ISSN: 0973-7510
E-ISSN: 2581-690X
Clinical presentation of bloodstream infection ranges from transitory bacteremia to fulminant septic shock with substantial mortality. Regular monitoring of bacteremia etiology helps rationalize therapy, revealing the spectrum of bacterial infections and their sensitivity pattern in a particular area. In this study, we investigated the prevalence and antimicrobial susceptibility profile of all bacterial pathogens isolated from blood cultures in a tertiary care hospital in Riyadh, Saudi Arabia. It was a retrospective study and data was collected over a period of one year from January 2022 to December 2022, with a total of 3,429 blood cultures requested. Culture positivity was found in 5% of suspected bacteremia cases, with a slight male predominance. Gram-positive bacteremia was 57%, mainly isolated from male patients. Methicillin-resistant Staphylococcus aureus (MRSA) was found in 38% of total Staphylococcus aureus (S. aureus) bacteremia cases, with all Gram-positive isolates susceptible to vancomycin and linezolid. Escherichia coli (E. coli) was the most common Gram-negative bacteria, while Pseudomonas and Stenotrophomonas species were the main non-fermenting pathogens, accounting for 66.7% of cases. Extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae accounts for 17.5% in total with the highest production in Proteus species, whereas 75% of Pseudomonas species were carbapenamase producers. Consequently, the prevalence of multidrug-resistance microorganisms in critically ill individuals may account as a significant threat in hospital settings. Continuous monitoring is essential for comprehending the prevalence of bacteremia and their susceptibility pattern to create successful empirical therapy options and antimicrobial stewardship.
Antimicrobial Susceptibility, Bacterial Pathogens, Bacteremia, Blood Culture, Methicillin-resistant Staphylococcus aureus
Bloodstream infection is a serious medical condition that needs to be treated without any delay because of the high mortality rate of 15% to 30%.1-3 Bloodstream infections are indeed severe medical conditions that can have devastating consequences. The most common causative agents are bacteria, with frequent isolates including Staphylococcus aureus, Escherichia coli, Klebsiella species, streptococci, and enterococci.4 Over time, there has been a notable shift from Gram-negative to Gram-positive organisms as predominant causes of BSIs.5 While bacteria are the primary pathogens, fungi also play a significant role, particularly Candida species, which rank among the top causes of BSIs in hospitalized patients. Other fungal pathogens include Cryptococcus, Trichosporon, and various molds.6 Viral infections can also cause sepsis, especially in immunocompromised patients. The spectrum of causative organisms has evolved, particularly in healthcare settings, with the emergence of multidrug-resistant organisms and new pathogens becoming increasingly prevalent.7,8
Bacteremia refers to the presence of viable bacteria in the bloodstream, while sepsis is a life-threatening condition characterized by a dysregulated host response to infection. Sepsis can progress to septic shock, which involves profound circulatory and metabolic abnormalities. Key symptoms include fever or hypothermia, changes in mental status, tachycardia, tachypnea, and hypotension.9 Bacterial bloodstream invasion can trigger severe systemic complications through a dysregulated immune response. When bacteria enter the bloodstream, they can cause sepsis, characterized by organ dysfunction and circulatory abnormalities. The key life threatening complications include shock, marked by profound hypotension resistant to fluid resuscitation, multiple organ failure due to reduced oxygen supply and inflammation-induced vascular dysfunction, and disseminated intravascular coagulation caused by disrupted coagulation cascades. These complications result from the body’s overwhelming inflammatory response, leading to impaired tissue perfusion, mitochondrial dysfunction, endothelial damage and microvascular thrombosis. Early recognition and prompt antimicrobial therapy are crucial, as mortality increases by 7% for each hour of delayed treatment.9
The rate of sepsis and resulting mortality is highly variable among different age groups of the patients and their geographical distribution.10 Whenever the clinicians suspect bloodstream infection based on the clinical presentation of the patient, without any further delay, they usually start empiric therapy with broad-spectrum antibiotics to cover both gram-positive and -negative organisms while waiting for the blood culture results from the lab. After the availability of the results, the empiric treatment therapy is modified according to the susceptibility of culture-positive pathogens.11 Prompt results of positive blood cultures foster diagnostic stewardship and perspective antibiotic use in clinical settings. Knowing the antimicrobial susceptibility profile of the isolated blood pathogen is critical for patient outcomes and shifting from empirical therapy to appropriate definitive therapeutic antibiotics.12
Factors increasing morbidity and mortality in bloodstream infections include patient-related factors such as age (>65 years), comorbidities like diabetes and immunosuppression; healthcare-associated factors like prolonged hospitalizations, central venous catheters, and prior broad-spectrum antibiotic use; pathogen-related factors like multidrug-resistant organisms and Gram-negative infections; and treatment-related factors such as delayed appropriate antimicrobial therapy and complications from immunosuppressive treatments.13-15
Bacterial resistance dissemination is a significant public health concern that might potentially contribute to bloodstream infections. Indeed, there has been a rise in the occurrence of bloodstream infections that are resistant to commonly used antimicrobial drugs in recent years. Additionally, the inappropriate use of medications has also had a role in the development of antibiotic resistance.16 One hospital-based study conducted in Saudi Arabia in 2010 documented the excessive use of antimicrobials in ICUs. Additionally, counter-sales of local drugs were prevalent in Saudi Arabia; thus, these factors were identified as significant contributors to the emergence and dissemination of antimicrobial resistance.17
To avoid the recurrence and complete recovery of bloodstream infections, it is necessary to administer antimicrobial treatment for an appropriate duration to eliminate bacterial pathogens from the blood. However, it is important to note that persistent use of antimicrobial therapy can lead to the emergence of antimicrobial resistance. As a result, the World Health Organization (WHO) issued a global action plan on antimicrobial resistance in 2015, and Saudi Arabia became a participant. To monitor antimicrobial resistance, WHO presented the Global Antimicrobial Resistance Surveillance System (GLASS) which includes surveillance for six priority blood pathogens including S. aureus, Streptococcus pneumoniae (among Gram-positive bacteria), E. coli, Klebsiella pneumoniae, Acinetobacter baumannii and Salmonella species (among Gram-negative bacteria).18 The global burden of bloodstream infection is recorded as 49 million people every year get this infection.19 According to a recent study, the burden of bloodstream infection is up to 11% reported by King Fahad Medical City, Riyadh, Saudi Arabia.20
Aims and Objectives
The main objectives of conducting this study were to ascertain the proportion of positive blood cultures that yielded bacterial growth and the overall count of cultures performed and to identify the antibiotic resistance patterns of WHO-priority pathogens along with other pathogens. Accurate and comprehensive hospital data on antibiotic resistance are crucial for monitoring the changing patterns of bacteria and their resistance. This information may then be used to improve infection prevention and control measures, as well as stewardship programs locally.
