ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms exhibit remarkable phenotypic plasticity in extreme environments, including the stratosphere. This domain poses significant challenges such as almost zero gravity and a pronounced deviation from the typical physical forces experienced by living cells. Studies have highlighted the resilience of terrestrial microorganisms, particularly bacteria, in conditions resembling outer space, exploring how bacteria acclimate to the harsh environment of the Earth’s stratosphere, marked by dryness, cryogenic temperatures, intense UV radiation, low pressure, and microgravity. This study encompasses various aspects of bacterial biology, including growth, morphology, physiology, and genetic profiles, to uncover the mechanisms behind their adaptation to microgravity. This exploration provides insights into the physiological adaptation of humans to space. This study involved exposing bacterial cells to microgravity conditions at an altitude of 33 km above Earth, achieved through a sealed container space capsule launched from a space balloon. This study highlights the capacity of terrestrial bacterial strains to thrive and adapt to such extraordinary conditions.
Outer Space, Microgravity, Bacteria, Growth, Adaptation, Stratosphere
The stratosphere is one of the harshest locations on Earth. It presents with high levels of ultraviolet radiation, low air pressure, cold temperatures, and conditions that resemble microgravity.1 Studying how microbes survive in space helps us to understand how life adapts to these conditions.
Microorganisms emerged billions of years ago, evolving to thrive in a diverse range of extreme environments, earning them the label “extremophiles”.2,3 Microorganisms exhibit exceptional resilience in the stratosphere, an extreme environment dominated by microgravity. This almost weightless environment challenges the conventional notions of bacterial adaptation.4
Extremophiles are microorganisms with distinct traits. These traits help them to live in harsh places with high salt, radiation, and little food.5 These adaptations make these organisms great for studying how microbes survive beyond Earth’s limits. Previous studies have shown that certain microbes can survive vacuum and radiation in space. This raises interesting questions about life on other planets.6 Soil microorganisms can be strong, but we do not know how they respond to near-space environments such as the stratosphere. The stratosphere offers a unique setting for studying microbial adaptation. In this environment, convective mixing is low and microgravity effects occur. This makes it a valuable model for space microbiological research.7 This study shows how bacteria change when exposed to near-space conditions at a height of 33 km. A CubeSat system was launched using a high-altitude balloon. This helped to check for bacterial growth, antibiotic resistance, and genomic changes in this special environment. These findings contribute to the current astrobiological literature. They demonstrate how microbes respond to weak convection and harsh atmospheric conditions.
Research has revealed the profound impact of microgravity on bacterial metabolism, with adaptations involving alterations in gene expression at the transcriptional level.8 However, microorganisms are too small to directly sense gravity. Instead, the primary influence of microgravity is its impact on the surrounding fluid environment, particularly in the absence of gravity-driven convective mixing. On Earth, convection currents facilitate the distribution of nutrients and gases within liquid media; however, under microgravity conditions, this mixing is significantly reduced.9 Consequently, diffusion becomes the dominant mode of molecular transport, potentially altering microbial access to nutrients, oxygen levels, and metabolic waste distribution. Notably, this adaptation results in increased production of microcystins, which is linked to increased photosynthetic pigment concentration.10
Within extraordinary terrestrial environments exist hot springs, saline lakes, deserts, deep ocean expanses, acidic and alkaline zones, polar frostiness, contaminated regions, and areas with scarce energy and nutrient provisions. Within these harsh domains, microorganisms adapt to shifting conditions and devise cellular, biochemical, and molecular strategies. They produce enzymes, molecules, and metabolites to safeguard against extreme salinity, pH, pressure, temperature, solar radiation, nutrient availability, oxygen levels, osmotic pressures, and gravitational variations.11 The existence of extremophiles raises the intriguing possibility of microbial endurance in extraterrestrial conditions, serving as model organisms for understanding biological systems in space.12 Although microorganisms have been included as model organisms, other extremophiles have been classified as astrobiological model organisms, such as Deinococcus radiodurans.
The presence of extremophiles has sparked the fascinating notion that microorganisms can withstand the challenges posed by extraterrestrial conditions. These resilient microorganisms can potentially act as model organisms, aiding our understanding of the fate of biological systems in such environments. This encompasses survival in space with its extreme radiation, vacuum pressure, fluctuating temperature, and microgravity.13-15
The stratosphere, defined by extreme aridity, fluctuating temperatures, intense UV radiation, low atmospheric pressure, and microgravity, is a unique environment that influences microbial growth and behavior.16 Similarly, studies on balloon-borne microbes have shown that extremophilic bacteria, including Deinococcus species, survive exposure to intense UV radiation and low pressure, suggesting that certain microbes may possess inherent resistance to near-space environments. Limited research has explored microbial responses to microgravity, highlighting the need for further investigations in this understudied domain.17-19 Simulation experiments simulating planetary conditions offer crucial insights into the adaptability of microorganisms to extraterrestrial environments.20,21
In some ground test experiments, microbial growth has been observed under laboratory-simulated planetary conditions. These conditions attempt to replicate the harsh environmental conditions of other celestial bodies such as Mars. These studies are crucial as they offer insights into the adaptability and potential survival strategies of microorganisms in extraterrestrial environments.22 Existing research in this area underscores the need for further investigation into how microorganisms respond to the complex interplay of conditions found in the stratosphere.23,24 Gaining a comprehensive understanding of these responses can illuminate the possibilities and limitations of life in outer space and enhance our knowledge of extremophiles on Earth.25,26
To understand the impacts of the space environment, particularly microgravity, on microorganisms, we carefully planned collaborative research with the Kepler Space Foundation (KSF) (Las Vegas, NV 89107, USA). The primary goal was to analyze how bacterial strains exposed to the challenges of outer space within the stratosphere were adapted. Our goal was to examine bacterial survival and physiological responses under near-space conditions. How microorganisms are able to adapt to and thrive in this challenging environment, particularly under microgravity conditions.
We aimed to reveal a range of insights into fundamental growth patterns to elaborate on physiological transformations. Additionally, our efforts have been extended to the realm of genetic adaptations. We sought to elucidate the underlying genomic changes that microorganisms undergo as they navigate space. By examining these aspects, we aimed to contribute to the lack of knowledge regarding the resilience of microorganisms in space. Our collective pursuit has the potential to unearth invaluable data that can inform future space missions and enhance our comprehension of extremophiles, both beyond our planet and on Earth. The study used CubeSat, a sealed container space capsule. It was launched using a high-altitude balloon. The CubeSat carried bacterial samples to 33 kilometers above sea level. This balloon platform provides a unique opportunity to study how microbes adapt to low-convection conditions. Near-space balloon missions differ from traditional space flights. In traditional missions, microorganisms encounter microgravity, vibrations, and radiation. But balloons create a unique environment. This setting revealed that microbial health, energy use, and gene changes were barely affected by convective mixing. The CubeSat was teamed up with a space balloon to transport the samples. It also created a controlled environment to aid our scientific studies.
In the exploration of the impact of microgravity on microbial behavior, studies have shed light on the intricate changes observed in bacteria within space environments. The microgravity environment induces a distinct genetic response that leads to increased bacterial growth, heightened virulence, and a noticeable shift in antibiotic susceptibility. Notably, there is a deviation in the gene expression related to glucose metabolism and acid resistance, which is attributed to the absence of gravity-driven forces that reshape nutrient transport mechanisms. A further study on the investigation of the same bacterial strains used in this study exposed to simulated microgravity conditions is needed to add to our understanding of the consequences of altered gravity on microbial health and physiological functions. This insight extends beyond immediate concerns for astronauts’ well-being during prolonged space missions, providing valuable insights into fundamental bacterial behavioral paradigms on Earth. The nuanced discussion on antibiotic resistance and adaptive strategies employed by microbes in space reveals a complex interplay between extraterrestrial conditions and microbial survival mechanisms. This convergence of research emphasizes the need for continued exploration of the intricacies of microbial responses under the influence of microgravity.
