ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms exhibit a complex relationship with heavy metals. While some of these metals are essential for microbial growth and function, others can be detrimental at elevated concentrations, inhibiting or even killing the organisms. In this study, free living diazotrophic bacteria were isolated from contaminated sites. The bacteria were investigated to resist different concentrations of Zn (II); 5-60 mg/L. The results revealed that the bacterial isolate was identified as Bacillus subtilis AUMC-B492 based on 16S rRNA gene sequencing. The bacteria were diazotrophic in nutrition and can fix atmospheric nitrogen for growth and replication in nitrogen free media. B. subtilis AUMC-B492 removed 30% of Zn (II) when initial concentration was 5 mg/L within 48 h in nitrogen free media. It could tolerate up to 100 mg/L of Zn (II) within 24 h. Therefore, strain AUMC-B492 could be used as a promising tool for bioremediation of heavy metals as well as holding potential for agricultural applications.
Bacteria, Bacillus, Heavy Metal, Resistance, Zinc Ions
Heavy metals, characterized by their high density, atomic weight, or atomic number, pose a threat to most living things because even small amounts can be toxic. Unlike many pollutants, heavy metals don’t break down or disappear over time, making them a persistent problem in the environment.1,2 Heavy metal pollution negatively affects both terrestrial and aquatic ecosystems.3,4 These pollutants can come from natural sources like rocks or mineral deposits, but human activities are also major culprits. This includes practices like farming, metalworking, energy generation, mining, and improper waste disposal.4
Some metals, like manganese, copper, iron, magnesium, and even zinc, are vital for living cells at low doses.5 They act like tiny partners, helping essential enzymes function properly. However, these same metals turn into toxins at high concentrations.6 These heavy metals overload disrupt cellular functions in various ways. It can damage the cell’s machinery (organelles), confuse vital enzymes (altering enzyme specificity), and tear apart the cell’s protective barrier (disrupt cell membranes). This chaos can even mess with cell division, potentially leading to cancer or cell death (apoptosis).7-10
A budget-friendly approach to treating heavy metal-laden wastewater involves using biological processes like biosorption and bioaccumulation.11 These methods leverage the natural abilities of living organisms to capture and concentrate the metals.12 The way heavy metals interact with microbes depends on several factors, such as the specific metal, the microbial species, and the surrounding environment.13 On the flip side, how the microbial cell structure and metabolic activity can also influence bioavailability, toxicity, and solubility of the metals.13 Many microbial species may exist in close contact with heavy metals because they have multiple resistance mechanisms. These resistance mechanisms against heavy metals, including:
- Special transport systems in their membranes can take up specific metals they can tolerate,14
- Microbial cells can trap toxic metals using special binding molecules, essentially locking them away,15
- They have powerful efflux systems encoded by their genes that literally throw toxic metals back out of the cell,16
- And some microbes have enzymes that can detoxify metal ions into less toxic forms.17
The certain nitrogen-fixing bacteria and the substances they produce can help remove heavy metals from the environment. For example, a study showed that a polysaccharide released by Azotobacter (extracellular polymer) could capture copper, zinc, and iron at concentrations between 15.5 and 25 milligrams per liter.18,19 Other research has explored how live cells of Azotobacter, along with Bacillus subtilis and Pseudomonas aeruginosa, can absorb chromium.20 Even better, some nitrogen-fixing bacteria (diazotrophs) can work together with plants to remove metals from contaminated areas.21 These metal-resistant bacteria, like Bacillus subtilis and Burkholderia sp., can help plants take up more heavy metals, accelerating the clean-up process.22,23
On this base, the main objective of this study is to isolate nitrogen fixing heavy metal resistant bacteria from different locations in Assiut governorate, Egypt.
Sample collection
Three different samples were collected from different areas in Assiut governorate, Egypt:
- Agricultural soil sample from Bani Zaid EL-Kurd near a water drain.
- Sludge sample from sewage treatment station in the Arab Al-Madabegh area.
