ISSN: 0973-7510
E-ISSN: 2581-690X
Honey bees (Apis cerana indica) play a crucial role in pollination and ecosystem stability, but their populations are increasingly threatened by pesticide exposure and environmental stressors. Lactic acid bacteria (LAB), particularly fructophilic LAB (FLAB), are key components of the honey bee gut microbiota, contributing to digestion, immune modulation, and pathogen resistance. This study investigates the diversity, phylogenetic relationships, and pesticide tolerance of LAB isolated from honey bee gut, pollen, and honey across four distinct agroecosystems in Tamil Nadu, India. A total of 41 LAB strains were identified using both morphological and molecular techniques, including Apilactobacillus kunkeei, Fructobacillus fructosus, A. apinorum, and Secundilactobacillus kimchicus. Notably, this study reports S. kimchicus in the honey bee gut for the first time, expanding the known microbiota diversity associated with honey bees. Phylogenetic analysis, based on molecular traits, revealed distinct clustering patterns, indicating regional adaptations and evolutionary divergence among isolates. Growth assays confirmed a strong preference for fructose-rich environments, consistent with their ecological niche in nectar and honey. Pesticide tolerance assays demonstrated that A. kunkeei exhibited the highest resilience to imidacloprid, dinotefuran, fipronil, and dimethoate, highlighting its potential role in mitigating pesticide-induced stress in honey bee colonies. These findings suggest the application of FLAB as probiotic candidates by degrading pesticides and enhance colony resilience by maintaining gut health. Future research should explore the functional mechanisms underlying pesticide detoxification and immune modulation to develop targeted probiotic formulations for sustainable apiculture.
Honey Bee Microbiota, Fructophilic Lactic Acid Bacteria, Pesticide Tolerance, Phylogenetic Analysis, Probiotics
Honey bees play a crucial role as agricultural pollinators, ensuring the reproduction of both wild plants and cultivated crops. However, global declines in honey bee populations have raised significant concerns, prompting extensive research to mitigate colony losses. As eusocial insects, honey bees exhibit complex behavioural structures and intricate social interactions, making them an excellent model organism for studying cognition, perception, and social dynamics.1 Additionally, their unique and highly specialized gut microbiota provides valuable insights into host-microbial interactions.
The honey bee gut microbiota is relatively simple, consisting of eight to ten dominant bacterial genera that collectively make up over 97% of the microbial community. Among these, Lactobacillus spp., Bifidobacterium spp., Snodgrassella alvi and Gilliamella apicola, play crucial roles in host health and these species constitute more than 99% of the microbial population in the honey bee gut.2,3 Experimental evidence highlights the role of microbiota in food digestion, immune system regulation, and pathogen resistance. Key bacterial taxa, including Lactobacillus, Bifidobacterium, Snodgrassella, Gilliamella, Apibacter, and Frischella, exhibit probiotic, antimicrobial, and symbiotic properties essential for maintaining gut homeostasis and overall bee health.4 Within this microbial community, Lactobacillus Firm-5 and Firm-4 are the most dominant groups, followed by Gilliamella, Snodgrassella, and Bifidobacterium.5 These bacteria occupy diverse nutritional niches and contribute to essential functions such as nutrient absorption, immune modulation, and pathogen resistance. Lactic acid bacteria (LAB), particularly Lactobacillus and Bifidobacterium, have been identified across various bee-related components, including honey (90.9%), pollen (74.6%), beebread (83.9%), royal jelly (93.3%), and the whole gut (30.0%).6 The long-standing symbiotic association between honey bees and their microbiota, facilitated by their eusocial lifestyle, has driven the genetic and functional diversification of these host-specific bacteria.7
While the gut microbiota of Apis mellifera is well-characterized, recent studies have also examined A. cerana, revealing differences in microbial composition. The gut microbiota of A. cerana includes Proteobacteria (70.7%), Actinobacteria (10.7%), Firmicutes (10.3%), and Bacteroidetes (8.4%), with LAB comprising 18.95% of the total microbiota. Key LAB species identified include Bifidobacterium asteroides, B. indicum, F. fructosus, A. apinorum, A. apis, A. helsingborgensis, A. kimbladii, A. kullabergensis, and A. kunkeei.8,9 Among these, A. kunkeei is particularly dominant and classified as a fructophilic lactic acid bacterium (FLAB) in honey bees.10,11
Recent research has drawn attention to the probiotic potential of LAB in honey bees, demonstrating their role in enhancing bee health, controlling diseases, and mitigating pesticide stress. LAB have been shown to degrade or sequester pesticides, reducing their absorption in the digestive tract, lowering toxicity, and decreasing honey bee mortality.12 In addition, LAB contribute to growth promotion, immune regulation, and intestinal homeostasis.13-15 LAB can degrade pesticides through enzymatic detoxification and also improve colony growth, immune health, honey production, and brood development.16 However, despite these benefits, limited studies have explored the diversity and pesticide tolerance of FLAB in different honey bee-associated sources such as the gut, pollen, and honey.
The identification and differentiation of FLAB within a microbiota are increasingly dependent on molecular-based techniques rather than phenotypic characteristics. One such approach involves sequencing rRNA genes, which has been widely used for LAB identification and phylogenetic classification.17-19 This molecular approach enables precise differentiation of FLAB species and their evolutionary relationships.
This study aims to bridge this knowledge gap by isolating and characterizing FLAB from the honey bee gut, pollen, and honey. Through morphological and molecular identification, phylogenetic analysis, and pesticide tolerance assays, we provide insights into the diversity, adaptive mechanisms, and functional roles of these bacteria. Understanding the evolutionary relationships and adaptive mechanisms of these FLAB strains is essential for developing targeted probiotic formulations to enhance honey bee resilience and support sustainable apiculture practices.
Sample collection
Samples were collected from four distinct locations in Tamil Nadu, India, representing diverse agricultural landscapes: Coimbatore (11.016953° N, 76.929275° E) from a cotton field, Erode (11.166276° N, 77.772489° E) from a coconut plantation, Dindigul (10.374818° N, 77.519624° E) from a mixed mango and coconut plantation, and Kanniyakumari (8.098276° N, 77.433700° E) from a rubber plantation (Figure 1). From each location, samples were collected from 10 honey bee colonies, and approximately 400-500 live adult worker honey bees were collected for subsequent analysis. In addition to the bees, approximately 20 mL of honey and 10 g of pollen were collected from the super chambers of each sampled colony, depending on availability. All samples were stored under appropriate conditions and transported to the laboratory for further processing.
Isolation of lactic acid bacteria
LAB were isolated from the honey bee gut using the following procedure. Live bees were first immersed in 70% ethanol for surface sterilization for two minutes. The ethanol was then removed by rinsing the bees with sterile distilled water to eliminate any contaminants. Using sterilized forceps, the guts of the honey bees were extracted aseptically. Each honey bee was clamped with forceps, and a slight incision was made at the tip of the last abdominal segment to remove the entire gut, including the crop, midgut, and hindgut. The dissected guts were placed in phosphate-buffered saline for further analysis. A total of 50 guts were collected for this study. The collected guts were homogenized by macerating them in sterile phosphate-buffered saline (PBS) using a sterile pestle and mortar under aseptic conditions. From the homogenate, 1 mL was used for 10-fold serial dilutions ranging from 10-1 to 10-6 to isolate LAB. Similarly, 1 g of pollen and 1 mL of honey were collected from the hives and subjected to serial dilution and plating techniques. For the pour plate technique, 1 mL of each serially diluted gut homogenate, pollen, and honey sample was added to sterile Petri plates. Subsequently, 15 mL of MRS agar medium (HiMedia Laboratories Private Limited), maintained at 40-45 °C, was poured into each plate. To inhibit fungal growth, 0.01% cycloheximide was added to the culture medium. The plates were incubated at 35 °C for 24 hours in a BOD incubator (LABTHERM Scientific Products, Serial No. BOD07/2021/018) under aerobic condition. Subculturing of presumptive LAB colonies was done thrice on fresh MRS agar to obtain pure cultures. The final pure cultures were stored at 4 °C for further analysis.20,21
Biochemical characterization of A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus
The biochemical properties of the isolated LAB strains were assessed using standard microbiological procedures.22 The following tests were conducted:
Catalase test
A loopful of fresh bacterial culture was placed on a clean glass slide, followed by the addition of a few drops of 3% hydrogen peroxide. Immediate bubble formation indicated a positive result (catalase activity), whereas the absence of bubbles indicated a negative result.
Oxidase test
This test was performed using oxidase reagent strips impregnated with tetramethyl-p-phenylenediamine. A bacterial colony was rubbed onto the reagent strip, and the appearance of a dark purple colour within 30 seconds indicated a positive result. No colour change or a persistent light coloration indicated a negative result.
Carbohydrate fermentation (Glucose, Lactose, and Sucrose)
Each sugar was tested using Phenol Red Carbohydrate Broth Base (HiMedia), supplemented with 1% of the respective sugar and a Durham tube to detect gas production. A colour change from red to yellow signified acid production, while the presence of gas bubbles in the Durham tube indicated gas production. Broth remaining red with no gas bubbles was considered negative.
Methyl red test
Isolates were inoculated into MR-VP broth (HiMedia) and incubated at 35 °C for 48 hours. After incubation, 5 drops of methyl red indicator were added. A red colour indicated a positive result, signifying mixed acid fermentation.
Growth at different temperatures (10 °C, 35 °C, and 45 °C)
The ability of isolates to grow at various temperatures was assessed in MRS broth. Cultures were incubated for 48 hours at the specified temperatures, and visible turbidity indicated positive growth.
Gas production from glucose
This test was performed in Phenol Red Glucose broth (HiMedia) containing a Durham tube. The presence of gas bubbles in the Durham tube indicated positive gas production.