Stratosphere Balloon Experiments/KSF Space Foundation
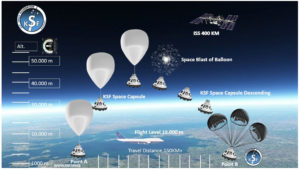
We deployed a high-altitude balloon carrying a sealed CubeSat system (Figure 1 and Figure 2). This helped us examine how bacteria adapted to near-space conditions. At that height, the bacterial samples were subjected to high radiation, low pressure, and microgravity-like conditions. Before launch, the balloon system underwent extensive tests to determine the integrity of the mission. The tests consisted of a cold soak test to mimic extreme thermal conditions. A drop test was conducted to evaluate mechanical strength. Operational tests to check data recording and tracking accuracy. The bacterial payload was placed in a pressurized chamber inside a CubeSat. This setup avoided contamination and let it face environmental stressors in the stratosphere.
Figure 1. A BalloonSat simple package designed by the KSF Space Foundation was used to carry out lightweight experiments in near space. The design is under weight and volume constraints, which allows for the inclusion of several BalloonSat during flight. The airframe material is lightweight Styrofoam or foam core, which provides reasonably good insulation (Source: KSF Space, Nevada, United States)
High-altitude balloons are useful for microbiological experiments in nearby spaces. They allow researchers to study how microbes react to extreme UV radiation, low atmospheric pressure, and low air mixing. In this study, a high-altitude balloon was used to carry bacterial samples at 33 km above sea level. The samples were exposed to near-space environmental stressors. The balloon system used temperature and pressure sensors. These sensors monitor the conditions during flight. This ensured that the experimental setup remained intact. The Kepler Space Foundation (KSF) tested the balloon system before flight. They wanted to evaluate their strength and stability in different environments. This process involved several tests. Cold soak tests checked how the payload withstood cryogenic temperatures. Drop tests were used to assess the impact resistance during the descent. Simulated pressure tests were conducted at high altitudes. These quality control steps kept the microbial samples sealed, uncontaminated, and thermally stable for the whole mission.
To investigate bacterial responses to near-space conditions, microbial cultures were housed within a sealed CubeSat payload specifically engineered to maintain sterile conditions while facilitating environmental exposure. The system was equipped to continuously monitor the temperature variations, radiation levels, and pressure changes during ascent and descent. This ensured that any observed alterations in microbial behavior could be directly linked to stratospheric microgravity conditions, eliminating the possibility of contamination or mechanical interference as contributing factors. The culmination of these sensors is an essential component in gathering invaluable data during flight. Before launch, rigorous testing protocols are necessary for BalloonSat to ensure their optimal performance and the successful acquisition of experimental data. These comprehensive tests encompass a series of evaluations, including the cold soak test, drop test, function test, and weighing procedure. The cold soak test was devised to replicate the harsh frigidity that the BalloonSat will encounter throughout its mission. The function of this test is to check the satellite’s resilience to extreme cold temperatures. Because spaceborne instruments must endure the stresses of launch and landing, a drop test is indispensable. This evaluation verified that the balloon sac remained structurally intact and continued to function seamlessly after abrupt descent. Meticulous attention given to these testing procedures ensures the reliability and operational capability of Balloon Sats, ultimately guaranteeing the delivery of meaningful experimental results.
Cultivation of bacterial strains and preparation for space capsule flight
Cultures of Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 29213), Bacillus subtilis (ATCC 6051), and Salmonella typhimurium (ATCC 14028) were cultivated under optimal laboratory conditions to ensure sampling during the logarithmic growth phase before exposure to near-space conditions. Each bacterial strain was cultured in sterile nutrient broth (NB) at 37 °C with continuous agitation at 200 rpm to maintain adequate aeration. Growth was monitored hourly by measuring the optical density at 600 nm (OD600) over 12 hours to ascertain the logarithmic growth phase. The growth kinetics of each strain were compared, identifying the logarithmic phase between 3 and 9 h post-inoculation, with peak exponential activity observed between 6 and 7 h. To ensure uniform growth conditions across all samples, cultures were harvested 6 h post-inoculation (mid-log phase) for exposure. To prepare for high-altitude balloon exposure, bacterial cultures were aliquoted into sterile 2 mL cryovials, which were subsequently sealed inside a sterile CubeSat payload. The payload was designed to maintain environmental integrity while permitting exposure to reduced convective mixing, low atmospheric pressure, and ultraviolet radiation at high altitudes. The constructed space capsule with the bacterial cultures was launched into the stratosphere using the high-altitude balloon (Figure 2).
B. subtilis is known to undergo sporulation in response to environmental stress, potentially affecting survival outcomes after exposure. Phase-contrast microscopy was employed before exposure to assess the presence of spores to differentiate between vegetative cells and spores. Post-exposure, samples underwent heat treatment at 80 °C for 10 min to eliminate vegetative cells, ensuring that survival measurements reflected the viability of spores exclusively. CFU counts were conducted before and after heat treatment to differentiate between the survival rates of spores and vegetative cells. These measures ensured that the observed differences in survival were attributed to the effects of near-space exposure rather than to inherent sporulation. These measures ensured that survival differences were attributed to near-space exposure rather than to natural sporulation. By combining phase-contrast microscopy, heat treatment, and CFU quantification, we effectively distinguished spore viability from vegetative cell survival, providing a precise assessment of the response of Bacillus subtilis to high-altitude conditions. After exposure, bacterial samples were promptly fixed with 2.5% glutaraldehyde in phosphate-buffered saline (PBS) at 4 °C to maintain cellular integrity for subsequent microscopy and biochemical analyses.
Determination of bacterial growth and survivability
A pure culture from each of the bacterial strains was obtained by streaking 10 µl of the (LB) broth into nutrient agar (NA) plates, and a single bacterial colony was picked up from the agar plate and used to inoculate 10 ml of (LB) broth. This procedure was implemented to prepare bacterial cultures for exposure to near-space conditions. Fresh bacterial cultures were streaked onto nutrient agar plates and incubated at 37 °C for 24 h to ensure active growth prior to inoculation into the liquid media for high-altitude balloon transport.
After vortexing, the culture was incubated for 24-48 h at 37 °C and 225 rpm. The physiological and morphological properties of the bacterial culture exposed to microgravity were examined both visually and microscopically and compared to the control ground strains. Microscopic analysis was performed to examine cellular morphology. Fixed samples, were treated with 2.5% glutaraldehyde in PBS to evaluate structural alterations induced by near-space exposure.
Bacterial growth and plate count for both the microgravity-exposed samples and controls were conducted by performing successive dilutions. This involved removing 1 ml of LB broth bacterial culture from each broth tube and then conducting 10-fold serial dilutions in fresh sterile LB. Diluted samples 100 µl of the diluted samples were spread on NA agar plates and incubated for 24 h-48 h at 37 °C, and bacterial colony growth was observed and recorded for up to 4 days. The antibacterial activity of the strains against antibiotics under microgravity and control conditions is shown in Figure 3 colonies were counted and expressed as log colony-forming units (CFU)/mL. The experiments were conducted three times (n = 3). We measured bacterial growth by counting the colony-forming units per milliliter (CFU/mL).
