, which may become detrimental to the prepared mash.47 In our current investigation, we determined that the optimal fermentation or incubation duration for bioethanol production is 72 hours for both sugar types. Beyond this threshold, extending the fermentation time resulted in a decrease in bioethanol yield. Our findings highlight that the maximum ethanol production was attained at the 72-hour, signifying the point at which the fermentation process becomes most efficient in converting fermentable sugars into alcohol. This observation is consistent with the outcomes of a study performed by Fasiku and Wakil, where optimizing the fermentation time to 72 hours yielded the highest ethanol production of 1.58 g/L using S. cerevisiae from pretreated maize straw hydrolysate, corroborating our own findings.45
The bioethanol production levels of E. mori SVIDE3 are 4.25 g/L and 2.69 g/L, which are lower compared to many microbes. However, it outperforms S. cerevisiae (1.289 g/L) utilizing water hyacinth as a substrate,48 as well as Pachysolen tannophilus MTCC 1077 (3.36 g/L) using sugarcane bagasse as a substrate.49 E. mori SVIDE3 exhibits superior bioethanol production compared to certain microbial counterparts, demonstrating exceptional sugar utilization and ethanol yield across diverse conditions. Its potential in lignocellulose-based bioethanol production makes it a valuable candidate for industrial applications. Genetic engineering can further enhance its ethanol production by optimizing carbon flux, minimizing by-products, and knocking out competing pathways such as lactate and acetate formation. Introducing ethanol-producing genes like adh (alcohol dehydrogenase) and pdc (pyruvate decarboxylase) from Zymomonas mobilis can improve fermentation efficiency, while engineering E. mori SVIDE3 to enhancing tolerance to inhibitors like furfural, acetic acid, and ethanol through stress response modifications increases strain robustness. Additionally, overexpressing sugar transporters improves substrate uptake, leading to higher ethanol yields, reinforcing E. mori SVIDE3’s promise as a strong candidate for second-generation bioethanol production.
In conclusion, our study successfully isolated an ethanologenic bacterium, identified it as Enterobacter mori SVIDE3, and subsequently optimized its fermentation parameters to enhance ethanol production efficiency. The OFAT approach identified optimal conditions for ethanol production from xylose and glucose, significantly improving yield. The robust ethanol-producing capabilities of E. mori SVIDE3 highlight its industrial feasibility for sustainable bioethanol production. Future research should explore genetic modifications to enhance ethanol tolerance and sugar utilization, as well as co-fermentation strategies to maximize efficiency. Scaling up production and integrating E. mori SVIDE3 into industrial bioethanol processes will further validate its potential for commercial applications.
ACKNOWLEDGMENTS
This study was supported by the Department of Biotechnology, Manonmaniam Sundaranar University, Tirunelveli, Tamil Nadu, India. The authors also thank the Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Kanyakumari, Tamil Nadu, India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Unsal SBE, Tufan HNG, Canatar M, Yatmaz HA, Turhan I, Yatmaz E. Ethanol production by immobilized Saccharomyces cerevisiae cells on 3D spheres designed by different lattice structure types. Process Biochem. 2023;125:104-112.
Crossref - Atitallah IB, Arous F, Louati I, et al. Efficient bioethanol production from date palm (Phoenix dactylifera L.) sap by a newly isolated Saccharomyces cerevisiae X19G2. Process Biochem. 2021;105:102-112.
Crossref - Xu J, Li M, Ni T. Feedstock for bioethanol production from a technological paradigm perspective. BioResources. 2015;10(3):6285-6304
- Tse TJ, Wiens DJ, Reaney MJT. Production of bioethanol—A review of factors affecting ethanol yield. Fermentation. 2021;7(4):268.
Crossref - Sunarno JN, Prasertsan P, Duangsuwan W, Cheirsilp B, Sangkharak K. Improve biotransformation of crude glycerol to ethanol of Enterobacter aerogenes by two-stage redox potential fed-batch process under microaerobic environment. Biomass and bioenergy. 2020;134:105503.
Crossref - Lin CW, Tran DT, Lai CY, Yet-Pole I, Wu CH. Response surface optimization for ethanol production from Pennisetum Alopecoider by Klebsiella oxytoca THLC0409. Biomass and Bioenergy. 2010;34(12):1922-1929.
Crossref - Lahiri D, Nag M, Mukherjee D, Garai S, Banerjee R, Ray RR. Recent trends in approaches for optimization of process parameters for the production of microbial cellulase from wastes. Environmental Sustainability. 2021;4(2):273-84.
Crossref - Pati S, Mohanty MK, Mohapatra S, Samantaray D. Bioethanol production by Enterobacter hormaechei through carbon monoxide rich syngas fermentation. Int J Energy Res. 2022;46(14):20096-106.