Salt tolerance (6.5% NaCl)
To assess salt tolerance, isolates were cultured in MRS broth supplemented with 6.5% NaCl (HiMedia) and incubated at 35 °C for 48 hours. Turbidity indicated growth (positive), while a clear broth denoted no growth (negative).
Nitrate reduction test
This test was carried out in nitrate broth (HiMedia). After 48 hours of incubation at 35 °C, 5 drops each of sulfanilic acid and a-naphthylamine were added. Development of a red colour indicated positive nitrate reduction.
Hydrogen sulfide (H2S) production
The production of H2S was assessed using Triple Sugar Iron (TSI) agar slants (HiMedia). After 24-48 hours of incubation at 35 °C, blackening of the butt portion of the slant indicated positive H2S production. The absence of black precipitate was considered negative.
Growth curve determination
For the growth curve assay, a single colony of each strain was inoculated into 10 mL of fresh MRS broth in a sterile 50 mL Erlenmeyer flask and incubated overnight at 35 °C. The overnight cultures were then diluted with sterile MRS broth to obtain an initial OD600 of 0.05. At regular time intervals (0, 2, 4, 6, 8, 10, 12, 16, 20, and 24 hours), a 1 mL aliquot of each bacterial culture was withdrawn from the flask, mixed gently, and transferred into a clean glass cuvette. OD600 was measured at 600 nm using a BioSpectrometer (Eppendorf AG Serial No.:6136GN903745) against a blank containing sterile MRS broth. The cuvette was thoroughly rinsed with distilled water between measurements to avoid cross-contamination. If the OD600 value exceeded 1.0, the sample was diluted appropriately (e.g., 1:2 or 1:5 with sterile MRS broth), and the final value was calculated by multiplying by the dilution factor. All measurements were performed in triplicates, and the average OD600 values were recorded for each time point. The data were plotted using Microsoft Excel (Microsoft Office LTSC Professional Plus 2024) to generate growth curves representing the bacterial growth phases.
Evaluation of fructophilic growth preference
The fructophilic nature of the isolated LAB was assessed by evaluating their growth in different media, following the methodology described by previous study with slight modifications.23 Four bacterial strains, A. kunkeei, F. fructosus, A. apinorum, and S. kimchicus were cultured in two different broth media: FYP broth and GYP broth. The composition of GYP broth was identical to FYP broth, except that it contained 10 g/L of D-glucose instead of D-fructose. The bacterial cultures were incubated at 30 °C for 48 hours, and turbidity was measured at 600 nm using a BioSpectrometer (Eppendorf AG Serial No.: 6136GN903745) to assess bacterial proliferation in each medium. Additionally, the growth of these bacterial strains was evaluated in MRS broth with and without 2% fructose supplementation.24 The bacterial cultures were inoculated into both MRS media (with and without fructose) and incubated under the same conditions. Growth was monitored by measuring optical density at 600 nm, and comparisons were made to determine the effect of fructose on bacterial proliferation.
Molecular identification of fructophilic lactic acid bacteria using 16S rRNA gene sequencing
Genomic DNA was extracted from FLAB isolates using a modified CTAB method. Bacterial pellets from 1 mL cultures were resuspended in 2% CTAB buffer, homogenized, and incubated at 65 °C for 1 hour. DNA purification was performed using phenol: chloroform: isoamyl alcohol extraction, followed by ethanol precipitation. DNA was quantified using a Nanodrop spectrophotometer (Thermo Scientific Nanodrop One, Serial No.: AZY2017038), and purity was assessed by the A260/A280 ratio. For bacterial identification, the nearly full-length 16S rRNA gene was amplified using universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′),25 PCR was carried out in a Veriti™ Thermal Cycler (Applied Biosystems, USA) with an initial denaturation at 95 °C for 10 min, followed by 35 cycles of 95 °C for 30s, 55 °C for 1 min, and 72 °C for 1.5 min, concluding with a final extension at 72 °C for 7 min.16 PCR products were analysed via 0.8% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light. Amplicons (40 µL) were sanger sequenced by Syngenome (OPC) Private Ltd., Coimbatore, India. The resulting nucleotide sequences were manually reviewed, edited, aligned, and trimmed using Sequencher 4.6 (Gene Codes Corporation, Ann Arbor, Michigan) and BioEdit v7.0.5. Trimmed consensus sequences were verified through homology searches against DNA databases using the BLASTn program available on the NCBI website (http://www.ncbi.nlm.nih.gov) and submitted.
Phylogenetic tree construction
The phylogenetic analysis of FLAB isolates was conducted using the MEGA software (11.0.13) with default settings.18,26 The 16S rRNA gene sequences A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus were aligned using the ClustalW algorithm implemented in MEGA, ensuring accurate sequence comparison.27 Phylogenetic relationships were inferred using the Neighbor-Joining method with 1,000 bootstrap replications to assess branch reliability.28,29 The evolutionary distances were computed using the Kimura 2-parameter model,30 which accounts for differences in transition and transversion rates. To ensure accurate rooting of the tree, Pseudomonas fluorescens was selected as an outgroup, allowing for better resolution of phylogenetic relationships among the LAB isolates. The final tree was visualized and edited in MEGA 11.0.13, and bootstrap values above 50% were displayed at the respective nodes to indicate confidence levels. Sequence accession numbers of closely related reference strains were retrieved from the NCBI GenBank database to enhance phylogenetic interpretation.
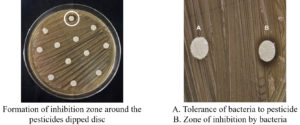
Pesticide tolerance assay
A preliminary pesticide tolerance assay was conducted to evaluate the ability of FLAB isolates to withstand exposure to four commonly used pesticides highly toxic to bees. The commercial formulations of fipronil 5% SC, dinotefuran 20% SG, imidacloprid 17.8% SL, and dimethoate 30% EC were used. Each pesticide was prepared at concentrations of 50 ppm, 100 ppm, 250 ppm, 500 ppm, and 1,000 ppm using sterile distilled water as the solvent.31 Prior to pesticide tolerance testing, overnight cultures of FLAB were grown in MRS broth and adjusted to an OD600 of 1.0 to ensure uniform bacterial inoculation.32 For the assay, MRS agar plates were inoculated by evenly swabbing LAB isolates onto the surface using a sterilized cotton swab. Sterile filter paper discs (6 mm diameter) were immersed in the respective pesticide solutions, air-dried under sterile conditions, and placed onto the inoculated plates using sterile forceps. The plates were then incubated at 35 °C for 96 hours.33 After incubation, the presence or absence of a zone of inhibition (ZOI) around the pesticide-impregnated discs was visually observed and measured in millimetres using a ruler. The formation of a clear inhibition zone around the disc indicated that the bacterial isolate was sensitive to the pesticide and could not tolerate its effects. Conversely, the absence of a zone of inhibition suggested that the isolate exhibited tolerance to the pesticide.34
This study focuses on the morphological characteristics, phylogenetic relationships, and pesticide tolerance of FLAB isolates obtained from honey bee gut, pollen, and honey. Detailed morphological observations provided insights into the growth patterns and colony characteristics of different LAB species, and phylogenetic analysis revealed the evolutionary relationships among the isolates and, offered insight on their genetic diversity and adaptation to the investigated honey bee ecosystem. Additionally, pesticide tolerance assays highlighted their ability to withstand agricultural chemicals.
Morphological characteristics of Apilactobacillus kunkeei
The taxonomic position of A. kunkeei is: Domain Bacteria, Phylum Firmicutes, Class Bacilli, Order Lactobacillales, Family Lactobacillaceae, Genus Apilactobacillus, Species Apilactobacillus kunkeei. This bacterium exhibits distinct morphological characteristics when cultured in MRS broth. It is a Gram-positive, non-motile, non-sporulating, rod-shaped, obligate heterofermentative bacterium that typically appears singly or in pairs or in chains. Growth in MRS broth results in turbidity with sediment formation, indicating active bacterial proliferation. As a facultative anaerobe, A. kunkeei can grow in both aerobic and anaerobic conditions, thriving best at a pH range of 5.5 to 6.5. It is catalase-negative, distinguishing it from other LAB that may exhibit catalase activity. On MRS agar, A. kunkeei forms smooth, circular, concave colonies that are creamy to white in appearance. The optimal growth temperature ranges between 15-37 °C. This bacterium is naturally found in the honey bee gut, pollen, and honey, where it plays a crucial role in maintaining hive health, fermenting nectar, and contributing to the microbial stability of bee-related environments. Its presence in these habitats highlights its adaptation to nutrient-rich yet fluctuating ecological niches within the honey bee colony (Figure 2A).
Figure 2. Four different fructophilic lactic acid bacteria isolated on MRS media (24 hours) from honey bee environment
Morphological characteristics of Apilactobacillus apinorum
The taxonomic position of A. apinorum is: Domain Bacteria, Phylum Firmicutes, Class Bacilli, Order Lactobacillales, Family Lactobacillaceae, Genus Apilactobacillus, Species Apilactobacillus apinorum. A. apinorum is a Gram-positive, rod-shaped, non-motile, and non-sporulating bacterium that occurs singly or in short chains. It is facultatively anaerobic, thrives under microaerophilic conditions, and is catalase-negative. When grown in MRS broth, it exhibits increasing turbidity over time, sometimes with sedimentation at the bottom. On MRS agar, it forms small, circular, opaque with smooth to rough surface, creamy white colonies. This species prefers mildly acidic conditions, with optimal growth around pH 5.5-6.5. Its carbohydrate fermentation ability varies, influencing its ecological role, particularly in honey bee-associated environments such as pollen and the honey bee gut (Figure 2B).