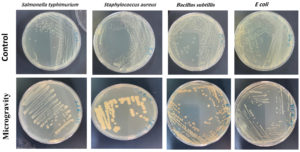
Figure 3. A visual representation of the observed variations offers a clear and accessible depiction of the phenotypic changes induced by the microgravity environment. The survival and pigmentation differences among (E. coli), (S. aureus), (B. subtilis), and (S. typhimurium) were observed after exposure, and above panel displays the control strains
Bacterial survival was determined by comparing the number of viable cells in the ground laboratory controls (not exposed to microgravity conditions) with the number of viable cells recovered from the tubes exposed to microgravity. Cell morphology, gram stain, and growth under different environmental conditions were also conducted.27
For pre-exposure preparation, individual bacterial colonies were transferred to sterile tubes containing nutrient broth and incubated until reaching the mid-logarithmic phase, approximately six hours post-inoculation. This procedure ensured synchronized growth before their placement in the CubeSat payload for launch. For post-exposure analysis, the retrieved bacterial cultures were immediately streaked onto nutrient agar plates and incubated under identical conditions to evaluate their viability and any morphological alterations.
Antibiotic resistance/susceptibility of the bacterial strains
To evaluate the susceptibility or resistance of bacterial strains exposed to microgravity compared to their Earth-bound counterparts, we conducted the Kirby-Bauer antibiotic sensitivity test protocol.28,29 This systematic procedure involves specific steps to evaluate the impact of microgravity on bacterial reactions to antibiotics. Initially, we prepared the medium using Mueller-Hinton agar with a pH range of 7.2 to 7.4. The agar medium was poured into plates, ensuring a consistent depth of 5 mm, and then chilled until it solidified. Overnight broth cultures of both the control bacterial strains and those exposed to microgravity were evenly distributed on the Mueller-Hinton agar plates. To assess antibiotic sensitivity, various antibiotic filter discs, including Azithromycin (AZM), Tetracycline (TE), Gentamicin (GEN), and Oxytetracycline (OT), were carefully placed at uniform intervals on the agar plates using forceps. Subsequently, all culture plates were inverted and incubated for a duration of 12-48 hours at a temperature of 37 °C. Following incubation, the plates were thoroughly examined for the presence or absence of a zone of inhibition around each antibiotic filter disc. Measurements were taken using a ruler calibrated in millimeters. This comprehensive testing process enabled us to understand how bacterial strains responded to antibiotics in a microgravity environment compared to their counterparts on Earth.
Molecular PCR analysis
We initiated growth in 25 ml flasks containing nutrient broth and incubated them for 24 hours at 37 °C. Subsequently, we took 10 µl of bacterial broth and streaked it onto nutrient agar plates (NA), followed by an overnight incubation at 37 °C. From each NA plate, we selected a single colony to represent one of the four bacterial strains. These colonies were inoculated into LB broth and placed in an incubator shaker at 37 °C, allowing genomic DNA extraction to take place over the course of the night. We utilized the Wizard Genomic DNA Extraction Kit (Promega Co., Madison, WI, USA) following the attached protocol (leaflet) to isolate total genomic DNA from each bacterial strain. The concentration of the isolated genomic DNA was assessed using nanodrop at 260 nm absorbance. For quality control, we performed gel electrophoresis on 1% agarose gel to ensure the stability of the extracted DNA. PCR reactions were conducted as per the protocols outlined in the references indicated in Table 1, taking into consideration the specific annealing temperatures for different genes and the sequences of the primers used. For instance, to amplify the 203 bp 16S rRNA gene for bacterial strain identification (Table 1), the PCR conditions were as follows: initial denaturation at 95 °C for 5 minutes, followed by 40 cycles of denaturation at 95 °C for 50 seconds, annealing at 64 °C for 50 seconds, extension at 72 °C for 30 seconds, and a final extension at 72 °C for 10 minutes. The PCR products (amplicons) were subsequently analyzed using 1% agarose gel electrophoresis.
Table (1):
Primer sequences and annealing temperatures for the different amplified sequences
Gene name |
Primer’s sequence |
Annealing temp. °C |
Reference |
|---|---|---|---|
16S rRNA gene |
V3F: 5’ CCAgACTCCTACGGGAGGCAG 3’ V3R: 5’ CGTATTACCGCGGCTGCTG 3’ |
64 |
66 |
16S rRNA gene |
V6F: 5’ TCGAtGCAACGCGAAGAA 3’ V6R: 5’ ACATtTCACaACACGAGCTGACGA 3` |
60 |
66 |
RecA gene |
RecA-F: 5’ CCTGAATCTTCYGGTAAAAC 3` RecA-R: 5’ GTTTCTGGGCTGCCAAACATTAC 3′ |
55 |
67 |
Dnak gene |
Dnak-F: 5’ TCTGGTTGGTCAGCCGGCTAA 3` Dnak-R: 5’CGTCGCCGTTATCAGCAGCAA3` |
62 |
68 |
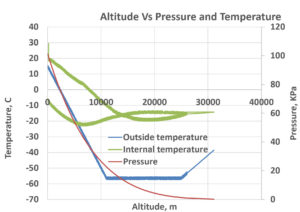
The bacterial samples on the high-flying balloon went to the stratosphere. They faced tough conditions. These included big temperature swings, low pressure, and higher UV and cosmic radiation. At a height of about 35 km, the outside temperature dropped to -55 °C. Inside the payload, it stayed around 5 °C, thanks to thermal insulation. The pressure dropped to 1.1 kPa, similar to that on Mars.
The study explored the impact of the microgravity space environment on bacterial strains, thoroughly examining and analyzing their characteristics at the phenotypic, growth, and molecular levels.
Survival and growth results of bacterial strains
The results indicate that Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Bacillus subtilis (B. subtilis), and Salmonella typhimurium (S. Typhimurium), exhibited differential survival rates under near-space condition, compared to the bacteria cultivated under standard ground conditions. The phenotypic variations of the strains are illustrated in Figure 4. It is crucial to note that the control group, representing bacteria grown under normal ground conditions, serves as a baseline for comparison. This control group was cultivated under normal growth conditions, ensuring a comprehensive understanding of the variations induced by the microgravity environment. E. coli showed the highest sensitivity, while B. subtilis demonstrated the greatest resistance due to its sporulation ability.
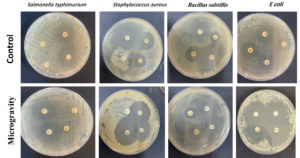
Figure 4. Inhibition zones surrounding antibiotic-impregnated filter discs for the various bacterial strains utilized in the study
Control cultures were incubated for 24 hours at 37 °C under standard laboratory conditions, whereas exposed samples were incubated immediately upon retrieval under identical conditions to evaluate survival and pigmentation alterations. Variations in incubation duration were considered in all comparative analyses. The pigmentation of S. aureus appears paler than typically observed, potentially due to metabolic adaptations in response to near-space conditions. Further validation through spectroscopic pigment analysis may provide quantitative confirmation. Colony-forming unit counts are detailed in Table 2.
Table (2):
The quantitative data in the table presents bacterial growth counts (CFU/ml) for strains exposed to microgravity and their corresponding control strains. A nuanced examination reveals that microgravity has a varying impact on different bacterial strains