- Agricultural soil sample from Al-Assara village. The soil and water samples were kept in sterilized plastic bags and bottles at 4 °C until use.
Physicochemical characterization of collected samples
Determination of pH of soil and water sample
A soil sample was mixed with water to create a 1:10 solution. This solution was shaken and allowed to settle overnight before measuring its pH. The extract was then filtered through Whatman filter paper, and the filtrate was centrifuged at 4000 rpm for 15-30 minutes. The electrode of the pH meter was then immersed directly in the supernatant.24 Before measuring the soil pH, the pH meter must be calibrated using standard buffer solutions of known pH values (pH 4, pH 7, and pH 10).
Determination of total dissolved salts (TDS%) and electrical conductivity (EC) of the soil sample
Conductivity measurements can be used to estimate the amount of dissolved solids in water, assuming most of these solids are ions. This relationship between ions and conductivity was studied according to Smith and Doran.25
Determination of heavy metals in the soil samples
Concentrations of Fe, Cu and Zn in soil samples were determined based on the method described by Ghosh et al.26 Using an Atomic Absorption Spectrophotometer (Spectra AA 240, Agilent Technologies, USA) using air-acetylene flame under the following operating conditions: wavelength 324.7 nm for Cu and 232 nm for Ni; slit width 0.5 nm for Cu and 0.2 nm for Ni. The lamp current for each metal was adjusted at 4.0 mA. Concentrations of heavy metals were determined against the blank after calibration with standards of known concentrations. Limit of detection (LOD) of Fe, Cu and Zn was determined three times and the mean standard deviation of absorbance of 10 replicates blank samples for each metal was found 0.01 mg L-1 for heavy metals.
Isolation of diazotrophic bacteria
10 grams of each soil sample were mixed with 90 mL of sterilized dist. water in a flask and shaken for 30 min. Nitrogen free (NF) medium containing per liter 20 g of sucrose, 0.2 g K2HPO4; 0.2 g NaCl; 0.2 g MgSO4.7H2O; 0.1 g K2SO4; 5.0 g CaCO3 and 20 g agar; pH 6.8 ± 0.2 was autoclaved at 121 °C for 20 min. Soil samples were diluted (10x) and 100 µL were placed on plates containing NF agar medium. The plates were incubated 28 ± 2 °C for 24-48 h. Diazotrophic bacterial colonies were selected based on their appearance and size.27,28
Molecular identification of bacterial isolates
The bacterial isolate was cultured in nutrient broth medium and incubated at 28 °C for 48 hours before being submitted for DNA extraction. Genomic DNA from the isolate was extracted using the method described by Hesham.29 The 16S rRNA gene sequencing was amplified using two universal primers namely 27F (5`-AGAGTTTGATCCTGGCTCAG-3`) and 1492R (5`-TACGGTTACCTTGTTACGACTT-3`). The PCR reaction and program condition were performed as per Mawad et al.30 The purified PCR products (amplicons) were reconfirmed using a size nucleotide marker (100 base pairs) by electrophoresis on 1% agarose gel. Purified amplicons were sequenced in the sense and antisense directions using 27F and 1492R primers with the incorporation of di-deoxynucleotides (dd NTPs) in the reaction mixture.29 Sequences were further analyzed using Basic Local Alignment Search Tool (BLAST) from the National Center of Biotechnology Information (NCBI) website. Phylogenetic tree was constructed as per Hesham et al.31
Screening Zn resistance of the isolated diazotrophs
A ZnSO4 stock solution was prepared by dissolving 1000 mg/L in double distilled water (ddH2O). This solution was then diluted to the needed concentration for the experiments. The isolated diazotrophic bacteria was assessed for their ability to grow at various Zn concentrations. This assay was conducted by using NF medium 30. An aliquot of pre-grown bacterial colony was streaked onto NF media supplemented with 5, 15, 30, 50 and 60 mg/L of ZnSO4. The plates were incubated at 28 ± 2 °C for 3 days. Bacteria that were able to grow in the presence of zinc were selected for further testing.