Morphological characteristics of Fructobacillus fructosus
The taxonomic position of F. fructosus is: Domain Bacteria, Phylum Firmicutes, Class Bacilli, Order Lactobacillales, Family Lactobacillaceae, Genus Fructobacillus, Species Fructobacillus fructosus. F. fructosus is a Gram-positive, non-motile, non-sporulating heterofermentative bacterium commonly found in fructose-rich environments such as pollen and honey bee gut. In MRS broth, F. fructosus exhibits weak growth due to the medium’s glucose-based formulation, as it primarily prefers fructose as a carbohydrate source. Morphologically, the cells appear as short rods or sometimes coccoid under certain growth conditions. The bacterium is facultatively anaerobic and exhibits weak turbidity in MRS broth, with better growth observed when supplemented with additional fructose. On MRS agar, it forms small, circular, convex, and smooth colonies. It is catalase-negative and ferments sugars to produce lactic acid, acetic acid, and ethanol. The optimal growth temperature ranges between 25-30 °C (Figure 2C).
Morphological characteristics of Secundilactobacillus kimchicus
The taxonomic position of S. kimchicus is as follows: Domain Bacteria, Phylum Firmicutes, Class Bacilli, Order Lactobacillales, Family Lactobacillaceae, Genus Secundilactobacillus, Species Secundilactobacillus kimchicus. This bacterium is a Gram-positive, rod-shaped, heterofermentative bacterium that is typically non-motile and non-spore forming. The cells of S. kimchicus are generally observed singly or in pairs or in short chains. Colonies of this bacterium appear circular, convex, and smooth with the entire edges, with a creamy to white coloration, measuring approximately 0.8-1.5 mm in diameter after 24-48 hours of incubation. As a facultatively anaerobic bacterium, S. kimchicus thrives in both aerobic and anaerobic conditions and exhibits a catalase-negative reaction. These morphological traits are consistent with those of S. kimchicus isolated from its original source, kimchi, a traditional fermented vegetable product. However, S. kimchicus was now first reported in honey bee gut from the Kanniyakumari region, may display slight variations due to their adaptation to the specific gut environment of the honey bee (Figure 2D).
Molecular identification was performed for all strains, and the sequences were submitted to the NCBI database. The accession numbers of the submitted sequences are provided in Table 1.
Table (1):
Accession numbers of fructophilic lactic acid bacteria isolated from the honey bee environment across different districts of Tamil Nadu, India
No. |
District |
Source |
Species |
Strain |
Accession Number |
|---|---|---|---|---|---|
1. |
Coimbatore |
Pollen |
Apilactobacillus kunkeei |
ACIP 01 |
PQ425901 |
2. |
Coimbatore |
Pollen |
Apilactobacillus kunkeei |
ACIP 02 |
PQ465585 |
3. |
Coimbatore |
Pollen |
Apilactobacillus kunkeei |
ACIP 03 |
PQ468037 |
4. |
Coimbatore |
Pollen |
Apilactobacillus apinorum |
ACIP 01 |
PQ468418 |
5. |
Coimbatore |
Honey |
Apilactobacillus kunkeei |
ACIH 04 |
PQ469951 |
6. |
Coimbatore |
Pollen |
Fructobacillus fructosus |
AMP 01 |
PQ112450 |
7. |
Coimbatore |
Larvae |
Apilactobacillus kunkeei |
AML1 01 |
PQ112451 |
8. |
Erode |
Pollen |
Apilactobacillus kunkeei |
ACIP 05 |
PQ469988 |
9. |
Erode |
Pollen |
Apilactobacillus kunkeei |
ACIP 06 |
PQ475853 |
10. |
Erode |
Pollen |
Apilactobacillus kunkeei |
ACIP 07 |
PQ472146 |
11. |
Erode |
Pollen |
Apilactobacillus kunkeei |
ACIP 08 |
PQ475867 |
12. |
Erode |
Honey |
Apilactobacillus kunkeei |
ACIH 09 |
PQ475893 |
13. |
Erode |
Honey |
Apilactobacillus kunkeei |
ACIH 10 |
PQ559814 |
14. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 11 |
PQ559823 |
15. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 12 |
PQ564691 |
16. |
Dindigul |
Gut |
Fructobacillus fructosus |
ACIG 1 |
PQ564753 |
17. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 24 |
PQ813545 |
18. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 13 |
PQ569344 |
19. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 14 |
PQ579878 |
20. |
Dindigul |
Gut |
Fructobacillus fructosus |
ACIG 2 |
PQ590175 |
21. |
Dindigul |
Gut |
Fructobacillus fructosus |
ACIG 3 |
PQ590316 |
22. |
Dindigul |
Gut |
Apilactobacillus kunkeei |
ACIG 25 |
PQ813578 |
23. |
Dindigul |
Gut |
Fructobacillus fructosus |
ACIG 4 |
PQ621274 |
24. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIP 15 |
PQ621275 |
25. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIP 16 |
PQ615307 |
26. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIP 17 |
PQ615309 |
27. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIP 18 |
PQ615322 |
28. |
Dindigul |
Pollen |
Fructobacillus fructosus |
ACIP 5 |
PQ615324 |
29. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIP 19 |
PQ615326 |
30. |
Dindigul |
Honey |
Apilactobacillus kunkeei |
ACIH 20 |
PQ615356 |
31. |
Dindigul |
Honey |
Apilactobacillus kunkeei |
ACIH 21 |
PQ615367 |
32. |
Dindigul |
Honey |
Apilactobacillus kunkeei |
ACIH 22 |
PQ615379 |
33. |
Dindigul |
Pollen |
Apilactobacillus kunkeei |
ACIH 23 |
PQ615409 |
34. |
Kanniyakumari |
Gut |
Secundilactobacillus kimchicus |
ACIG 1 |
PQ615442 |
35. |
Kanniyakumari |
Gut |
Secundilactobacillus kimchicus |
ACIG 2 |
PQ615446 |
36. |
Kanniyakumari |
Gut |
Secundilactobacillus kimchicus |
ACIG 3 |
PQ615638 |
37. |
Kanniyakumari |
Gut |
Secundilactobacillus kimchicus |
ACIG 4 |
PQ813579 |
38. |
Kanniyakumari |
Pollen |
Apilactobacillus kunkeei |
ACIP 26 |
PQ813613 |
39. |
Kanniyakumari |
Honey |
Apilactobacillus kunkeei |
ACIH 27 |
PQ813615 |
40. |
Kanniyakumari |
Honey |
Apilactobacillus kunkeei |
ACIH 28 |
PQ814164 |
41. |
Kanniyakumari |
Honey |
Apilactobacillus kunkeei |
ACIH 29 |
PQ814165 |
Biochemical characterization of A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus
All tested bacteria (A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus) were catalase- and oxidase-negative. They fermented glucose and sucrose, while lactose fermentation was positive for all except F. fructosus. Methyl Red test was positive for all species. Growth was observed at 10 °C and 35 °C in A. kunkeei, A. apinorum, and F. fructosus, whereas S. kimchicus grew at 35 °C and 45 °C. Gas production from glucose was positive for all, but salt tolerance (6.5% NaCl) was observed only in S. kimchicus. None of the species showed nitrate reduction or H2S production (Table 2).
Table (2):
Biochemical test results of A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus
Biochemical Test |
A. kunkeei |
A. apinorum |
F. fructosus |
S. kimchicus |
|---|---|---|---|---|
Catalase test |
– |
– |
– |
– |
Oxidase test |
– |
– |
– |
– |
Glucose fermentation |
+ |
+ |
+ |
+ |
Lactose fermentation |
+ |
+ |
– |
+ |
Sucrose fermentation |
+ |
+ |
+ |
+ |
Methyl Red test |
+ |
+ |
+ |
+ |
Growth at 10 °C |
+ |
+ |
+ |
– |
Growth at 35 °C |
+ |
+ |
+ |
+ |
Growth at 45 °C |
– |
– |
– |
+ |
Gas from glucose |
+ |
+ |
+ |
+ |
Salt tolerance 6.5% NaCl |
– |
– |
– |
+ |
Nitrate reduction |
– |
– |
– |
– |
H2S production TSI test |
– |
– |
– |
– |
Growth kinetics of isolated LAB
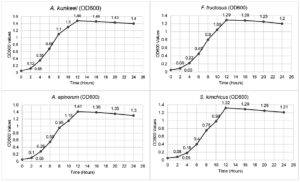
The growth of A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus was monitored over 24 hours using OD600 measurements. All strains exhibited a steady increase in optical density, reaching their peak at 12 hours (A. kunkeei: 1.48, A. apinorum: 1.41, F. fructosus: 1.29, S. kimchicus: 1.32). Growth stabilized thereafter, with a slight decline observed at 24 hours (Figure 3).
Fructophilic preference of isolated LAB
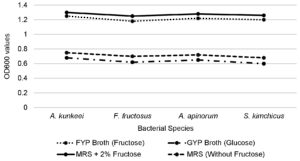
To determine their fructophilic nature, bacterial growth was evaluated in different media. All four bacterial strains (A. kunkeei, F. fructosus, A. apinorum, and S. kimchicus) exhibited significantly higher turbidity in FYP broth compared to GYP broth, indicating a preference for fructose over glucose. Specifically, the OD600 values in FYP broth ranged from 1.25 ± 0.05 (A. kunkeei), followed by 1.18 ± 0.04 (F. fructosus), 1.22 ± 0.05 (A. apinorum) and 1.20 ± 0.02 (S. kimchicus), while in GYP broth, the values were significantly lower, ranging from 0.68 ± 0.04 (A. kunkeei) followed by 0.62 ± 0.03 (F. fructosus), 0.65 ± 0.02 (A. apinorum) and 0.60 ± 0.03 (S. kimchicus) (Figure 4).