| Bacterial strain | Colony counts (CFU/ml) | |
|---|---|---|
| Salmonella typhimurium | Control | 5.0 x 107 |
| Microgravity | 4.0 x 107 | |
| Staphylococcus aureus | Control | 9.0 x 107 |
| Microgravity | 7.0 x 107 | |
| Bacillus subtilis | Control | 8.0 x 107 |
| Microgravity | 1.3 x 107 | |
| Escherichia coli | Control | 7.7 x 107 |
| Microgravity | 6.5 x 107 |
S. typhimurium: Microgravity shows a slightly lower impact on colony count compared to the control.
S. aureus: Microgravity results in a lower colony count compared to control.
B. subtilis: Microgravity significantly reduces the colony count compared to the control.
E. coli: Microgravity results in a slightly lower colony count compared to control.
The impact of microgravity on microbial behavior is evident through the phenotypic, morphological, and color changes observed in bacterial strains exposed to the microgravity outer space environment, compared to their non-exposed counterparts on Earth. Table 2 provides a detailed insight into the adaptability, survival, and growth of bacterial strains exposed to microgravity, particularly on LB medium after their return to Earth. Remarkably, there were no significant changes in growth, measured by colony-forming units per milliliter (CFU/mL), between the bacterial strains exposed to microgravity and the control strains. This suggests that these microorganisms survived in the challenging microgravity environment of the stratosphere. Microscopic analyses revealed that E. coli cells elongated significantly. Additionally, B. subtilis showed a greater tendency to form biofilm-like clusters. These findings match earlier studies. They show that microgravity changes bacterial cytoskeleton structure and affects extracellular matrix synthesis. This may impact colony growth and how bacteria stick to surfaces.28,29
The pigment differences in S. aureus suggest changes in the production of its secondary metabolites. Microgravity’s effect on microbial pigment biosynthesis is still unclear. However, past space missions show that it boosts carotenoid and protective pigment production. This increase likely helps combat oxidative and radiation stress.30,31 Past studies showed similar changes in pigment composition under high radiation and oxidative stress. This supports the idea that these secondary metabolites help microbes adapt to stress.32 Future metabolomic studies may show if the changes come from gene expression in pigment pathways or from stress adaptation methods.
The adaptability and resilience demonstrated by these bacteria in response to extreme conditions, as illustrated by their performance on LB medium, offer valuable The phenotypic variations of the strains are illustrated in Figure 3. Furthermore, while the section mentions remarkable adaptability on LB medium, additional details about specific traits or behaviors observed would enhance the scientific depth of the discussion. Clarifying these aspects will contribute to a more comprehensive interpretation of the results.
The results presented in Table 2 reveal that the bacterial strains have demonstrated remarkable adaptability, survival, and growth on the LB medium upon the return of the balloon carrying these bacteria to Earth. Moreover, it’s noteworthy that there were no significant changes observed in the growth, as measured by colony-forming units per milliliter (CFU/mL), between the bacterial strains exposed to microgravity and the control strains. This implies that these microorganisms were able to thrive even in the challenging microgravity environment of the stratosphere. Such resilience and adaptation of these bacteria to extreme conditions provide valuable insights into their capabilities and potential applications in various contexts, including space exploration.
Antibiotic susceptibility/resistance results
The antibacterial activity of the strains against antibiotics under microgravity and control conditions is shown in Figure 4. The antibiotic resistance profiles are summarized in Table 3.
Table (3):
Antibiotic resistance or sensitivity of the bacterial strains exposed to outer space in microgravity when compared to that of control bacterial strains
| Bacterial strain and sensitivity to antibiotics (*S, I, R) | Antibiotic concentration (30 μg/ml) and diameter of inhibition zone | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Azithromycin | Gentamycin | Tetracycline | Oxytetracycline | ||||||
| Salmonella typhimurium | Control | S | 33 | S | 27 | R | 0 | R | 1 |
| Microgravity | S | 35 | S | 37 | R | 1 | R | 1.3 | |
| Staphylococcus aureus | Control | S | 34 | S | 35 | S | 27 | I | 24 |
| Microgravity | S | 36 | S | 30 | S | 34 | S | 35 | |
| Bacillus subtilis | Control | I | 27 | S | 35 | S | 21 | R | 17 |
| Microgravity | S | 30 | S | 36 | S | 22 | R | 15 | |
| Escherichia coli | Control | S | 33 | S | 28 | I | 19.5 | R | 17 |
| Microgravity | S | 38 | S | 36 | S | 36 | S | 50 | |
Examining antibiotic resistance or sensitivity of bacterial strains in microgravity compared to control conditions yields the following insights: S. typhimurium: Microgravity has a mixed impact on sensitivity across antibiotics.
S. aureus: Generally, microgravity seems to enhance sensitivity across antibiotics.
B. subtilis: Microgravity appears to improve sensitivity to some antibiotics not to all.
E. coli: Microgravity consistently enhances sensitivity across all tested antibiotics.
*S = Sensitive, I = Intermediate, R = Resistance
Molecular and sequencing analysis (blastn analysis)
The selected DNA sequences underwent successful amplification and alignment through BLASTN Entrez (NCBI), a methodology chosen for its precision. In Table S1 (Appendix), we present the alignment of partial 16S rRNA gene sequences alongside sequences of specific genes (RecA, Dnak). The inclusion of BLASTN Entrez aligns with our commitment to methodological rigor. Our study adopts a holistic approach by considering not only 16S rRNA gene sequences but also specific genes (RecA, Dnak).33-35 The rationale for selecting these genes lies in their role in the challenging conditions of microgravity and harsh environments, the RecA and Dnak genes play central roles in microbial survival. The RecA gene encodes a recombinase enzyme, taking a pivotal position in DNA repair and recombination. The heightened vulnerability to DNA damage in microgravity and extreme conditions requires robust repair mechanisms, making RecA crucial for maintaining genomic integrity. Its involvement in homologous recombination enables microorganisms to effectively address DNA lesions, preserving vital genetic information. Conversely, the Dnak gene, responsible for a molecular chaperone, becomes essential in sustaining protein homeostasis amidst environmental stress. Microgravity and harsh conditions often induce protein misfolding and aggregation, posing a threat to cellular functionality. Dnak acts as a protector, facilitating proper protein folding and preventing aggregation, contributing to microbial adaptation and resilience. The coordinated functions of RecA and Dnak underscore their roles as molecular guardians, orchestrating genomic stability and protein quality control to ensure microbial survival in challenging extraterrestrial and terrestrial settings, contributing to a more comprehensive understanding of genetic variations. Table S1 showcases bacterial strains before and after exposure to microgravity and stratosphere conditions, emphasizing differences in substitutions and gaps. A nuanced evaluation includes a detailed breakdown of these variations, supported by specific examples. To fortify our scientific prowess, we have researched the interpretation of observed genetic variations, particularly in the context of stress adaptation. Our data emphasizes the importance of studying stress-related genes throughout the section, ensuring a seamless integration into the detailed breakdown of genetic variations. This highlights their direct relevance to the health of astronauts exposed to microgravity conditions, establishing a clearer connection between our genetic findings and their potential impact on astronaut well-being. We extracted genomic DNA from bacterial strains to study the changes caused by microgravity. We took samples before and after exposure. We amplified the 16S rRNA, RecA, and dnaK genes using PCR. Then, we compared the sequencing results with BLASTN alignment. Control and microgravity-exposed samples were compared. They showed slight differences in the RecA gene of Escherichia coli. The sequence similarity was 99%, with 4 mismatches in a 391-bp stretch. Similarly, dnaK sequence alignment from S. aureus showed a single synonymous substitution. No major changes were seen in the gene sequences of Bacillus subtilis or S. typhimurium (Table S1, Supplementary Data). These findings suggest that microgravity exposure didn’t cause major genomic changes. However, small genetic changes might happen in stress response genes. So, more research is needed with whole-genome sequencing and transcriptomic studies.