Crossref - Choonut A, Saejong M, Sangkharak K. The production of ethanol and hydrogen from pineapple peel by Saccharomyces cerevisiae and Enterobacter aerogenes. Energy Procedia. 2014;1 (52):242-249.
Crossref - Mohseni M, Ebrahimi H, Chaichi MJ. Isolation and optimization of ethanol producing bacteria from natural environments of mazandaran province in Iran. J Genet Resour. 2015;1(1):35-44.
- Ningthoujam R, Jangid P, Yadav VK, et al. Fermentation of rice straw hydrolyzates for bioethanol production and increasing its yield by applying random physical and chemical mutagenesis. Waste Biomass Valor. 2024;15(9):5105-5123.
Crossref - Ningthoujam R, Jangid P, Yadav VK, Sahoo DK, Patel A, Dhingra HK. Bioethanol production from alkali-pretreated rice straw: effects on fermentation yield, structural characterization, and ethanol analysis. Front Bioeng Biotechnol. 2023;3:11:1243856.
Crossref - Bitew D, Tesfaye A, Andualem B. Isolation, screening and identification of ethanol producing yeasts from Ethiopian fermented beverages. Biotechnol Reports. 2023;40:e00815.
Crossref - Babiye B. Isolation and identification of bacteria from fresh fruit juice prepared in cafeterias and restaurants, Axum Town, Ethiopia. Biosci Biotechnol Res Asia. 2017;14(1):307-313.
Crossref - Promon SK, Kamal W, Rahman SS, Hossain MM, Choudhury N. Bioethanol production using vegetable peels medium and the effective role of cellulolytic bacterial (Bacillus subtilis) pre-treatment. F1000Research. 2018;7:271.
Crossref - Seelye RN, Turney TA. The iodoform reaction. J Chem Educ. 1959;36(11):572.
Crossref - Sarkar D, Prajapati S, Poddar K, Sarkar A. Production of ethanol by Enterobacter sp. EtK3 during fruit waste biotransformation. Int Biodeterior Biodegrad. 2019;145:104795.
Crossref - Weerasinghe WMLI, Madusanka DAT, Manage PM. Isolation and identification of cellulase producing and sugar fermenting bacteria for second-generation bioethanol production. Int J Renew Energy Dev. 2021;10(4):699-711.
Crossref - Jin G, Jiranek V, Hayes AM, Grbin PR. Isolation and characterization of high-ethanol-tolerance lactic acid bacteria from Australian Wine. Foods. 2022;11(9):1231.
Crossref - Biswas R, Vivekanand V, Saha A, Ghosh A, Sarkar A. Arsenite oxidation by a facultative chemolithotrophic Delftia spp. BAs29 for its potential application in groundwater arsenic bioremediation. Int Biodeterior Biodegradation. 2019;136:55-62.
Crossref - Seo HB, Kim HJ, Lee OK, Ha JH, Lee HY, Jung KH. Measurement of ethanol concentration using solvent extraction and dichromate oxidation and its application to bioethanol production process. J Ind Microbiol Biotechnol. 2009;36(2):285-292.
Crossref - Sriariyanun M, Mutrakulcharoen P, Tepaamorndech S, Cheenkachorn K, Rattanaporn K. A rapid spectrophotometric method for quantitative determination of ethanol in fermentation products. Orient J Chem. 2019;35(2).
Crossref - Toor M, Kumar SS, Malyan SK, et al. An overview on bioethanol production from lignocellulosic feedstocks. Chemosphere. 2020;242:125080.
Crossref - Kang A, Lee TS. Converting sugars to biofuels: ethanol and beyond. Bioengineering. 2015;2(4):184-203.
Crossref - Gencturk E, Ulgen KO. Understanding HMF inhibition on yeast growth coupled with ethanol production for the improvement of bio-based industrial processes. Process Biochem. 2022;121:425-438.
Crossref - Finneran KT, Popovic J. Solvent production from xylose. Appl Microbiol Biotechnol. 2018;102:8707-8715.
Crossref - Jagtap SS, Rao C V. Microbial conversion of xylose into useful bioproducts. Appl Microbiol Biotechnol. 2018;102:9015-9036.
Crossref - Nasreen Z, Jabeen S, Shafique M, et al. Production of alcohol by yeast isolated from apple, orange and banana. Int J Food Nutr Sci. 2014;1(2):16-19.
- Nitiyon S, Keo-Oudone C, Murata M, et al. Efficient conversion of xylose to ethanol by stress-tolerant Kluyveromyces marxianus BUNL-21. Springerplus. 2016;5:1-12.