Similarly, growth was enhanced in MRS broth supplemented with 2% fructose, with OD600 values ranging from 1.30 ± 0.03 (A. kunkeei), followed by 1.25 ± 0.04 (F. fructosus), 1.28 ± 0.02 (A. apinorum) and 1.26 ± 0.03 (S. kimchicus), compared to lower values in MRS without fructose 0.75 ± 0.02 (A. kunkeei) followed by 0.70 ± 0.03 (F. fructosus), 0.72 ± 0.04 (A. apinorum) and 0.68 ± 0.02 (S. kimchicus). These findings confirm that the tested LAB exhibit a strong fructophilic growth preference (Figure 4).
Phylogenetic analysis and evolutionary divergence
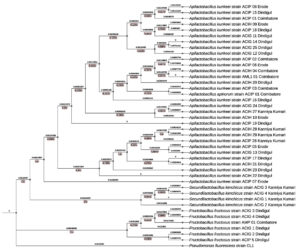
A Neighbor-Joining tree was constructed based on the 16S rRNA gene sequences of LAB isolates, including species from the genera Apilactobacillus, Secundilactobacillus, and Fructobacillus. The phylogenetic relationships were inferred using a Kimura 2-parameter model, and bootstrap analysis (1,000 replicates) was performed to assess the robustness of the tree topology. Pseudomonas fluorescens strain CL1 was used as an outgroup, exhibiting a significant branch length (0.222), confirming its distant phylogenetic relationship to the LAB isolates (Figure 5).
Figure 5. Phylogenetic analysis of lactic acid bacteria isolated from the honey bee environment based on 16S rRNA gene sequences
Apilactobacillus kunkeei clade
The A. kunkeei clade in the phylogenetic tree shows strong clustering with high bootstrap support values, suggesting a well-resolved evolutionary relationship among the isolates. The bootstrap values within this clade range from 0.90 to 0.99, indicating strong confidence in the phylogenetic grouping. The branch lengths vary, with shorter branches pointing to close evolutionary relationships and longer branches indicating greater genetic divergence among certain isolates. The A. kunkeei isolates cluster into distinct subgroups based on their geographic origin.
The Dindigul cluster (ACIP 15, ACIP 18, ACIG 11, ACIG 14, ACIG 25, and ACIG 12) is characterized by a high bootstrap value of 0.96 and short branch lengths (0.0019523 to 0.00297718). This minimal genetic divergence suggests a recent evolutionary split or strong selective pressures maintaining genetic stability in this region. In contrast, the Kanyakumari cluster (ACIH 27, ACIH 28, ACIH 29, and ACIP 26) has a slightly lower bootstrap value of 0.91 and longer branch lengths (0.0132 to 0.0162). The greater genetic divergence observed here may be attributed to regional environmental factors influencing microbial evolution. The Coimbatore cluster, which is positioned basally in the phylogenetic tree, consists of ACIP 01, ACIP 02, ACIH 04, and AML1 01. With a bootstrap value of 0.94 and short branch lengths (0.00045 to 0.00148), this cluster appears to be more ancestral, suggesting it represents a core population from which other regional strains have diverged. Lastly, the divergent isolates (ACIP 07, ACIP 05, and ACIG 13) have a bootstrap value of 0.90 and the longest branch lengths (0.0361 to 0.0461). This indicates significant evolutionary changes, likely driven by adaptation to unique ecological niches such as high sugar concentrations in nectar or the antimicrobial properties of honey.
The overall topology of the A. kunkeei clade reveals a combination of conserved and regionally adapted strains. The high bootstrap values (>0.90) reinforce the reliability of these evolutionary relationships. The basal position of the Coimbatore isolates suggests that they may be ancestral, while the longer branch lengths in certain isolates highlight evolutionary pressures that have led to functional divergence. The genetic divergence observed among distant isolates may reflect adaptive mechanisms that enable A. kunkeei to thrive in different environmental conditions within the honey bee microbiome.
Apilactobacillus apinorum sub-clade
The A. apinorum Sub-Clade is a distinct subgroup within the A. kunkeei cluster contained A. apinorum strain ACIP 01 (Coimbatore), forming a separate lineage with a bootstrap value of 85%. The branch length separating this sub-clade from the main A. kunkeei group was 0.0026, suggesting minor genetic differentiation.
Fructobacillus fructosus clade
In F. fructosus Clade, the isolates from Dindigul and Coimbatore grouped within the F. fructosus lineage. This clade exhibited high bootstrap support (99%), with longer branch lengths (0.012 to 0.045), suggesting a higher degree of evolutionary divergence. The F. fructosus isolates were phylogenetically distinct from Apilactobacillus and Secundilactobacillus, forming a separate lineage. The greater genetic divergence in F. fructosus isolates suggests greater evolutionary variability, potentially reflecting adaptations to different environmental niches.
Secundilactobacillus kimchicus clade
In S. kimchicus Clade isolates from Kanniyakumari (strains ACIG 1, 2, 3, and 4) formed a well-supported independent clade (bootstrap 88%). The branch lengths within this group ranged from 0.0022 to 0.046, indicating greater genetic variability compared to the Apilactobacillus cluster. The phylogenetic placement of S. kimchicus as a distinct clade supports its classification as a separate evolutionary lineage within LAB.
The Neighbor-Joining tree clearly shows that A. kunkeei isolates from different locations form a tight clade, while Fructobacillus and Secundilactobacillus are more genetically diverse. The bootstrap values (ranging from 85% to 99%) provide statistical confidence in the phylogenetic grouping. The branch length variations (ranging from 0.0006 to 0.222) suggest different levels of evolutionary divergence among the isolates. The short branch lengths within the A. kunkeei group suggest recent diversification from a common ancestor. The greater genetic divergence in F. fructosus indicates potential adaptations to different ecological niches. The high bootstrap values (85-99%) provide statistical confidence in the phylogenetic groupings. The variation in branch lengths (0.0006 to 0.222) suggests differing rates of evolutionary divergence among the LAB isolates. Overall, the Neighbor-Joining tree clearly delineates the phylogenetic relationships among LAB isolates, highlighting the close genetic relatedness of A. kunkeei while emphasizing the greater diversity of Fructobacillus and Secundilactobacillus.
Tolerance of lactic acid bacteria to pesticides
The ability of 41 FLAB isolates to tolerate four pesticides such as imidacloprid, dinotefuran, fipronil, and dimethoate was assessed at varying concentrations (50, 100, 250, 500, and 1,000 ppm). The isolates exhibited different tolerance levels, categorized as very low, low, moderate, high, and very high based on the presence or absence of growth inhibition zones (Figure 6).
Tolerance to imidacloprid
Among the tested LAB species, A. kunkeei strains demonstrated the highest tolerance, with multiple isolates (ACIP 02, ACIH 04, AML1 01, ACIP 05, ACIP 08, ACIG 12, ACIG 24, ACIG 13, ACIG 14, ACIP 15, ACIP 17, ACIP 18, ACIP 19, ACIH 20, ACIH 22, ACIH 23, ACIP 26, ACIH 27, ACIH 28, ACIH 29) exhibiting very high tolerance, even at the highest concentration (1,000 ppm). On the other hand, A. apinorum (ACIP 01) and F. fructosus (AMP 01, ACIG 1, ACIG 3, ACIG 4, ACIP 5) displayed moderate tolerance, with growth inhibition observed at 500 ppm. S. kimchicus (ACIG 3) was the least tolerant, showing inhibition at 250 ppm (Table 3).
Table (3):
Tolerance of lactic acid bacteria to different concentrations of imidacloprid pesticide
| LAB Species | Strain | Inhibition zone diameter (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | 50 ppm | 100 ppm | 250 ppm | 500 ppm | 1,000 ppm | Tolerant | ||
| A. kunkeei | ACIP 01 | 0 | 0 | 0 | 0 | 0 | 7.1 | High |
| A. kunkeei | ACIP 02 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 03 | 0 | 0 | 0 | 0 | 6.2 | 8.1 | Moderate |
| A. apinorum | ACIP 01 | 0 | 0 | 0 | 0 | 6.3 | 7.5 | Moderate |
| A. kunkeei | ACIH 04 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | AMP 01 | 0 | 0 | 0 | 0 | 6.8 | 8.1 | Moderate |
| A. kunkeei | AML1 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 05 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 06 | 0 | 0 | 0 | 0 | 6.7 | 7.6 | Moderate |
| A. kunkeei | ACIP 07 | 0 | 0 | 0 | 0 | 0 | 7.1 | High |
| A. kunkeei | ACIP 08 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 09 | 0 | 0 | 0 | 0 | 0 | 7.0 | High |
| A. kunkeei | ACIH 10 | 0 | 0 | 0 | 0 | 0 | 7.5 | High |
| A. kunkeei | ACIG 11 | 0 | 0 | 0 | 0 | 0 | 7.1 | High |
| A. kunkeei | ACIG 12 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 1 | 0 | 0 | 0 | 0 | 6.5 | 7.8 | Moderate |
| A. kunkeei | ACIG 24 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 13 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 14 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 6.9 | High |
| F. fructosus | ACIG 3 | 0 | 0 | 0 | 0 | 6.8 | 7.2 | Moderate |
| A. kunkeei | ACIG 25 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 4 | 0 | 0 | 0 | 0 | 6.2 | 7.2 | Moderate |
| A. kunkeei | ACIP 15 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 16 | 0 | 0 | 0 | 0 | 7.2 | 8.0 | Moderate |
| A. kunkeei | ACIP 17 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 18 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIP 5 | 0 | 0 | 0 | 0 | 7.1 | 7.9 | Moderate |
| A. kunkeei | ACIP 19 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 20 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 21 | 0 | 0 | 0 | 0 | 0 | 7.6 | High |
| A. kunkeei | ACIH 22 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 23 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 1 | 0 | 0 | 0 | 0 | 6.9 | 8.2 | Moderate |
| S. kimchicus | ACIG 2 | 0 | 0 | 0 | 0 | 6.8 | 7.2 | Moderate |
| S. kimchicus | ACIG 3 | 0 | 0 | 0 | 6.3 | 6.5 | 8 | Less |
| S. kimchicus | ACIG 4 | 0 | 0 | 0 | 0 | 6.5 | 7.3 | Moderate |
| A. kunkeei | ACIP 26 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 27 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 28 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 29 | 0 | 0 | 0 | 0 | 0 | 6.7 | High |
*The experiment was performed with 3 replications
Tolerance to dinotefuran
A similar pattern was observed with dinotefuran, where A. kunkeei strains demonstrated high tolerance. In contrast, A. apinorum (ACIP 01) and some F. fructosus isolates (AMP 01, ACIG 4, ACIP 5) exhibited moderate tolerance, showing inhibition at 500 ppm. Notably, A. kunkeei (ACIH 21) and S. kimchicus (ACIG 3) were highly sensitive, as they were inhibited at just 100 ppm (Table 4).