Results from KSF space foundation
Flight data including temperature and pressure measurements are presented in Figure 5. including the internal temperature (green line), the external temperature (blue line), pressure (orange-red line), of the CubeSat and payload. At sea level, both the outside and internal temperatures were approximately 25 °C. The pressure is at approximately 0 kPa (kilopascal) as the CubeSat ascends in altitude, the outside temperature drops at a steady pace until the CubeSat reaches 10,100 meters. Internal temperature also drops to -25 °C at this altitude and slowly starts to increase as the CubeSat starts to descend. The pressure also decreases upon ascending because at higher altitudes the air is less dense causing the pressure to drop. Weather balloons operate effectively due to the decrease in atmospheric pressure as they ascend in altitude. This pressure decrease causes the helium inside the balloon to expand outward, allowing the balloon to ascend higher. For instance, at 3:22:22 hours, the pressure measured in kilopascals (kPa) at an altitude of 25,871.0 meters was 1.03. The temperature outside the platform was recorded as -54.2 °C, while inside it was -16.2 °C. Additionally, the gravity experienced at heights between 26 and 30 kilometers was 9.76 m/s2. We assessed bacterial viability by counting colony-forming units (CFU/mL) following exposure. We also examined their shape. E. coli, S. aureus, B. subtilis, and S. typhimurium showed no significant reduction in CFU/mL. This shows they are highly resistant to short-term stratospheric stress. Distinct phenotypic changes were seen in S. aureus and B. subtilis. They showed pigmentation changes. This suggests that stress triggers adaptations in how they produce secondary metabolites. Also, previous studies showed that B. subtilis forms more biofilm in microgravity. This affects bacterial adhesion and the production of the extracellular matrix.36, 37 These findings show that bacteria can survive tough environmental stress. They hint at adaptive methods that need more study.
Stress affects how S. aureus and B. subtilis change color. This also impacts their production of secondary metabolites. These changes might relate to higher oxidative stress and radiation. Past studies show that environmental stress increases genes for carotenoid and flavonoid biosynthesis.38 Microgravity changes gene expression linked to pigment production. In Bacillus species, spaceflight boosts phenoloxidase and melanin genes. Our results support these findings. They suggest that bacteria change pigmentation to adapt to space-like conditions. More transcriptomic and metabolomic studies are needed to identify the regulatory pathways involved.
Amidst the hostile expanse of outer space, featuring extreme cold, solar and cosmic radiation, solar wind, cosmic radiation, low pressures, and microgravity, microorganisms emerge as the most resilient life forms. They have developed mechanisms to either evade or repair the damage caused by these environmental stressors.39 This emphasizes the pivotal role of microorganisms as valuable subjects for understanding the complex mechanisms of survival and adaptation in the challenging conditions of outer space.
The CubeSat containing the bacterial samples was not equipped with an active temperature control system, resulting in the exposure of bacterial cultures to the external temperature fluctuations of the stratosphere. At the maximum altitude of approximately 35 km, temperatures decreased to -55 °C, while the internal payload temperatures stabilized around 5 °C due to insulation. Although microgravity was the primary variable of interest, the extreme cold and increased radiation exposure may have contributed to microbial stress responses.
To address this, parallel Earth control experiments were conducted, wherein bacterial cultures were maintained in a laboratory incubator at 5 °C to replicate the CubeSat’s internal temperature. However, the Earth-based control was not subjected to the same radiation levels as the high-altitude samples. While no significant reduction in CFU/mL was observed between the microgravity-exposed and control samples, it remains plausible that low temperatures influenced stress response pathways. Future experiments should incorporate a fully temperature-matched control, along with radiation shielding comparisons, to isolate the specific effects of microgravity on microbial adaptation.
Moreover, investigating microbial life within the microgravity setting of space and analogous conditions on Earth carries substantial potential for future advancements in biotechnology and commercial applications. This research realm is positioned to drive innovative and practical developments across various fields. Leveraging microgravity as a research tool, in conjunction with molecular technology approaches, presents the opportunity to provide researchers with invaluable insights into microbial life at the cellular, molecular, and evolutionary levels. This subject demands further research and attention, especially in exploring the effects of microgravity on microbes. These insights extend beyond microorganisms and have implications for higher organisms, including humans. Understanding adaptation and survival in low-gravity environments holds vital importance, and studying microbial life in microgravity conditions offers substantial potential for future advancements in biotechnology and commercial applications. While similar studies on bacterial, fungal, and archaeal responses to microgravity exist, there have been reports of inconsistent results, particularly in cellular growth rates. Our study, however, revealed phenotypic and morphological changes in bacterial growth, highlighting the adaptability of bacterial cells to space microgravity conditions.40,28 In our study, the enumeration of bacterial cell counts revealed a slight increase in growth, indicating the adaptability of bacterial cells to microgravity stress conditions, including extreme low temperatures during their journey to outer space.
Understanding these variations requires considering the microbial strains used and the culture methods employed. Suspension cultures, where cells experience continuous “free fall,” are more likely to encounter microgravity conditions. This study reshapes our understanding of microbial responses to space conditions and their practical applications. Balloon altitudes create a unique setting. This helps us understand how microbes respond when fluid dynamics shift in low-gravity settings. In controlled labs, gravity helps evenly distribute nutrients and gases for microbes. However, in a sealed balloon, microorganisms face limited mixing of air and nutrients. This specific setup removes convection effects. As a result, it reveals key insights into cell growth driven by diffusion. Cell growth in tightly controlled conditions is crucial for future bioengineering advances. This is especially important for efforts to keep microbes alive during space missions. Future studies could expand on this model. They might create open-air reactors to grow microorganisms in balloon altitudes. This setup lets us monitor how these organisms react to stress in low-gravity fluid environments in real-time.
In our study, when the enumeration of the bacterial cell counts population was done of the bacterial suspension cultures exposed to microgravity verses the ground control ones, as an indicator of growth rate, a slight increase in growth was found indicating that the bacterial cells were able to adapt and tolerate the microgravity stress conditions and other stress factors such as extreme low temperatures.
Some studies have reported that certain microbes exposed to simulated microgravity exhibited slower growth compared to the control group41 while other studies reported that no significant differences were found in the growth rates of microbes subjected to spaceflight and those of the ground control group.42 Probably the discrepant results of such studies may depend largely on the microbes and strains used. Additionally, the choice of culture method employed can have a significant impact.
Certain microorganisms have evolved mechanisms at the genomic level to repair damage induced by the harsh conditions of space. Studies have demonstrated that exposure to ionizing radiation can instigate alterations in microbial behavior. For instance, a study led by Nicholson et al.43 highlighted changes in the morphology and motility of Bacillus subtilis spores, which exhibit resistance to radiation. In our study, when we assessed the survival and growth capabilities of bacterial strains exposed to microgravity under frigid temperatures, specifically those experienced during their journey to outer space via the space balloon, intriguing results emerged. This resilience aligns with the findings presented by Onofri et al.,44 who also documented the adaptability of certain microorganisms to the rigors of space, enabling them to endure prolonged exposure to exceedingly cold temperatures. The temperature fluctuations experienced in space can significantly influence microbial growth and behavior. These observations shed light on the impressive adaptive capabilities of microorganisms in the face of challenging conditions, which holds significant implications for space microbiology and the broader study of extremophiles. Our findings support earlier studies on high-altitude balloon microbiology. They show how microbes can survive and adapt to conditions in near-space. The E-MIST study found that bacterial spores can live through long-term exposure to stratospheric UV radiation and low pressure.45 Additionally, the research confirmed that microbes from the upper atmosphere have unique adaptations. For example, they show improved DNA repair methods. These help them survive in extreme conditions. High-altitude balloon platforms are great for testing how microbes respond to space environments. This supports our findings’ importance in astrobiology. Our research adds to this literature. We show that key bacteria, Staphylococcus aureus, and E. coli, can handle stress from bad airflow and near-space conditions.46,47
Various theories have emerged to elucidate the physiological changes observed in bacteria during space travel. One prevailing hypothesis posits that, in the absence of gravity, the rate of molecular activity both within and outside the bacterial cell is severely constrained. This forces the bacteria into a kind of starvation mode, where they develop distinct characteristics suited to low-gravity environments. However, it’s important to note that not all types of bacteria will respond in the same way.4,10,48
The influence of the space environment on microorganisms is undeniably intricate and multifaceted. It is conceivable that microgravity may increase mutation rates among microbes by affecting their DNA repair mechanisms.4 One of the most critical yet often overlooked factors influencing microbial behavior in microgravity is the role of fluid dynamics. Under terrestrial conditions, convective mixing driven by gravitational forces ensures the even distribution of nutrients and waste within liquid environments.47 However, in microgravity, convective forces are nearly absent, and diffusion becomes the primary mode of molecular transport. This shift can lead to the formation of concentration gradients around microbial cells, impacting nutrient uptake, metabolic byproduct accumulation, and gas exchange. Such changes may explain the variations in bacterial growth, morphology, and antibiotic susceptibility observed in spaceflight experiments.49 Additionally, reduced convective mixing may influence quorum sensing and biofilm formation, further altering microbial behavior.