Determination of Maximum Tolerable Concentrations (MTCs)
The colorimetric method was used to determine maximum tolerable concentration (MTC) of diazotrophic bacteria for Zn. Nitrogen-free broth media supplemented with 50-120 mg/L of ZnSO4 were inoculated with an overnight grown culture (10%, OD600 = 0.3) of diazotrophic isolate and incubated at 28 ± 2 °C at 120 rpm for 72 hrs. Growth was determined by measuring the absorbance at 600 nm using 5010 UV-spectrophotometer. Samples showing zero absorbance were further assessed for growth by determining the total viable count. Maximum tolerable concentration (MTC) of heavy metal was identified as the highest concentration of Zn that allowed growth after incubation period.31 All the experiments were performed in triplicate and the results indicate mean values.
Determination of diazotrophic growth and zinc removal
Growth curves in the presence of zinc ions was determined using a 2 mL exponentially growing bacterial culture (OD600 = 0.3). The bacteria were suspended in 100 mL NF broth media containing Zn (II) (5-60 mg/L) and incubated for 72 h at 28 ± 2 °C and 150 rpm in an orbital shaker. Bacterial growth was determined by measuring optical density at 600 nm using a spectrophotometer (Shimadzu UV- 2500, Japan). A control experiment was conducted without adding bacteria to determine whether the heavy metals would precipitate or disappear on their own.
Zinc ions concentration in the supernatant was determined using atomic absorption spectrometry (AAS, PerkinElmer 2380) within the 72 h of growth and the standard curve of Zn (II) was plotted according to standard methods.32 The removal efficiency of Zn (II) was calculated based on equation (1) as follows.
The removal efficiency = [ C0 – Ce / C0 ] × 100 …(1)
where C0 and Ce are the initial and equilibrium metal concentration (mg/L), respectively.
Statistical analysis
The data were analyzed via One-Way Analysis of Variance (One-way ANOVA) on Minitab. Results were expressed as standard deviation with statistical difference when p < 0.05.
Sample characterization
The physiochemical characterization of the collected samples in Table showed that they were alkaline samples with the pH ranged from 7.4 to 10.4. The texture of the collected samples was muddy. The TDS was highest (34.2 %) in sample 1, which also had the highest EC (68.4 mS/m). Sample 3 showed the highest sodium and potassium content 624 and 2.31 mg g-1, respectively. Fe (III), Cd (II), Cu (II) and Zn (II) were the highest levels in sample 1; 1, 11.9 and 117.2 mg kg-1, respectively. On the other hand, sample 2 contained of the highest level of Ni and Fe, 0.33 and 31.4 mg kg-1, respectively. It was noticed that Zn was the predominant heavy metal present in the collected samples. Based upon this finding, the bacteria were screened for Zn ion tolerance.
Table:
Physicochemical characterization of collected samples
| Soil characteristic | Contaminated mg/kg | ||
|---|---|---|---|
| Sample 1 | Sample 2 | Sample 3 | |
| pH | 9.1 | 7.4 | 10.4 |
| Texture | mud | mud | mud |
| TDS (%) | 34.2 | 33.3 | 30.1 |
| EC (mS/m) | 68.4 | 66.6 | 60.2 |
| Na (mg g-1) | 45.4 | 42.2 | 624 |
| K (mg g-1) | 0.48 | 0.82 | 2.31 |
| Cd (mg kg-1) | 1 | 0.24 | 0.109 |
| Ni (mg kg-1) | 0.25 | 0.33 | 0.026 |
| Fe (mg kg-1) | 26.6 | 31.4 | 11.3 |
| Cu (mg kg-1) | 11.9 | 8.9 | 7.2 |
| Zn (mg kg-1) | 117.2 | 77.2 | 32.2 |
TDS: total dissolved salts, EC: electron conductivity
The normal range of pH of water is between 6.0 and 8.0.33 The collected samples had a high pH when compared to normal water, indicating the alkaline nature of the effluent due to the presence of high concentrations of salts of sodium, potassium, and carbonate. The presence of a higher level of total dissolved salts in the effluent may be attributed to the presence of insoluble organic matter and unused inorganic salts.34 Fe, Ni, Cu, Zn and Cd are among the most hazardous components of the industrial effluents. The excessive use of chemicals during industrial activities results in high levels of these pollutants in effluents. This heavy metal contamination of the environment creates selective pressure, fostering the development of microorganisms resistant to these metals in the soil and water surrounding industrial areas.35
Isolation of Zn (II) resistant diazotrophic bacteria
A total of fifty diazotrophic bacterial isolates were isolated from the collected samples. These isolates were characterized by their capability to grow on nitrogen free medium and can fix atmospheric nitrogen. Among them, fifteen isolates could grow on Zn (II) ion. The isolated AUMC B-492 exhibited the highest resistance with rapid growth on Zn (II), therefore, it was selected for further study.