Table (4):
Tolerance of lactic acid bacteria to different concentrations of dinotefuran pesticide
| LAB Species | Strain | Inhibition zone diameter (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | 50 ppm | 100 ppm | 250 ppm | 500 ppm | 1,000 ppm | Tolerant | ||
| A. kunkeei | ACIP 01 | 0 | 0 | 0 | 0 | 0 | 7.3 | High |
| A. kunkeei | ACIP 02 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 03 | 0 | 0 | 0 | 0 | 6.9 | 9.2 | Moderate |
| A. apinorum | ACIP 01 | 0 | 0 | 0 | 6.4 | 7.2 | 10.8 | Moderate |
| A. kunkeei | ACIH 04 | 0 | 0 | 0 | 0 | 6.8 | 8.3 | Moderate |
| F. fructosus | AMP 01 | 0 | 0 | 0 | 0 | 0 | 7.5 | High |
| A. kunkeei | AML1 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 05 | 0 | 0 | 0 | 0 | 6.7 | 8.9 | Moderate |
| A. kunkeei | ACIP 06 | 0 | 0 | 0 | 0 | 6.6 | 9.2 | Moderate |
| A. kunkeei | ACIP 07 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 08 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 09 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 10 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 11 | 0 | 0 | 0 | 0 | 6.9 | 9.4 | Moderate |
| A. kunkeei | ACIG 12 | 0 | 0 | 0 | 0 | 0 | 7.8 | High |
| F. fructosus | ACIG 1 | 0 | 0 | 0 | 0 | 6.5 | 8.7 | Moderate |
| A. kunkeei | ACIG 24 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 13 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 14 | 0 | 0 | 0 | 0 | 0 | 7.9 | High |
| F. fructosus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 8.1 | High |
| F. fructosus | ACIG 3 | 0 | 0 | 0 | 0 | 0 | 8.0 | High |
| A. kunkeei | ACIG 25 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 4 | 0 | 0 | 0 | 0 | 6.9 | 8.3 | Moderate |
| A. kunkeei | ACIP 15 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 16 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 17 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 18 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIP 5 | 0 | 0 | 0 | 0 | 0 | 7.7 | High |
| A. kunkeei | ACIP 19 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 20 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 21 | 0 | 0 | 6.5 | 7.4 | 8.2 | 10.6 | Very less |
| A. kunkeei | ACIH 22 | 0 | 0 | 0 | 0 | 0 | 7.7 | High |
| A. kunkeei | ACIH 23 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 1 | 0 | 0 | 0 | 0 | 6.9 | 7.6 | Moderate |
| S. kimchicus | ACIG 2 | 0 | 0 | 0 | 0 | 6.8 | 8.2 | Moderate |
| S. kimchicus | ACIG 3 | 0 | 0 | 0 | 6.7 | 7.9 | 8.0 | Less |
| S. kimchicus | ACIG 4 | 0 | 0 | 0 | 0 | 7.1 | 8.6 | Moderate |
| A. kunkeei | ACIP 26 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 27 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 28 | 0 | 0 | 0 | 6.5 | 7.8 | 10.8 | Less |
| A. kunkeei | ACIH 29 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
*The experiment was performed with three replications
Tolerance to fipronil
Most A. kunkeei isolates showed very high tolerance to fipronil, except for ACIP 19 and ACIH 22, which exhibited lower tolerance levels. F. fructosus (ACIP 5) and A. apinorum (ACIP 01) displayed moderate tolerance, with inhibition occurring at 500 ppm. Interestingly, S. kimchicus (ACIG 1, ACIG 2, ACIG 3, ACIG 4) exhibited very high tolerance, suggesting that this species may possess mechanisms for resistance (Table 5).
Table (5):
Tolerance of lactic acid bacteria to different concentrations of fipronil pesticide
| LAB Species | Strain | Inhibition zone diameter (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | 50 ppm | 100 ppm | 250 ppm | 500 ppm | 1,000 ppm | Tolerant | ||
| A. kunkeei | ACIP 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 02 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 03 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. apinorum | ACIP 01 | 0 | 0 | 0 | 0 | 6.8 | 8.2 | Moderate |
| A. kunkeei | ACIH 04 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | AMP 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | AML1 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 05 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 06 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 07 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 08 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 09 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 10 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 11 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 12 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 1 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 24 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 13 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 14 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 3 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 25 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 4 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 15 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 16 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 17 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 18 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIP 5 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 19 | 0 | 0 | 0 | 0 | 6.9 | 7.6 | Moderate |
| A. kunkeei | ACIH 20 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 21 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 22 | 0 | 0 | 0 | 0 | 6.5 | 8.3 | Moderate |
| A. kunkeei | ACIH 23 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 1 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 3 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 4 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 26 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 27 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 28 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 29 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
*The experiment was performed with three replications
Tolerance to dimethoate
A similar pattern was observed for dimethoate, with A. kunkeei strains showing very high tolerance. However, S. kimchicus strains (ACIG 1, ACIG 2, ACIG 3, ACIG 4) exhibited high tolerance, with slight inhibition observed at 1,000 ppm. A. apinorum (ACIP 01) and F. fructosus (ACIP 5, ACIG 4) showed moderate tolerance, being inhibited at 500 ppm (Table 6).
Table (6):
Tolerance of lactic acid bacteria to different concentrations of dimethoate pesticide
| LAB Species | Strain | Inhibition zone diameter (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | 50 ppm | 100 ppm | 250 ppm | 500 ppm | 1,000 ppm | Tolerant | ||
| A. kunkeei | ACIP 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 02 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 03 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. apinorum | ACIP 01 | 0 | 0 | 0 | 6.8 | 7.5 | 7.9 | Moderate |
| A. kunkeei | ACIH 04 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | AMP 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | AML1 01 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 05 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 06 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 07 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 08 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 09 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 10 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 11 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 12 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 1 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 24 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 13 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 14 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 3 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIG 25 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIG 4 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 15 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 16 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 17 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 18 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| F. fructosus | ACIP 5 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIP 19 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 20 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 21 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 22 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 23 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| S. kimchicus | ACIG 1 | 0 | 0 | 0 | 0 | 0 | 7.1 | High |
| S. kimchicus | ACIG 2 | 0 | 0 | 0 | 0 | 0 | 6.8 | High |
| S. kimchicus | ACIG 3 | 0 | 0 | 0 | 0 | 0 | 7.6 | High |
| S. kimchicus | ACIG 4 | 0 | 0 | 0 | 0 | 0 | 8.0 | High |
| A. kunkeei | ACIP 26 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 27 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 28 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
| A. kunkeei | ACIH 29 | 0 | 0 | 0 | 0 | 0 | 0 | Very high |
*The experiment was performed with three replications
Among the tested LAB strains, A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus exhibited varying levels of tolerance to the four pesticides—imidacloprid, dinotefuran, fipronil, and dimethoate. Several A. kunkeei strains (ACIP 02, AML1 01, ACIP 05, ACIP 08, ACIG 12, ACIG 24, ACIG 13, ACIG 14, ACIP 15, ACIP 17, ACIP 18, ACIP 19, ACIH 20, ACIH 23, ACIP 26, ACIH 27, ACIH 28, and ACIH 29) demonstrated very high tolerance to all four pesticides, with no inhibition observed even at the highest concentration (1,000 ppm). Similarly, F. fructosus (ACIG 1, ACIG 2, ACIG 3, ACIG 4, ACIP 5) and S. kimchicus (ACIG 1, ACIG 2, ACIG 3, ACIG 4) also exhibited very high tolerance, suggesting potential resistance mechanisms. Other A. kunkeei strains, including ACIP 01, ACIP 07, ACIH 09, ACIH 10, ACIG 11, and ACIH 22, exhibited high tolerance, with slight inhibition at 1,000 ppm.
Some LAB isolates showed moderate tolerance to pesticides. For example, A. apinorum (ACIP 01), F. fructosus (AMP 01, ACIG 1, ACIG 3, ACIG 4, ACIP 5), and A. kunkeei (ACIP 03, ACIP 06, ACIG 25, ACIG 16) were able to withstand exposure up to 500 ppm before inhibition occurred. However, certain strains displayed much lower tolerance. A. kunkeei (ACIH 21) was inhibited at just 100 ppm of dinotefuran, while S. kimchicus (ACIG 3) was the most sensitive, showing the least resistance to both imidacloprid and dinotefuran. These results emphasize the wide range of pesticide tolerance among LAB isolates, with A. kunkeei emerging as the most resilient species.