These interrelated factors underscore the complex interplay between microorganisms and their space environment, creating a captivating realm for continuous exploration and revelation in the field of space microbiology. This area of study promises to unravel profound insights into the adaptive strategies employed by microorganisms when confronted with the challenges of outer space, with implications for both fundamental science and practical applications.
Mutated biological phenotypes of microorganisms, such as pathogenicity and antibiotic resistance, and the corresponding genes have been observed in response to microgravity. 50,51 A study has been done by Morokuma et al.52 on Planarian flatworms shipped to space under microgravity conditions when analyzed immediately after returning to earth, that some whole worms showed immediate unusual behavior.
In our study regarding bacterial antibiotic sensitivity tests, our results indicate a varied response among bacterial strains when exposed to microgravity. Some strains displayed increased resistance, while others became more sensitive to certain antibiotics. For instance, Escherichia coli and Staphylococcus aureus strains exhibited heightened sensitivity to Azithromycin, Gentamycin, Tetracycline, and Oxytetracycline when compared to the control strains. Additionally, Bacillus subtilis demonstrated sensitivity to most of the antibiotics tested, except for Oxytetracycline, to which it showed resistance. These findings may offer valuable insights into the potential variations in drug responses within microbial communities exposed to space conditions.
Following microbial exposure to the space environment, significant changes in drug-resistance genes and their associated regulatory transcription factors have been observed.53 These findings hold immense promise for advancing drug development and strategies in combating refractory infections. However, to bolster the scientific depth of our understanding, it’s crucial to delve into the specific molecular mechanisms underpinning these observed changes.
To provide a more nuanced understanding, we should explore the specific factors driving this acceleration and the molecular processes that result in the synthesis of these secondary metabolites.54 Moreover, reversible increases in antibiotic resistance have been noted during short-term spaceflight.55 While this observation is intriguing, we should aim to elucidate the underlying mechanisms. Exploring existing theories or proposing new hypotheses will enhance our understanding of microbial responses to space conditions and their potential implications for drug development and infection management.
The BLASTN analysis summarized in table S1(supplementary Data) of the PCR products from the different strains of bacteria along with the selected amplified sequences, including partial 16S rRNA (partial sequences), some selected genes (RecA, dnak), demonstrates alterations in the bacterial selected sequences following their exposure to the stratosphere. The RecA gene is crucial for DNA repair. In microgravity, where traditional repair mechanisms might be disrupted, the RecA gene plays a pivotal role in ensuring the integrity of the microbial genome. The increased potential for genetic mutations in microgravity underscores the importance of an efficient DNA repair system.56,57 RecA is involved in homologous recombination, a process critical for genetic diversity. In microgravity, the selective pressure may lead to the evolution of microbial populations. The RecA gene facilitates genetic exchange, allowing microorganisms to adapt and survive in changing environments.58 On the other hand, Dnak is a chaperone protein that assists in proper protein folding and stability. In microgravity, the absence of gravitational forces can impact cellular processes, including protein folding. The Dnak gene becomes essential for maintaining the correct three-dimensional structures of proteins, ensuring their functionality.59,60 Microgravity induces stress on microbial cells. The Dnak gene is involved in stress response mechanisms, helping microorganisms cope with environmental challenges. This stress response is crucial for microbial survival and adaptation to the unique conditions of a microgravity environment.61,62 The RecA gene plays an important role in DNA repair, particularly under severe environmental stress. Its role in homologous recombination helps keep bacterial genomes stable. In microgravity, oxidative stress and radiation increase mutation rates.21,58 This is very important. The dnaK gene makes a stress protein. This protein helps proteins fold correctly when under stress. Research shows that the dnaK gene increases in heat-shocked and oxidative stress-treated bacteria. This suggests it may help microbes adapt to microgravity.57 Sequence variations in these genes may show how microbes adapt to stress in space-like environments. More work with transcriptomics and proteomics is needed to see how these genetic changes affect function. Detailed sequencing results, including observed nucleotide variations in RecA, and Comprehensive sequencing results, which include observed nucleotide variations in RecA and dnaK, are presented in Table S1 (Supplementary Data). Sequence identity was consistently high (99%) across all bacterial strains, with minor substitutions identified in E. coli and S. aureus., which are provided in Table S1 (Supplementary Data). Sequence identity remained high (>99%) for all bacterial strains, with minor substitutions detected in E. coli and S. aureus. The lack of significant sequence variations in B. subtilis and S. typhimurium indicates that exposure to microgravity may elicit strain-specific genomic responses, potentially influenced by mechanisms of oxidative stress resistance (Table S1).
Shedding light on the adaptability of bacterial cells to the harsh microgravity environment and thus offers valuable insights into the potential implications of stratospheric exposure on genetic evolution. These findings not only expand our understanding of the resilience of microbial life forms but also contribute to the broader scientific discourse on the impact of high-altitude environments on genetic profiles and could be relevant to understanding the adaptation and evolution of life beyond earth.
Numerous studies have yielded valuable insights into the molecular and genetic alterations that occur when microorganisms are cultivated under simulated microgravity conditions. For instance, Arunasri et al.63 observed significant molecular changes in genes related to stress adaptation and DNA replication when exposing the Escherichia coli strain (K12 MG1655) to simulated microgravity. Similarly, Ott et al.64 conducted research with the bacterial strain Deinococcus radiodurans R1 and discovered molecular alterations in specific proteins associated with DNA, such as Dnak, PolA, and DNA ligase when exposed to simulated microgravity conditions. These findings underscore the profound impact of microgravity on the genetic and molecular composition of microorganisms, shedding light on the adaptive mechanisms these organisms employ when confronted with environments resembling those of space. Such research is instrumental in enhancing our understanding of the intricacies of space microbiology and its implications for various scientific and practical applications.
Gravity serves as a constant and fundamental force that has played a pivotal role in the ongoing process of biological evolution on Earth. Studying how living organisms adapt to the absence of this ever-present force not only provides us with valuable insights into its profound influence on life on our planet but also equips us with essential knowledge for formulating strategies to mitigate the negative consequences of microgravity encountered during space missions.
While various bacteria have exhibited changes because of spaceflight, the precise mechanisms underlying these alterations remain elusive, which will become increasingly crucial for aerospace technology, particularly in its applications. Understanding these mechanisms is instrumental in ensuring the safety and success of space missions and in harnessing the unique features of microgravity for beneficial purposes.
The findings of our study may hold the potential to enhance our understanding of the microbial communities involved in spaceflight and their intricate interactions with humans. This knowledge may, in turn, prove invaluable in advancing our comprehension of the origins of life and the processes of biological evolution. The application of microgravity as a research tool, combined with cutting-edge molecular technology, offers researchers a unique opportunity to delve into how variations in this physical force impact microbial life at the cellular, molecular, and evolutionary levels.
Gravity’s constant force has played a fundamental role in biological evolution on Earth. Understanding how microorganisms adapt to the absence of this force provides insights into its profound influence on life. The findings of our study contribute to advancing our understanding of microbial communities in spaceflight and their interactions with humans. This knowledge holds promise in elucidating the origins of life and the processes of biological evolution.65
While our study provides valuable insights, there remains a considerable gap in our explicit knowledge regarding the molecular mechanisms enabling the survival and adaptation of microorganisms exposed to the harsh outer space environment. Additional research and investigations are imperative to uncover these mechanisms and further our understanding of the remarkable resilience exhibited by microorganisms in the challenging conditions of space.