Molecular characterization of zinc resistance diazotroph
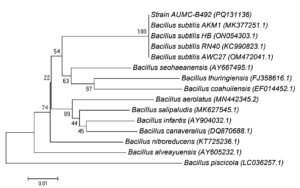
AUMC B-492 was characterized through phylogenetic analysis based on 16S rRNA gene sequence comparison. The alignment revealed a 100% similarity between the 16S rRNA sequence of AUMC B-492 and that of Bacillus subtilis. To further solidify AUMC B-492’s phylogenetic position, a phylogenetic tree was constructed using 16S rRNA sequences of various Bacillus species retrieved from the GenBank database. The tree revealed that the bacterial strain showed 100% identity coverage with several strains of Bacillus subtilis including the B. subtilis strain AKM1, HB, RN40 and AWC27 with GenBank accession no. (MK377251.1, ON054303.1, KC990823.1 and OM 472041 respectively) as demonstrated in Figure 1.
Figure 1. 16S rRNA gene-based phylogenetic analysis of Bacillus subtilis AUMC B-492. Bootstrap values and scale bar are indicated. The phylogenetic tree constructed by the neighbor-joining method showing the position of strain AUMC B-492
The 16S rRNA gene nucleotide sequence of the isolated strain AUMC B-492 has been deposited in the GenBank nucleotide sequence database under accession number PQ1311.36
Industrial activities and agricultural practices contribute to increased zinc levels in soil and water, leading to selective pressure on bacterial populations. This pressure drives the evolution of zinc resistance in bacteria.36 Bacillus species are widely recognized for their diverse metabolic capabilities, which enable them to interact with and transform a range of environmental pollutants, including heavy metals like zinc.37 Several Bacillus species, including B. subtilis, B. thuringiensis, B. sterothermophilus, B. megaterium, B. cereus, B. pumilus, B. licheniformis, and B. jeotgali have demonstrated exceptional abilities in removing heavy metals.38 Bacillus species employ two primary mechanisms for heavy metal removal: bioaccumulation, where metals are absorbed into the cell interior, and biosorption, where metals bind to the external surface of the cell.39
Impact of initial Zn (II) concentrations on the bacterial cell growth
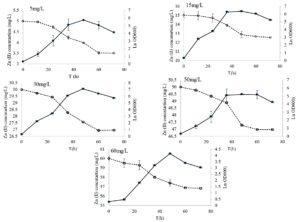
The results in Figure 2 showed that the time it took for bacteria to start growing (lag phase) depended on the concentration of zinc was initially added to the solution. When there was more than 50 mg/L of Zn (II), the bacteria took longer to start growing. As illustrated in Figure 2, at 60 mg/L of Zn (II), the lag phase was 12 hours. However, when the Zn (II) concentration was between 5 and 50 mg/L of Zn (II), the bacteria grew quickly, indicating that low levels of Zn (II) did not inhibit their growth.