Honey bees depend extensively on their gut microbial communities to combat pests, pathogens, environmental toxins, and nutrient-deficient food sources.13,35 In most animals, gut microflora plays a vital role in nutrient assimilation and immune function.36 Similarly, in honey bees, these microbial communities enhance resistance to pesticides by facilitating detoxification and strengthening immune defenses. However, pesticide exposure can disrupt these microbial populations, rendering bees more vulnerable to pathogens. Disruptions caused by these stressors have significantly contributed to the high rates of colony losses observed in recent years. The decline in honey bee populations is a complex issue, with pesticide exposure and disease outbreaks being major contributing factors.37
Among the beneficial microbes residing in the honey bee gastrointestinal tract, LAB play a crucial role. The most well-known LAB species associated with honey bees belong to the genera Lactobacillus and Bifidobacterium.38 Traditionally, culture-based methods were employed to isolate and identify these LABs.39 A study identified A. kunkeei as the most prevalent Lactobacillus species among Apis species.10 FLAB such as A. kunkeei and Fructobacillus spp. have been isolated from honey bee guts and honey produced by stingless bees.40,41 These bacteria thrive in fructose-rich environments, showcasing their adaptability and potential probiotic properties.42 Consistent with these findings, our study also identified A. kunkeei, F. fructosus, A. apinorum, and S. kimchicus from the honey bee environment, further highlighting the presence of beneficial microbial communities associated with honey bees.
A. kunkeei, a key member of the honey bee gut microbiota, plays a crucial role in maintaining gut health and is also significant in wine fermentation due to its fructophilic nature it is commonly referred to as ‘ferocious lactobacilli’.43 However, in the honey bee ecosystem, this species has co-evolved with bees and functions as a crucial probiotic, playing a vital role in protecting both bees and their hives from pathogens. The presence of these beneficial microbes underscores the importance of maintaining a stable gut microbiota in honey bees to mitigate the adverse effects of environmental stressors and promote colony health.
In this study, we aimed to isolate FLAB from the honeybee gut with potential probiotic benefits. We successfully obtained 30 strains of A. kunkeei from the gut, pollen, and honey similar to the previous reports,6,44-46 one strain of A. apinorum from pollen,47 six F. fructosus strains from the gut and pollen,24 and four S. kimchicus strains from the gut of A. cerana indica. This is the first report of S. kimchicus in gut of A. cerana indica honey bee, collected from rubber plantations. The observed LAB diversity may be influenced by the floral composition of nectar, extrafloral nectaries, and pollen, as well as microbial exposure from flowers. These findings are consistent with, previous findings suggest that LAB species distribution in bees can vary geographically and seasonally.10 Although prior studies have reported stable honeybee microbiota across regions, our findings indicate that specific environmental factors, such as nectar sources in rubber plantations, may contribute to LAB diversity in the honeybee gut.39 Previous research has shown that members of Lactobacillus and Leuconostoc are associated with honeybee health and colony size.21 In this study, we successfully isolated and characterized 41 bacterial strains belonging to four LAB species: A. kunkeei, A. apinorum, F. fructosus, and S. kimchicus. Further research is needed to assess their potential probiotic roles and interactions within honeybee colonies.
This study demonstrated that all tested bacterial strains, including A. kunkeei, F. fructosus, A. apinorum, and S. kimchicus, exhibited significantly growth in fructose-rich environments, with OD values consistently higher than in glucose-based media, confirming its fructophilic nature. Similarly, enhanced growth was observed in MRS supplemented with 2% fructose compared to MRS without fructose, further reinforcing their preference for fructose. Recent study reported similar trends, indicating that A. kunkeei and F. fructosus grew well in FYP broth but poorly in GYP broth under anaerobic conditions, classifying them as FLAB.24 The fructophilic strains F. fructosus, A. kunkeei, F. pseudoficulneus, and F. durionis exhibited a preference for D-fructose over D-glucose.48 These bacteria thrive in fructose-rich environments, showcasing their adaptability and potential probiotic properties.42 The above studies confirm the fructophilic nature of specific FLAB, particularly A. kunkeei and F. fructosus and this study expands this classification by including A. apinorum and S. kimchicus, which also displayed fructophilic tendencies. The similarity in results strengthens the conclusion that these bacteria thrive in fructose-rich environments, a characteristic that may be ecologically relevant for their natural habitats, such as honey and nectar.
Phylogenetic analysis of FLAB isolates is crucial for understanding their evolutionary relationships, ecological distribution, and functional adaptation. The 16S rRNA gene is commonly used for bacterial taxonomy due to its conserved nature and ability to reveal subtle genetic differences.49 The phylogenetic analysis revealed clear distinctions among the FLAB species isolated from the honey bee environment. A. kunkeei and A. apinorum clustered closely, indicating a shared evolutionary lineage, whereas S. kimchicus and F. fructosus formed distinct clades, suggesting greater divergence. Notably, F. fructosus exhibited the longest branch length, implying higher genetic differentiation. The bootstrap values further validated these relationships, with strong support (>90%) for the clustering of A. kunkeei and A. apinorum, while S. kimchicus and F. fructosus showed moderate to high support values, confirming their distinct phylogenetic placement. Isolates from Coimbatore, Erode, and Dindigul formed a highly supported monophyletic clade (bootstrap >90%), suggesting a strong evolutionary relationship among these strains. The short branch lengths (0.0006-0.0098) within this group indicate low genetic divergence, consistent with previous findings on A. kunkeei being highly conserved in honeybee microbiomes.50 Interestingly, A. kunkeei isolates from different regions clustered more closely than expected, suggesting potential microbial dispersal via environmental factors such as bees, pollen, and nectar transfer.10 In a phylogenetic tree comprising A. kunkeei, A. apinorum, and F. fructosus, all three species cluster within the FLAB group. A. kunkeei and A. apinorum are closely related within the Apilactobacillus genus. The sub-cluster containing A. apinorum strain ACIP 01 branched distinctly within the A. kunkeei clade, supported by a bootstrap value of 85%. The observed branch length (0.0026) suggests incipient genetic divergence, which might indicate ongoing evolutionary separation or ecological adaptation.51 Both phylogenetic trees of the previous and present study reveal a close branching relationship between A. kunkeei and A. apinorum.46 This highlights a significant evolutionary link between these species, suggesting a common ancestral lineage. Their close proximity in the phylogenetic tree implies that they share key genetic and functional similarities, potentially due to adaptation to similar ecological niches, such as honeybee-associated environments. The clustering of these species may reflect their involvement in comparable symbiotic or metabolic pathways within their host organisms. Additionally, their minimal genetic divergence could indicate niche specialization or subtle functional differentiation over evolutionary time. These findings provide valuable insights into their evolutionary trajectory and potential functional roles within their microbiome, encouraging further research into their genomic and ecological characteristics.
In the phylogenetic analysis conducted by previous study, a distinct relationship is observed among A. kunkeei, F. fructosus and S. kimchicus cluster closely, suggesting a shared evolutionary lineage or functional similarity.51 This proximity is indicative of conserved traits, such as sugar metabolism and acid tolerance, which are crucial for survival in sugar-rich and acidic environments. Despite their ecological differences, A. kunkeei which is predominantly associated with the honey bee gut microbiota and S. kimchicus which is commonly found in fermented foods, appear to share adaptive strategies for thriving in challenging conditions. In contrast, F. fructosus emerges as a more distantly related lineage within the broader clade, suggesting an earlier evolutionary divergence likely driven by specialization in fructose-rich floral environments.
The present study provides complementary yet distinct insights into the phylogenetic relationships of these FLAB. The isolates from Kanniyakumari (ACIG 1-4) formed an independent lineage (bootstrap 88%) within Secundilactobacillus, highlighting moderate genetic variability (branch lengths 0.0022-0.046). The phylogenetic tree constructed in this study reflects both similarities and differences compared to the previous study tree.51 A. kunkeei clusters with other honey bee-associated microbes, underscoring its evolutionary adaptation to the honey bee gut microbiota and its specialized functional role within this niche. Conversely, S. kimchicus in the investigated tree showed genetic proximity to other Lactobacillus species, likely reflecting shared traits such as acid tolerance mechanisms essential for survival in both gut environments and fermented foods. Notably, F. fructosus formed a distinct lineage in our phylogenetic tree, consistent with its classification under the Fructobacillus genus. Isolates from Dindigul and Coimbatore clustered within the F. fructosus lineage (bootstrap 99%), with branch lengths ranging from 0.012 to 0.045, suggesting higher evolutionary divergence compared to Apilactobacillus isolates. This separation further emphasizes a closer evolutionary relationship between the two Apilactobacillus species (A. kunkeei and S. kimchicus) compared to F. fructosus.
The observed differences between the previous study tree and the present study’s tree may stem from several methodological factors.51 These include the use of geographically diverse strains, differences in datasets, the genes targeted for analysis, or variations in phylogenetic reconstruction models. The differences between the previous phylogenetic tree and our phylogenetic analysis of FLAB isolated from the honey bee environment provide key insights into LAB evolution and function.51 One significant distinction lies in the origin of isolates – while previous study likely included LAB from diverse environments, our study focuses specifically on LAB from the honey bee gut, pollen, and honey, making it directly relevant to honey bee microbiome function.51 These differences in isolate sources may influence phylogenetic placements, revealing whether certain LAB strains have co-evolved with honey bees or possess unique adaptations to their host. Additionally, if our tree shows distinct clustering patterns or novel lineage divergence, it could indicate honey bee-specific evolutionary pressures shaping FLAB diversity. Functional implications also emerge from these differences, as phylogenetic variations can correlate with traits such as pesticide tolerance, probiotic potential, and symbiotic roles within the honey bee gut. Unlike broader phylogenetic analyses, our study provides region-specific insights, considering FLAB from honey bees in Tamil Nadu, India, which may highlight biogeographical influences on FLAB evolution. Understanding these differences helps clarify how FLAB contribute to honey bee health and informs the selection of potential probiotic strains for mitigating environmental stressors, including pesticide exposure. For instance, while the recent study tree highlights a closer evolutionary relationship between A. kunkeei and S. kimchicus, our tree offers finer resolution of their evolutionary trajectories, shaped by their respective ecological niches and environmental pressures. The broader implications of these findings underscore the complex interplay between shared evolutionary traits and niche-specific adaptations in shaping microbial communities. P. fluorescens strain CL1, used as an outgroup, exhibited a significantly longer branch length (0.222), confirming its phylogenetic distinctiveness from LAB isolates and validating tree topology. Together, these results emphasize the role of ecological and functional pressures in shaping the evolutionary pathways of lactic acid bacteria.