Research on microbial life in the stratosphere is becoming more important. These microbes show great adaptability and can survive extreme conditions. This is key for space microbiology. The high-altitude balloon platform helps us study how microbes survive in space. It also lets us observe microbial behavior when there’s less convection. Future research should examine long-term microbial growth in these conditions. This will show us how microbes interact with fluids in space and in biotechnological environments on Earth. This area of study has unveiled the i tenacity of microorganisms in the face of the formidable challenges presented by extraterrestrial settings. The stratosphere, acting as a unique experimental arena, provides valuable insights into the fundamental attributes that enable microorganisms to acclimate to harsh environmental circumstances. Furthermore, the rapid reproduction rates of microorganisms accelerate the assessment of adaptive changes, making them optimal candidates for deciphering the complexities of biological systems in space. A key focus of scientific exploration lies in understanding how space environments affect microorganisms, encompassing their reactions to microgravity. Researchers are keen to understand how alien environments impact microorganisms. They focus on how these tiny life forms react to microgravity. Microorganisms can’t sense gravity directly. The changes we observe are probably due to fluid dynamics around them, not because of microgravity itself. In microgravity, there is no convective mixing. This impacts how nutrients are delivered, oxygen diffuses, and waste is removed. As a result, microbial growth, metabolism, and gene expression are also affected. Understanding these indirect effects is key. It helps us interpret microbial behaviors in space. It also aids in designing controlled experiments for future space microbiology research.
In summary, the intricate interplay between microorganisms and their space environment offers an enthralling arena for ongoing research and discovery. Research shows that microorganisms change at the molecular and genetic levels in simulated microgravity. Our findings and past research from balloon flights show that microorganisms can survive and adapt to tough near-space conditions. Future research should focus on how microbial populations adapt over time in these environments. This is important because it might affect policies on how we protect planets and the life-support systems for habitats beyond Earth.
Understanding the molecular changes affecting genes responsible for stress adaptation, DNA replication, and critical proteins in microorganisms opens doors to innovative applications in biotechnology and beyond. Space microbiology continues to evolve as a burgeoning field with profound implications for space exploration and biotechnology. The lessons derived from the investigation of microorganisms in the stratosphere and under simulated microgravity conditions hold promise for a wide array of scientific and commercial applications. As we progress, further research and interdisciplinary collaboration will remain imperative. To unlock the full potential of these remarkable microorganisms in space and on Earth.
Our findings align with previous microgravity studies using random positioning machines (RPMs) and clinostats to minimize sedimentation and convective mixing. Research with these systems has shown increased biofilm formation, cell shape changes, and altered antibiotic resistance in bacteria. Our balloon experiment confirmed bacterial adaptation to microgravity, but comparisons with real spaceflight data are needed to identify specific cellular and molecular changes. Ground-based simulators and stratospheric vehicles can improve our understanding of microbial survival in space habitats.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HIM, AKA and AAAA conceptualized the study. AA and HJA applied research methodology and performed data collection. AKA, AAAA, AA and HJA performed experiments. MK, and MRR performed data analysis. AKA, AAAA, AA and HJA performed data validation. HIM funding acquisition. HIM, AKA and AAAA wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by the Deanship of Research & Graduate Studies at Yarmouk University, Jordan, with project number 7/2021.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Mortfitt R, Van Der Werff R. Navigating the Stratosphere. 2025. SDSU Data Science Symposium (https://openprairie.sdstate.edu/datascience_symposium/2025/sessions/38/ ).
- Anitori RP. Extremophiles: Microbiology and Biotechnology: Caister Academic Press. 2012.
- Rothschild LJ, Mancinelli RL. Life in extreme environments. Nature. 2001;409(6823):1092-1101.
Crossref - Prasad B, Richter P, Vadakedath N, et al. Exploration of space to achieve scientific breakthroughs. Biotechnol Adv. 2020;43:107572.
Crossref - Ashaolu TJ, Malik T, Soni R, Prieto MA, Jafari SM. Extremophilic Microorganisms as a Source of Emerging Enzymes for the Food Industry: A Review. Food Sci Nutr. 2025;13(1):e4540.
Crossref - Wani AK, Akhtar N, Sher F, Navarrete AA, Americo-Pinheiro JHP. Microbial adaptation to different environmental conditions: molecular perspective of evolved genetic and cellular systems. Archives of Microbiology. 2022;204(2):144.
Crossref - Fierer N, Wood SA, Bueno de Mesquita CP. How microbes can, and cannot, be used to assess soil health. Soil Biol Biochem. 2021;153:108111.
Crossref - Seckbach J, Oren A, Stan-Lotter H. Polyextremophiles: Life Under Multiple Forms of Stress: Springer Netherlands. 2013.
Crossref - Thakur N, Singh SP, Zhang C. Microorganisms under extreme environments and their applications. Curr Res Microb Sci. 2022;3:100141.
Crossref - Sharma G, Curtis PD. The Impacts of Microgravity on Bacterial Metabolism. Life. 2022;12(6).
Crossref - Pikuta EV, Hoover RB, Tang J. Microbial extremophiles at the limits of life. Crit Rev Microbiol. 2007;33(3):183-209.
Crossref - Cao Z, Liu H, Zhao B, Pang Q, Zhang X. Extreme Environmental Stress-Induced Biological Responses in the Planarian. Biomed Res Int. 2020;2020:7164230.
Crossref - Horneck G, Klaus DM, Mancinelli RL. Space microbiology. Microbiol Mol Biol Rev. 2010;74(1):121-156.
Crossref - Simoes MF, Antunes A. Microbial Pathogenicity in Space. Pathogens. 2021;10(4).
Crossref - Yamagishi A, Kawaguchi Y, Shin-ichi Y, et al. Environmental Data and Survival Data of Deinococcus aetherius from the Exposure Facility of the Japan Experimental Module of the International Space Station Obtained by the Tanpopo Mission. Astrobiology. 2018;18(11):1369-1374.
Crossref - Tirumalai MR, Karouia F, Tran Q, et al. The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic. NPJ Microgravity. 2017;3:15.
Crossref - Huang B, Li D-G, Huang Y, Liu C-T. Effects of spaceflight and simulated microgravity on microbial growth and secondary metabolism. Mil Med Res. 2018;5(1):1-14.
Crossref - Smith DJ, Sowa MB. Ballooning for Biologists: Mission Essentials for Flying Life Science Experiments to Near Space on NASA Large Scientific Balloons. Gravit Space Res. 2017;5(1):52-73.
Crossref - Thombre RS, Kaur K, Jagtap SS, Dixit J, Vaishampayan PV. Chapter 6 – Microbial life in space. In R. Thombre & P. Vaishampayan (Eds.). New Front Astrobiol. 2022:135-166.
Crossref - Fajardo-Cavazos P, Morrison MD, Miller KM, Schuerger AC, Nicholson WL. Transcriptomic responses of Serratia liquefaciens cells grown under simulated Martian conditions of low temperature, low pressure, and CO2-enriched anoxic atmosphere. Sci Rep. 2018;8(1):14938.
Crossref - Schuerger AC, Nicholson WL. RETRACTED: Twenty-Three Species of Hypobarophilic Bacteria Recovered from Diverse Ecosystems Exhibit Growth under Simulated Martian Conditions at 0.7 kPa. Astrobiology. 2016;16(5):335-347.
Crossref - Bijlani S, Stephens E, Singh NK, Venkateswaran K, Wang CCC. Advances in space microbiology. Iscience. 2021;24(5).
Crossref - Cavicchioli R. Extremophiles and the search for extraterrestrial life. Astrobiology. 2002;2(3):281-292.