Figure 2. Growth curves of B. subtilis AUMC-B492 and metal removal efficiencies under different Zn (II) concentrations. Error bars represent the standard deviation at n = 3
Growth curves of AUMC-B492 were determined in the presence of five different concentrations of Zn (II) and the removal efficiency (%) of the metals was estimated (Figure 2). After a short delay, the bacteria began to grow rapidly, even when the metals were present. The stationary phase level was significantly higher (p < 0.05) for lower concentration of Zn (II) (15 mg/L, Figure 2), while higher metal concentration (60 mg/L, Figure 2) resulted in lower final cell densities. Higher metal concentrations Zn (II) (60 mg/L) resulted in a transitory decrease before the bacteria recovered. A prolonged stationary phase (36-60 hours) was observed. On the other hand, for the remaining concentrations, the stationary phase ended after 48 h of cultivation (Figure 2).
When bacteria were exposed to heavy metals, they experienced a longer delay before starting to grow rapidly. This delay was caused by the bacteria needing extra time to adjust their internal physiological and metabolic processes and be able to survive in the presence of these metals.40 Many reported discuss the role of various Bacillus species in removal of Zn. Huang et al.40 reported that B. cereus RC-1, was able to remove Zn (II) (38.3% maximum removal efficiency). Moreover, it has been reported that B. licheniformis and Salmonella typhi were reported to remove more than 90% of Zn from contaminated samples, whereas Pseudomonas fluorescens and E. coli were effective in removing more than 96% and 93% of Zn, respectively via adsorption mechanism.41,42 On the other hand, B. subtilis and B. pumilus were also able to facilitate the accumulation Zn (32,500 mg kg-1 dry soil) and other heavy metals in tissues of Zea mays and Sorghum bicolor.43,44 In addition to He et al.45 showed that the root and shoot biomass of Orychophragmus violaceus were significantly enhanced when inoculated with B. subtilis and B. cereus due to accumulation of Zn (II) when compared with non-inoculated one.
Zn (II) removal curves
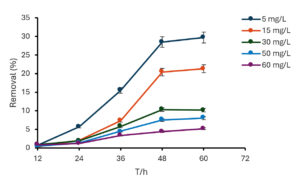
The amount of zinc removed by the bacteria depended on the initial concentration of added zinc. When there was 20 mg/L of zinc, the bacteria removed the most zinc when they were growing quickly (exponential phase) (Figure 3). However, with higher concentrations (50 and 100 mg/L), the bacteria removed the most zinc when they stopped growing (stationary phase). This suggests that the best growth phase for zinc removal depends on the initial zinc ions concentration. Additionally, at a concentration of 50 mg/L, the amount of zinc remaining in the solution slightly increased during the stationary phase. This could be because some zinc that was attached to the bacteria’s surface or inside the cells was released.
Figure 3. The removal percentage of Zn (II) via B. subtilis AUMC-B492 at pH = 7.3, along 60 hours of incubation at 30 °C
The bacteria were most effective at removing zinc when there was a low concentration of zinc in the solution. The maximum removal was 30.2% for zinc at an initial concentration of 5 mg/L. When there was more zinc, the bacteria were less effective (Figure 3). This could be because there were fewer places on the bacteria for the zinc to attach, or because the bacteria were not as active.41 Moreover, the capability of bacteria to remove Zn (II) in nitrogen free medium suggested that, this bacterium could be applied for bioremediation of heavy metal and for agricultural purposes.
Viable B. subtilis D215 strain exhibited superior zinc removal, achieving a 63.73% bioremoval rate under optimal conditions of 30 °C and pH 7 after 48 hours of incubation. In contrast, dead B. subtilis cells demonstrated a lower zinc removal efficiency, with only 26.83% bio-removal.42 A B. thuringiensis strain, ISI, demonstrated a promising ability to remove zinc from agricultural wastewater, achieving a 54% reduction initially. However, this efficiency declined to 31% after four days.43 These findings suggest that strain AUMC-B492 may not be optimal for a robust, long-term bioremediation system, it still holds potential for agricultural applications.