Among the tested 41 LAB strains, A. kunkeei, F. fructosus, and S. kimchicus exhibited varying levels of pesticide tolerance. Several A. kunkeei strains (ACIP 02, AML1 01, ACIP 05) demonstrated exceptionally high tolerance to all four pesticides (imidacloprid, dinotefuran, fipronil, and dimethoate), showing no inhibition even at 1,000 ppm. Similarly, F. fructosus and S. kimchicus displayed considerable tolerance, indicating the presence of resistance mechanisms. Some A. kunkeei strains exhibited slight inhibition at 1,000 ppm, while others, like A. apinorum and F. fructosus, showed moderate tolerance. A few strains, such as A. kunkeei (ACIH 21) and S. kimchicus (ACIG 3), demonstrated low tolerance at lower pesticide concentrations. Overall, A. kunkeei proved to be the most resilient species. Tolerant isolates can be further investigated for their potential ability to degrade pesticides in future studies.
Among the LAB species tested, A. kunkeei exhibited remarkable tolerance to imidacloprid, suggesting potential resistance mechanisms. This aligns with findings of recent study reported that L. plantarum and L. acidophilus could grow in the presence of imidacloprid up to 100 ppm.52 In vitro tests reported that sublethal doses of imidacloprid did not affect the bacterial diversity in the gut of honeybees.53 A. kunkeei, isolated from beebread, was found to reduce the mortality effect of acetamiprid on honeybees.54 This finding was same as the previous study, showed that honeybee gut bacteria could thrive in the presence of imidacloprid under in vitro conditions.55 These studies emphasize the critical role of the microbiome in maintaining honeybee health. Our results suggest that A. kunkeei may possess similar potential, making it a promising candidate for bioremediation of pesticide-contaminated environments.
Interestingly, while the tolerance of FLAB strains to fipronil was significant, there are no known studies reporting its direct degradation by LAB. Recent studies have identified Pseudomonas sp. and Rhodococcus sp. as potent degraders of fipronil.56 The presence of fipronil in soil samples has been shown to significantly alter bacterial community composition, highlighting the selective pressure exerted by fipronil on microbiota.57 Further research is required to explore whether LAB strains possess similar degradation capabilities.
The findings of this study revealed significant variation in pesticide tolerance among FLAB isolates, with A. kunkeei demonstrating the highest resilience. A similar observation was reported that Lactobacillus plantarum strains isolated from food sources can tolerate and degrade organophosphorus pesticides such as omethoate, phorate, and dimethoate.58 Additionally, Pediococcus acidilactici isolated from honey bee environments was highly resistant to imidacloprid, exhibiting increased bacterial growth.59 Certain LAB strains have also been capable of growing in the presence of high concentrations of chlorpyrifos.60 For instance, L. plantarum RS60 and P. acidilactici D15 exhibited the highest cypermethrin removal capacity and good pesticide tolerance capacity.61
FLAB strains namely A. kunkeei and S. kimchicus exhibited tolerance to dimethoate in this study. Similar studies have demonstrated the role of LAB strains in pesticide degradation. L. plantarum could degrade organophosphorus pesticides,58 while recent report showed that L. plantarum could degrade up to 81.28% of dimethoate.62 There is a decrease in the half-life of various pesticides, including dimethoate, phorate, and trichlorphon, during lactic acid fermentation.63 L. plantarum was effective in pesticide removal during black olive fermentation, achieving significant reductions in pesticide residues.64
A recent study explored the effects of pesticide exposure on L. plantarum Pb3, revealing increased antioxidant capacity in the presence of chlorpyrifos.52 Several LAB species, including Lactococcus lactis, Leuconostoc mesenteroides, and Lactobacillus rhamnosus, have been reported to degrade organophosphate pesticides.65 LAB strains, including L. bulgaricus and L. plantarum, were effective in degrading various pesticides in milk.66 These studies collectively highlight the diverse potential of LAB strains in pesticide tolerance and degradation.
The ability of A. kunkeei, F. fructosus, and S. kimchicus strains to tolerate high concentrations of pesticides, particularly dimethoate, suggests potential applications in bioremediation and the development of safer, pesticide-free food products. Further research is needed to explore the mechanisms underlying this tolerance and to assess the potential for these strains in bioremediation strategies. Future studies should investigate their functional roles in pesticide detoxification and immune modulation.
This study analysed the diversity, phylogenetic relationships, and pesticide tolerance of FLAB in the honey bee environment. It significantly advances the understanding of honey bee microbiota by reporting S. kimchicus for the first time from the honey bee gut and identifying A. kunkeei as highly tolerant to major bee-toxic pesticides. The isolation and characterization of these key bacterial species highlight their roles in maintaining gut health, with A. kunkeei showing remarkable pesticide resilience. Fructophilic growth preferences further confirm their ecological adaptation to nectar, pollen, and honey environments. These findings support the development of FLAB-based probiotic strategies to enhance honey bee resilience, promote colony health, mitigate pesticide impacts, and contribute to sustainable apiculture.
ACKNOWLEDGMENTS
The authors express gratitude to the Department of Agricultural Entomology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India, for providing the necessary facilities to conduct the research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SP performed bacteria isolation, morpho-molecular identification, phylogenetic analysis, biochemical tests and pesticide tolerance test. YSJTE and MS performed project administration. YSJTE, SM, MS and NMB supervised the study. SP wrote the original draft. YSJTE, SM, and NMB wrote the manucript. YSJTE, SM, NMB, VRS and AS reviewed the manuscript. YSJTE, SM, and NMB approved the manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
Raw sequence reads of A. kunkeei, F. fructosus, A. apinorum, and S. kimchicus have been deposited in the NCBI GenBank database. All sequences are publicly available through the NCBI GenBank database at https://www.ncbi.nlm.nih.gov/genbank.
ETHICS STATEMENT
Not applicable.
- Scheiner R, Abramson CI, Brodschneider R, et al. Standard methods for behavioural studies of Apis mellifera. J Apic Res. 2013;52(4):1-58.
Crossref - Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14(6):374-384.
Crossref - Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7(4):e36393.
Crossref - Hariprasath K, Mohankumar S, Sudha M, Saranya N, Saminathan VR. The role of honeybee gut and honey microbiome in sustainable bee and human health. J Pure Appl Microbiol. 2025;19(1):19-33.
Crossref - Zheng H, Steele MI, Leonard SP, Motta EVS, Moran NA. Honey bees as models for gut microbiota research. Lab Anim (NY). 2018;47(9):317-325.
Crossref - Asama T, Arima TH, Gomi T, et al. Lactobacillus kunkeei YB38 from honeybee products enhances IgA production in healthy adults. J Appl Microbiol. 2015;119(3):818-826.
Crossref - Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109(27):11002-11007.
Crossref - Duong BTT, Lien NTK, Thu HT, et al. Investigation of the gut microbiome of Apis cerana honeybees from Vietnam. Biotechnol Lett. 2020;42(11):2309-2317.
Crossref - Khan KA, Ganeshprasad DN, Sachin HR, Shouche YS, Ghramh HA, Sneharani AH. Gut microbial diversity in Apis cerana indica and Apis florea colonies: A comparative study. Front Vet Sci. 2023;10:1149876.
Crossref - Vasquez A, Forsgren E, Fries I, et al. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS ONE. 2012;7(3):e33188.
Crossref - Lanh PT, Duong BTT, Thu HT, T Hoa NT, Quyen DV. Comprehensive analysis of the microbiome in Apis cerana honey highlights honey as a potential source for the isolation of beneficial bacterial strains. PeerJ. 2024;12:e17157.
Crossref - Trinder M, McDowell TW, Daisley BA, et al. Probiotic Lactobacillus rhamnosus reduces organophosphate pesticide absorption and toxicity to Drosophila melanogaster. Appl Environ Microbiol. 2016;82(20):6204-6213.
Crossref - Daisley BA, Pitek AP, Chmiel JA, et al. Lactobacillus spp. attenuate antibiotic-induced immune and microbiota dysregulation in honey bees. Commun Biol. 2020;3(1):534.
Crossref - Lin CF, Lin MY, Lin CN, et al. Potential probiotic Lactobacillus strains isolated from the intestinal tracts of pigs and feces of dogs with antibacterial activity against multidrug-resistant pathogenic bacteria. Arch Microbiol. 2020;202(7):1849-1860.
Crossref - Partrick KA, Rosenhauer AM, Auger J, et al. Ingestion of probiotic (Lactobacillus helveticus and Bifidobacterium longum) alters intestinal microbial structure and behavioral expression following social defeat stress. Sci Rep. 2021;11(1):3763.
Crossref - Pradeep S, Edward YSJT, Suganthi A, Senthilkumar M, Saminathan VR, Boopathi NM. Lactic Acid Bacteria: A probiotic to mitigate pesticide stress in honey bee, Probiotics Antimicrob Proteins. 2025.