Crossref - Mavrommati M, Papanikolaou S, Aggelis G. Improving ethanol tolerance of Saccharomyces cerevisiae through adaptive laboratory evolution using high ethanol concentrations as a selective pressure. Process Biochemistry. 2023;1(124):280-289.
Crossref - Saini P, Beniwal A, Kokkiligadda A, Vij S. Response and tolerance of yeast to changing environmental stress during ethanol fermentation. Process Biochem. 2018;1(72):1-2.
Crossref - Tesfaw A, Oner ET, Assefa F. Optimization of ethanol production using newly isolated ethanologenic yeasts. Biochem Biophys reports. 2021;25:100886.
Crossref - Xia J, Yang Y, Liu CG, Yang S, Bai FW. Engineering Zymomonas mobilis for robust cellulosic ethanol production. Trends Biotechnol. 2019;37(9):960-972.
Crossref - Zhang K, Lu X, Li Y, Jiang X, Liu L, Wang H. New technologies provide more metabolic engineering strategies for bioethanol production in Zymomonas mobilis. Appl Microbiol Biotechnol. 2019;103:2087-2099.
Crossref - Maintinguer SI, Lazaro CZ, Pachiega R, Varesche MBA, Sequinel R, de Oliveira JE. Hydrogen bioproduction with Enterobacter sp. isolated from brewery wastewater. Int J Hydrogen Energy. 2017;42(1):152-160.
Crossref - Rachman MA, Furutani Y, Nakashimada Y, Kakizono T, Nishio N. Enhanced hydrogen production in altered mixed acid fermentation of glucose by Enterobacter aerogenes. J Ferment Bioeng. 1997;83(4):358-363.
Crossref - Lee KJ, Skotnicki ML, Tribe DE, Rogers PL. The effect of temperature on the kinetics of ethanol production by strains of Zymomonas mobilis. Biotechnol Lett. 1981;3:291-296.
Crossref - Hayashi T, Kato T, Furukawa K. Respiratory chain analysis of Zymomonas mobilis mutants producing high levels of ethanol. Appl Environ Microbiol. 2012;78(16):5622-5629.
Crossref - Sivamani S, Baskar R. Process design and optimization of bioethanol production from cassava bagasse using statistical design and genetic algorithm. Prep Biochem Biotechnol. 2018;48(9):834-841.
Crossref - Periyasamy S, Venkatachalam S, Ramasamy S, Srinivasan V. Production of bio-ethanol from sugar molasses using Saccharomyces cerevisiae. Mod Appl Sci. 2009;3(8):32-37.
Crossref - Ado SA, Olukotun GB, Ameh JB, Yabaya A. Bioconversion of cassava starch to ethanol in a simultaneous saccharification and fermentation process by co-cultures of Aspergillus niger and Saccharomyces cerevisiae. Sci World J. 2009;4(1).
Crossref - Banerjee S, Sen R, Pandey RA, et al. Evaluation of wet air oxidation as a pretreatment strategy for bioethanol production from rice husk and process optimization. Biomass and Bioenergy. 2009;33(12):1680-1686.
Crossref - Pinilla L, Torres R, Ortiz C. Bioethanol production in batch mode by a native strain of Zymomonas mobilis. World J Microbiol Biotechnol. 2011;27:2521-2528.
Crossref - Sharma S, Jha PK, Panwar A. Production of bioethanol from wheat straw via optimization of co-culture conditions of Bacillus licheniformis and Saccharomyces cerevisiae. Discov Energy. 2021;1:1-8.
Crossref - Adedayo Fasiku S, Monilola Wakil S. Pretreatment of maize straw with Pleurotus ostreatus and Lentinus squarrosulus for bioethanol production using Saccharomyces cerevisiae. Nov Res Microbiol J. 2021;5(6):1480-1493.
- Nikel PI, Ramirez MC, Pettinari MJ, Mendez BS, Galvagno MA. Ethanol synthesis from glycerol by Escherichia coli redox mutants expressing adhE from Leuconostoc mesenteroides. J Appl Microbiol. 2010;109(2):492-504.
Crossref - Tenkolu GA, Kuffi KD, Gindaba GT. Optimization of fermentation condition in bioethanol production from waste potato and product characterization. Biomass Convers Biorefinery. 2022:1-19.
Crossref - Zhang Q, Weng C, Huang H, Achal V, Wang D. Optimization of bioethanol production using whole plant of water hyacinth as substrate in simultaneous saccharification and fermentation process. Front Microbiol. 2016;6:166358.
Crossref - Sasikumar E, Viruthagiri T. Simultaneous saccharification and fermentation (SSF) of sugarcane bagasse-kinetics and modeling. Int J Chem Biol Eng. 2010;3(2):57-64.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.