Determination of maximum tolerable concentration of zinc (II)
The ability of microbes to survive in the presence of heavy metals was measured by the MTC. It is the maximum concentration that allows growth after 24 hours.31,44 The AUMC-B492 isolate showed a high degree of resistance to Zn (II) reached to 100 mg/L.45 reported that the minimum inhibitory concentration of (MIC) for Zn (II) was determined to be 20 mM by Bacillus altitudinis MT422188. Liquid-based toxicity testing offers an efficient way to evaluate metal toxicity in polluted environments like industrial discharges and leachate from sewage sludge.46 Liquid-based toxicity testing is different from solid-based toxicity, where the conditions of diffusion, complexation, and availability of metals are different from those in solid medium.46
The ability to grow in nitrogen-free medium indicates that AUMC-B492 can convert atmospheric nitrogen (N₂) into usable forms like ammonia (NH₃). This is crucial for its own growth and can also benefit surrounding ecosystems. This characteristic makes the bacterium potentially valuable for agricultural applications via reducing the need for synthetic nitrogen fertilizers. It also enhances its potential for bioremediation in environments where nitrogen availability may be limited. This strain also exhibited high resistance to Zn (II). Its removal from aqueous solutions reached its maximum after approximately 60 hours of incubation. Removal efficiencies varied based on the initial Zn (II) concentration, with a maximum removal of 30% observed at an initial concentration of 5 mg/L. A maximum tolerable concentration of 100 mg/L indicates the upper limit of zinc concentrations that AUMC-B492 can survive in. Therefore, the combined nitrogen-fixing and zinc-removing abilities make AUMC-B492 a promising candidate for bioremediation, particularly in environments contaminated with both heavy metals and nitrogen deficiency.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Mitra S, Chakraborty AJ, Tareq AM, et al. Impact of heavy metals on the environment and human health: Novel therapeutic insights to counter the toxicity. J King Saud Univ Sci. 2022;34(3):101865.
Crossref - Islam MS, Kormoker T, Khan R, et al. Strategies for heavy metals remediation from contaminated soils and future perspectives. In: Shit, P.K Adhikari, PP Bhunia, GS Sengupta, Springer, Chem. Soil Health and Environmental Sustainability: Application of Geospatial Technology. 2022:615-644.
Crossref - Ali MM, Hossain D, Al-Imran, Khan MS, Begum M, Osman MH. Environmental pollution with heavy metals: A public health concern. Heavy metals-their environmental impacts and mitigation 2021;771-783.
Crossref - Talukder P, Ray R, Sarkar M, Das A, Chakraborty S. Adverse effects of mining pollutants on terrestrial and aquatic environment and its remediation. Environmental Quality Management. 2024;33(4):595-610.
Crossref - Jomova K, Makova M, Alomar SY, et al. Essential metals in health and disease. Chem Biol Interact. 2022;367:110173.
Crossref - Khalef RN, Hassan AI, Saleh HM. Heavy metal’s environmental impact. Environmental Impact and Remediation of Heavy Metals. IntechOpen. 2022.
Crossref - Baj J, Flieger W, Barbachowska A, et al. Consequences of disturbing manganese homeostasis. Int J Mol Sci. 2023;24(19):14959.
Crossref - Kiran, Bharti R, Sharma R. Effect of heavy metals: An overview. Materials Today: Proceedings. 2022;51(Part 1)880-885.
Crossref - Sun Q, Li Y, Shi L, et al. Heavy metals induced mitochondrial dysfunction in animals: Molecular mechanism of toxicity. Toxicology. 2022;469:153136.
Crossref - Fu Z, Xi S. The effects of heavy metals on human metabolism. Toxicol Mech Methods. 2020;30(3):167-176.
Crossref - Kanamarlapudi SLRK, Chintalpudi VK, Muddada S. Application of biosorption for removal of heavy metals from wastewater. Biosorption. 2018;18(69):70-116.