Crossref - Settanni L, van Sinderen D, Rossi J, Corsetti A. Rapid differentiation and in situ detection of 16 sourdough Lactobacillus species by multiplex PCR. Appl Environ Microbiol. 2005;71(6):3049-3059.
Crossref - Tajabadi N, Mardan M, Manap MYA, Mustafa S. Molecular identification of Lactobacillus spp. isolated from the honeycomb of the honey bee (Apis dorsata) by 16S rRNA gene sequencing. J Apic Res. 2013;52(5):235-241.
Crossref - Collins MD, Rodrigues U, Ash C, et al. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett. 1991;77(1):5-12.
Crossref - Elzeini HM, Ali ARA, Nasr NF, Elenany YE, Hassan AAM. Isolation and identification of lactic acid bacteria from the intestinal tracts of honey bees, Apis mellifera L., in Egypt. J Apic Res. 2020;60(2):349-357.
Crossref - Mathialagan M, Edward YSJT, David PMM, Senthilkumar M, Srinivasan MR, Mohankumar S. Isolation, characterization, and identification of probiotic lactic acid bacteria (LAB) from honey bees. Int J Curr Microbiol Appl Sci. 2018;7(4):849-906.
Crossref - Amanullah S, Kabir M, Rahman M, Halder P, Hossain SMJ, Samad M. Isolation, identification and biochemical characterization of lactic acid bacteria from selected yogurt samples. Bangladesh J Livest Res. 2021;27(1-2):64-72.
Crossref - Endo A, Futagawa-Endo Y, Dicks LM. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst Appl Microbiol. 2009;32(8):593-600.
Crossref - Filannino P, Di Cagno R, Addante R, Pontonio E, Gobbetti M. Metabolism of fructophilic lactic acid bacteria isolated from the Apis mellifera L. bee gut: Phenolic acids as external electron acceptors. Appl Environ Microbiol. 2016;82(23):6899-6911.
Crossref - Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461-2470.
Crossref - Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022-3027.
Crossref - Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-4680.
Crossref - Saitou N, Nei M. The Neighbor-Joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111-120.
Crossref - Verma JP, Jaiswal DK, Maurya PK. Screening of bacterial strains for developing effective pesticide-tolerant plant growth-promoting microbial consortia from rhizosphere soils of vegetable fields of eastern Uttar Pradesh, India. Energy Ecol Environ. 2016;1:408–418.
Crossref - Liu Y, Lehnert T, Gijs MAM. Effect of inoculum size and antibiotics on bacterial traveling bands in a thin microchannel defined by optical adhesive. Microsyst Nanoeng. 2021;7(1):86.
Crossref - Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15(3):e2001861.
Crossref - El Khoury S, Giovenazzo P, Derome N. Endogenous honeybee gut microbiota metabolize the pesticide clothianidin. Microorganisms. 2022;10(3):493.
Crossref - Wu Y, Zheng Y, Chen Y, Chen G, Zheng H, Hu F. Apis cerana gut microbiota contribute to host health through stimulating host immune system and strengthening host resistance to Nosema ceranae. R Soc Open Sci. 2020;7(5):192100.
Crossref - Sandine WE. Role of Lactobacillus in the intestinal tract. J Food Prot. 1979;42(3):259-262.
Crossref - Cuesta-Mate A, Renelies-Hamilton J, Kryger P, Jensen AB, Sinotte VM, Poulsen M. Resistance and vulnerability of honeybee (Apis mellifera) gut bacteria to commonly used pesticides. Front Microbiol. 2021;12:717990.
Crossref - Tajabadi N, Mardan M, Saari N, Mustafa S, Bahreini R, Manap MYA. Identification of Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus fermentum from the honey stomach of honeybee. Braz J Microbiol. 2013b;44(3):717-722.
Crossref - Gilliam M. Microbiology of pollen and bee bread: the genus Bacillus. Apidologie. 1979;10(3):269-274.
Crossref - Ahmad W, Khaliq S, Akhtar N, et al. Whole genome sequence analysis of a novel Apilactobacillus species from giant honeybee (Apis dorsata) gut reveals occurrence of genetic elements coding prebiotic and probiotic traits. Microorganisms. 2022;10(5):904.
Crossref - Andrade-Velasquez A, Harnandez Sanchez H, Dorantes-Alvarez L, et al. Honey characterization and identification of fructophilic lactic acid bacteria of fresh samples from Melipona beecheii, Scaptotrigona pectoralis, Plebeia llorentei, and Plebeia jatiformis hives. Front Sustain Food Syst. 2023;7.
Crossref - Bal M, Onlu H, Osmanagaoglu O. Isolation and In vitro Probiotic Characterization of Fructophilic Lactic Acid Bacteria from Different Plants and The Digestive System of Bees. J Inst Sci Tech. 2024;14(3):1013-1030.
Crossref - Bisson LF, Walker G, Ramakrishnan V, et al. The two faces of Lactobacillus kunkeei: Wine spoilage agent and bee probiotic. Catalyst. 2017;1(1):1-11.
Crossref - Lanh PT, Duong BTT, Thu HT, et al. The gut microbiota at different developmental stages of Apis cerana reveals potential probiotic bacteria for improving honeybee health. Microorganisms. 2022;10(10):1938.
Crossref - Crovadore J, Chablais R, Raffini F, et al. Draft genome sequences of 3 strains of Apilactobacillus kunkeei isolated from the bee gut microbial community. Microbiol Resour Announc. 2021;10(13):e00088.
Crossref - Olofsson TC, Alsterfjord M, Nilson B, Butler E, Vasquez A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov., and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int J Syst Evol Microbiol. 2014;64(Pt 9):3109-3119.
Crossref - Dyrhage K, Garcia-Montaner A, Tamarit D, et al. Genome evolution of a symbiont population for pathogen defense in honeybees. Genome Biol Evol. 2022;14(11):evac153.
Crossref - Sakandar HA, Kubow S, Sadiq FA. Isolation and in vitro probiotic characterization of fructophilic lactic acid bacteria from Chinese fruits and flowers. LWT. 2019;104:70-75.
Crossref - Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J Clin Microbiol. 2007;45(9):2761-2764.
Crossref - Endo A, Salminen S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst Appl Microbiol. 2013;36(6):444-448.
Crossref - Maeno S, Nishimura H, Tanizawa Y, Dicks L, Arita M, Endo A. Unique niche-specific adaptation of fructophilic lactic acid bacteria and proposal of three Apilactobacillus species as novel members of the group. BMC Microbiol. 2021;21(1):41.
Crossref - Palanisamy M, Vijila K. Potential protective role of probiotic strains of lactobacilli against pesticide toxicity. Madras Agric J. 2022;109(1-3):51-55.
Crossref - Hui-Ru J, Yan-Yan W, Ping-Li D, Qiang W, Ting Z. Effects of the sublethal doses of imidacloprid on the bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae). Acta Entomol Sin. 2015;58(2):139-146.
Crossref - Liu P, Niu J, Zhu Y, et al. Apilactobacillus kunkeei alleviated toxicity of acetamiprid in honeybee. Insects. 2022;13(12):1167.
Crossref - Raymann K, Motta EVS, Girard C, Riddington IM, Dinser JA, Moran NA. Imidacloprid decreases honey bee survival rates but does not affect the gut microbiome. Appl Environ Microbiol. 2018;84(13):e00545-18.
Crossref - Jaiswal A, Tripathi A, Dubey SK. Biodegradation of fipronil: Molecular characterization, degradation kinetics, and metabolites. Res Square. 2023;rs.3.rs-2885549.
Crossref - Guima SES, Piubeli F, Bonfa MRL, Pereira RM. New insights into the effect of fipronil on the soil bacterial community. Microorganisms. 2023;11(1):52.
Crossref - Li C, Ma Y, Mi Z, et al. Screening for Lactobacillus plantarum strains that possess organophosphorus pesticide-degrading activity and metabolomic analysis of phorate degradation. Front Microbiol. 2018;9:2048.
Crossref - Leska A, Nowak A, Miskiewicz K, Rosicka-Kaczmarek J. Binding and detoxification of insecticides by potentially probiotic lactic acid bacteria isolated from the honeybee (Apis mellifera L.) environment-An in vitro study. Cells. 2022;11(23):3743.
Crossref - Harishankar MK, Sasikala C, Ramya M. Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech. 2012;3(2):137-142.
Crossref - Zhang M, Ming Y, Guo H, et al. Screening of lactic acid bacteria for their capacity to bind cypermethrin in vitro and the binding characteristics and its application. Food Chem. 2021;347:129000.
Crossref - Yuan S, Yang F, Yu H, Xie Y, Guo Y, Yao W. Biodegradation of the organophosphate dimethoate by Lactobacillus plantarum during milk fermentation. Food Chem. 2021;360:130042.
Crossref - Bo LY, Zhang YH, Zhao XH. Degradation kinetics of seven organophosphorus pesticides in milk during yoghurt processing. J Serb Chem Soc. 2011;76(3):353-362.
Crossref - Kumral AY, Kumral NA, Kolcu A, Maden B, Artik B. Simulation study for the degradation of some insecticides during different black table olive processes. ACS Omega. 2020;5(26):14164-14172.
Crossref - Yuan S, Li C, Yu H, Xie Y, Guo Y, Yao W. Screening of lactic acid bacteria for degrading organophosphorus pesticides and their potential protective effects against pesticide toxicity. LWT. 2021a;147:111672.
Crossref - Zhao X, Wang J. A brief study on the degradation kinetics of seven organophosphorus pesticides in skimmed milk cultured with Lactobacillus spp. at 42 °C. Food Chem. 2012;131(1):300-304.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.