Crossref - Olsson-Francis K, Cockell CS. Experimental methods for studying microbial survival in extraterrestrial environments. J Microbiol Methods. 2010;80(1):1-13.
Crossref - Koike J, Oshima T, Koike KA, et al. Survival rates of some terrestrial microorganisms under simulated space conditions. Adv Space Res. 1992;12(4):271-274.
Crossref - Nandhini B, Ramesh A, Monisha M, Krishna KS. An overview of astrobiology and microbial survival in space. Int J Inn Sci Res Technol. 2021;6(3):21-28.
- James G. Cappuccino, C. W. Microbiology: A Laboratory Manual. 2017.
- Zea L, Larsen M, Estante F, et al. Phenotypic changes exhibited by E. coli cultured in space. Front Microbiol. 2017;8:1598.
Crossref - Tirumalai MR, Karouia F, Tran Q, et al. The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic. NPJ Microgravity. 2017;3:15.
Crossref - Abdelaziz AA, Kamer AMA, Al-Monofy KB, Al-Madboly LA. Pseudomonas aeruginosa’s greenish-blue pigment pyocyanin: its production and biological activities. Microbial Cell Fact. 2023;22(1):110.
Crossref - Antonic V, Stojadinovic A, Zhang B, Izadjoo MJ, Alavi M. Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect Drug Resist. 2013;6:175-186.
Crossref - Azzam EI, Jay-Gerin J-P, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Letters. 2012;327(1):48-60.
Crossref - Li H, Lu J, Zhao H, et al. The impact of space environment on gene expression in Arabidopsis thaliana seedlings. Sci China Technol Sci. 2017;60(6):902-910.
Crossref - Puig J, Knodlseder N, Quera J, Algara M, Guell M. DNA Damage Protection for Enhanced Bacterial Survival Under Simulated Low Earth Orbit Environmental Conditions in Escherichia coli. Front Microbiol. 2021;12:789668.
Crossref - Zhao L, Zhang G, Tang A, Huang B, Mi D. Microgravity alters the expressions of DNA repair genes and their regulatory miRNAs in space-flown Caenorhabditis elegans. Life Sci Space Res (Amst). 2023;37:25-38.
Crossref - Agarwal H, Bajpai S, Mishra A, et al. Bacterial Pigments and Their Multifaceted Roles in Contemporary Biotechnology and Pharmacological Applications. Microorganisms. 2023;11(3):11030614.
Crossref - Orlandi VT, Martegani E, Giaroni C, Baj A, Bolognese F. Bacterial pigments: A colorful palette reservoir for biotechnological applications. Biotechnol Appl Biochem. 2022;69(3):981-1001.
Crossref - Rampelotto PH. Resistance of Microorganisms to Extreme Environmental Conditions and Its Contribution to Astrobiology. Sustainability. 2010;2(6):1602-1623.
Crossref - Mileikowsky C, Cucinotta FA, Wilson JW, et al. Natural transfer of viable microbes in space: 1. From Mars to Earth and Earth to Mars. Icarus. 2000;145(2):391-427.
Crossref - Karouia F, Tirumalai MR, Nelman-Gonzalez MA, et al. Long-term exposure of bacterial cells to simulated microgravity. Methods, and Missions for Astrobiology. 2012.
Crossref - Fang A, Pierson DL, Mishra SK, Demain AL. Relief from glucose interference in microcin B17 biosynthesis by growth in a rotating-wall bioreactor. Lett Appl Microbiol. 2000;31(1):39-41.
Crossref - Kacena MA, Todd P, Landis WJ. Osteoblasts subjected to spaceflight and simulated space shuttle launch conditions. In Vitro Cell Dev Biol Anim. 2003;39(10):454-459.
Crossref - Nicholson WL, Ricco AJ. Nanosatellites for biology in space: in situ measurement of Bacillus subtilis spore germination and growth after 6 months in low earth orbit on the O/OREOS mission. Life. 2019;10(1):1.
Crossref - Onofri S, Selbmann L, de Hoog GS, et al. Evolution and adaptation of fungi at boundaries of life. Adv Space Research. 2007;40(11):1657-1664.
Crossref - Smith DJ, Sowa MB. Ballooning for Biologists: Mission Essentials for Flying Life Science Experiments to Near Space on NASA Large Scientific Balloons. Gravit Space Res. 2017;5(1):52-73.
Crossref - DasSarma P, DasSarma S. Survival of microbes in Earth’s stratosphere. Curr Opin Microbiol. 2018;43:24-30.
Crossref - Klomchitcharoen S, Wechakarn P, Tangwattanasirikun T, et al. High-altitude balloon platform for studying the biological response of living organisms exposed to near-space environments. Heliyon. 2024;10(6):e27406.
Crossref - Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev. 2004;68(2):345-361.
Crossref - Gonzalez JM, Aranda B. Microbial Growth under Limiting Conditions-Future Perspectives. Microorganisms. 2023;11(7).
Crossref - Rosenzweig JA, Abogunde O, Thomas K, et al. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl Microbiol Biotechnol. 2010;85(4):885-891.
Crossref - Wilson J, Ott CM, Zu Bentrup KH, et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci. 2007;104(41):16299-16304.
Crossref - Morokuma J, Durant F, Williams KB, et al. Planarian regeneration in space: Persistent anatomical, behavioral, and bacteriological changes induced by space travel. Regeneration (Oxf). 2017;4(2):85-102.
Crossref - Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resistance Updates. 2000;3(5):303-311.
Crossref - Taylor P. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect Drug Resist. 2015;8:249-262.
Crossref - Dey D. Space microbiology: modern research and advantages for human colonization on Mars. Int J Res Appl Sci Biotechnol. 2019;6.
Crossref - Jones DL, Baxter BK. DNA Repair and Photoprotection: Mechanisms of Overcoming Environmental Ultraviolet Radiation Exposure in Halophilic Archaea. Front Microbiol. 2017;8:1882.
Crossref - Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage l. Microbiol Mol Biol Rev. 1999;63(4):751-813, table of contents.
Crossref - Johnston CHG, Hope R, Soulet AL, Dewailly M, De Lemos D, Polard P. The RecA-directed recombination pathway of natural transformation initiates at chromosomal replication forks in the pneumococcus. Proc Natl Acad Sci U S A. 2023;120(8):e2213867120.
Crossref - Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta (BBA). 2010;1803(6):650-661.
Crossref - Schonfeld HJ, Schmidt D, Schroder H, Bukau B. The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J Biol Chem. 1995;270(5):2183-2189.
Crossref - Dahl J-U, Gray MJ, Jakob UProtein quality control under oxidative stress conditions. J Mol Biol. 2015;427(7):1549-1563.
Crossref - Senatore G. Impact of the exposure to simulated microgravity conditions on the probiotic lactobacillus reuteri DSM 17938. 2020;Thesis.
- Arunasri K, Adil M, Venu Charan K, Suvro C, Himabindu Reddy S, Shivaji S. Effect of simulated microgravity on E. coli K12 MG1655 growth and gene expression. PLoS One. 2013; 8(3): e57860.
Crossref - Ott E., Fuchs FM., Moeller R. et al. Molecular response of Deinococcus radiodurans to simulated microgravity explored by proteometabolomic approach. Sci Rep. 2019; 9 (18462).
Crossref - Afshinnekoo E, Scott RT, MacKay MJ, et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell. 2020;183(5):1162-1184.
Crossref - Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330-339.
Crossref - Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PloS One. 2010;5(4):e10034.
Crossref - Seyer K, Lessard M, Piette G, Lacroix M, Saucier L. Escherichia coli heat shock protein DnaK: production and consequences in terms of monitoring cooking. Appl Environ Microbiol. 2003;69(6):3231-3237.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.