Crossref - Abd Elnabi MK, Elkaliny NE, Elyazied MM, et al. Toxicity of heavy metals and recent advances in their removal: a review. Toxics. 2023;11(7):580.
Crossref - Essa AMM, Al Abboud MA, Khatib SI. Metal transformation as a strategy for bacterial detoxification of heavy metals. J Basic Microbiol. 2018;58(1):17-29.
Crossref - Porcheron G, Garenaux A, Proulx J, Sabri M, Dozois CM. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol. 2013;3:90.
Crossref - Pal A, Bhattacharjee S, Saha J, Sarkar M, Mandal P. Bacterial survival strategies and responses under heavy metal stress: A comprehensive overview. Crit Rev Microbiol. 2021;48(3):327-355.
Crossref - Thakur S, Chandra A, Kumar V, Bharti S. Environmental Pollutants: Endocrine Disruptors/Pesticides/Reactive Dyes and Inorganic Toxic Compounds Metals, Radionuclides, and Metalloids and Their Impact on the Ecosystem. In Biotechnology for Environmental Sustainability (pp. 55-100). Singapore: Springer Nature Singapore 2025.
- Alotaibi BS, Khan M, Shamim S. Unraveling the underlying heavy metal detoxification mechanisms of Bacillus species. Microorganisms. 2021;9(8):1628.
Crossref - Kapoore RV, Wood EE, Llewellyn CA. Algae biostimulants: A critical look at microalgal biostimulants for sustainable agricultural practices. Biotechnol Adv. 2021;49:107754.
Crossref - Rasulov BA, Yili A, Aisa HA. Biosorption of metal ions by exopolysaccharide produced by Azotobacter chroococcum XU1. Indian J Environ Prot. 2013;4(09):989-993.
Crossref - Kurniawan SB, Imron MF, Purwanti IF. Biosorption of chromium by living cells of Azotobacter s8, Bacillus subtilis and Pseudomonas aeruginosa using batch system reactor. J Ecol Eng. 2019;20(6):184-189.
Crossref - Lawal I. Nitrogen fixing bacteria and their application for heavy metal removal: a mini review. J Biochem Microbiol Biotechnol. 2021;9(2):43-47.
Crossref - Wang Y, Narayanan M, Shi X, et al. Plant growth-promoting bacteria in metal-contaminated soil: Current perspectives on remediation mechanisms. Front Microbiol. 2022;13:966226.
Crossref - Saleem S, Mushtaq NU, Shah WH, Rasool A, Rehman RU. Microbial and plant-assisted bioremediation of heavy metal polluted environments. Heavy Metal Toxicity in Plants. 2021:139-156.
Crossref - Thomas GW. Soil pH and soil acidity. Methods of Soil Analysis: Part 3 Chemical Methods. 1996;5:475-490.
Crossref - Smith JL, Doran JW. Measurement and use of pH and electrical conductivity for soil quality analysis. Methods for Assessing Soil Quality. 1997;49:169-185.
Crossref - Ghosh A, Ali S, Mukherjee SK, Saha S, Kavira A. Bioremediation of copper and nickel from freshwater fish Cyprinus carpio using rhiozoplane bacteria isolated from Pistia stratiotes. Environ Process. 2020;7:443-461.
Crossref - Dobereiner J. Isolation and identification of root associated diazotrophs. In: Skinner, F.A., Boddey, R.M., Fendrik, I. (eds) Nitrogen Fixation with Non-Legumes. Developments in Plant and Soil Sciences, 1989;35. Springer, Dordrecht.
Crossref - Kifle MH, Laing MD. Isolation and Screening of Bacteria for Their Diazotrophic Potential and Their Influence on Growth Promotion of Maize Seedlings in Greenhouses. Front Plant Sci. 2016;6:1225.
Crossref - Hesham AEl-L. New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications. J Pure Appl Microbiol. 2014;8(1):383-388.
- Mawad AMM, Hesham AE-L, Mostafa YM, Shoriet A. Pyrene degrading Achromobacter denitrificans ASU-035: growth rate, enzymes activity, and cell surface properties. Rend Fis Acc Lincei. 2016;27:557-563.
Crossref - Hesham AE-L, Mostafa YS, AlSharqi LEO. Optimization of citric acid production by immobilized cells of novel yeast isolates. Mycobiology. 2020;48(2):122-132.
Crossref - Raffi MM, Charyulu PBBN. Nitrogen fixation by the Native Azospirillum spp. Isolated from Rhizosphere and Non-Rhizosphere of foxtail Millet. Asian J Biol Life Sci. 2012;1(3):213-218.
- Schmidt T, Schlegel HG. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176(22):7045-54.
Crossref - Vellaichamy S, Palanivelu K. Preconcentration and separation of copper, nickel and zinc in aqueous samples by flame atomic absorption spectrometry after column solid-phase extraction onto MWCNTs impregnated with D2EHPA-TOPO mixture. Journal of Hazardous Materials. 2011;185(2-3):1131-1139.
Crossref - Rahman A, Jahanara I, Jolly YN. Assessment of physicochemical properties of water and their seasonal variation in an urban river in Bangladesh. Water Science and Engineering. 2021;14(2):139-148.
Crossref - Moghannem SA, Refaat BM, El-Sherbiny GM, et al. Characterization of heavy metal and antibiotic-resistant bacteria isolated from polluted localities in Egypt. Egypt Pharm J. 2015;14(3):158-165.
Crossref - Caracciolo AB, Terenzi V. Rhizosphere microbial communities and heavy metals. Microorganisms. 2021;9(7):1462.
Crossref - Ghosh S, Bhattacharya J, Nitnavare R, Webster TJ. Heavy metal removal by Bacillus for sustainable agriculture. In: Islam, M.T., Rahman, M., Pandey, P. (eds) Bacilli in Agrobiotechnology. Bacilli in Climate Resilient Agriculture and Bioprospecting. Springer, Cham. 2022:1-30.
Crossref - Velasquez L, Dussan J. Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J Hazard Mater. 2009;167(1-3):713-716.
Crossref - Reilly M, Dinsdale R, Guwy A. Mesophilic biohydrogen production from calcium hydroxide treated wheat straw. Int J Hydrogen Energy. 2014;39(30):16891-16901.
Crossref - Huang F, Li K, Wu R-R, Yan YJ, Xiao RB. Insight into the Cd2+ biosorption by viable Bacillus cereus RC-1 immobilized on different biochars: roles of bacterial cell and biochar matrix. J Clean Prod. 2020;272:122743.
Crossref - Sabae SZ, Hazaa M, Hallim SA, Awny NM, Daboor SM. Bioremediation of Zn, Cu and Fe using Bacillus subtilis d215 and Pseudomonas putida biovar ad 225. Biosci Res. 2006;3(1):189-204
- Kumar V, Singh S, Kashyap N, et al. Bioremediation of heavy metals by employing resistant microbial isolates from agricultural soil irrigated with industrial waste water. Orient J Chem. 2015;31(1):357-361.
Crossref - Maity S, Sarkar D, Poddar K, Patil P, Sarkar A. Biofilm-mediated heavy metal removal from aqueous system by multi-metal-resistant bacterial strain Bacillus sp. GH-s29. Appl Biochem Biotechnol. 2023;195(8):4832-4850.
Crossref - Khan M, Ijaz M, Chotana GA, Malik A, Shamim S. Bacillus altitudinis MT422188: A potential agent for zinc bioremediation. Bioremediat J. 2022;26(3):228-248.
Crossref - Molaey R, Yesil H, Calli B, Tugtas AE. Enhanced heavy metal leaching from sewage sludge through anaerobic fermentation and air-assisted ultrasonication. Chemosphere. 2021;279;130548.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.