Study Design and Settings: This was a retrospective study including all the suspected bacteremia cases among febrile patients attending our tertiary care hospital (450 beds) and blood cultures were requested for them. Blood culture specimen collection and inoculation into the BacT/ALERT® 3D system was done by using standard protocols according to the manufacturer’s guidelines and SOPs of the hospital. All the patient details were taken and entered into the electronic system of the hospital.
Samples were processed for microbial identification and antimicrobial susceptibility testing in the Department of Microbiology, Medical Diagnostic Lab (MD Lab), Riyadh, KSA. Patient samples from a tertiary care hospital were sent to the MD lab after they appeared to flag positive in an automated blood culture machine (BacT/ALERT 3D system).
Data collection
Over a period of one year from January 2022 to December 2022, the data was collected and analyzed.
Ethical consideration
The study was approved by the Research Committee of Dr. Sulaiman Al Habib Medical Group Research Center, Riyadh, Kingdom of Saudi Arabia (IRB study number RC23.11.18).
Inclusion criteria
All non-duplicate samples requested for blood culture and sensitivity from all age groups of the patients visiting this hospital either registered Inpatient or Outdoor patients were included in this study.
Exclusion criteria
All repeat, duplicate, and follow-up cultures (within 7 days of the first culture request) were excluded from this study.
Statistical analysis
Data was statistically analyzed using Microsoft Excel and IBM SPSS version 20. All p-values less than 0.05 were considered statistically significant.
Study protocol
Blood samples require careful handling to ensure both safety and specimen integrity. Blood cultures were collected using a strict aseptic technique, with thorough skin preparation using 70% alcohol followed by 2% chlorhexidine antiseptic, into appropriate culture bottles (aerobic and anaerobic for adults, pediatric bottles for children). Appropriate volume of the samples were collected by trained nurses and phlebotomists according to the standard protocols of blood culture sample collection. Samples were immediately transported to the microbiology lab at room temperature where these blood culture bottles were incubated in automated BacT/ALERT 3D system without any delay.
Following the detection of positive results in the bottles, a Gram stain was done on the broth to preliminarily identify the type of microorganisms. All work-up with positive blood culture bottles was done in Class II biological safety cabinet. After processing, the positive blood culture bottles were kept in the incubator, for another 7 days in case if had any discrepancy. All blood culture samples and associated materials (e.g., culture bottles and gloves) were disposed in biohazard waste containers that are puncture-resistant and leak-proof. These properly labeled biohazard bag were then taken by Saudi Environmental and Protection Company (SEPCO) for further discarding. The sub-cultures were made by inoculating blood agar, chocolate agar, and MacConkey agar plates, which were then placed in an incubator set at a temperature of 37 °C for a duration of 18 to 24 hours. The mature bacterial colonies were distinguished based on their colony morphology, and Gram stain. If there was no growth observed within a period of 7 days, the culture was reported as sterile (negative for bacterial growth). All isolates were subjected to species identification and antimicrobial susceptibility testing by the Vitek 2 system. Antimicrobial susceptibility testing was also done by the Standard Kirby-Bauer disc diffusion method according to CLSI guidelines.
The choice of antibiotics arises from commonly prescribed drugs from the physicians locally and CLSI was used as a reference guide to select the antibiotics to be tested. Reference strains of S. aureus (ATCC 25923) for disc diffusion, S. aureus (ATCC 29213) for Vitek 2, E. coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853) were used as quality control of the antimicrobial susceptibility testing for disc diffusion and Vitek 2 system. Results were interpreted according to the breakpoints and zone of inhibition given by CLSI guidelines.
A total of 3,429 blood cultures were requested during the study period. The highest number of blood cultures were requested from adults (age group above 12), followed by neonates (0-1 month). Table 1 displays the occurrence of bacteremia based on the age groups of the patients, providing details on the number of suspected bacteremia cases, and the number and percentage of positive blood cultures in the respective age groups. Overall culture positivity was seen in 5% of all the suspected bacteremia cases in all age groups. In our study, the occurrence of bacteremia was highest in the age group of 1 month to 1 year (8.5%). A rate of 7.2%, 2.8%, and 2.0% in the age groups over 12 years, 0-1 month, and 1-12 years, respectively, followed. This association of positive bacteremia cases with the age of the patients was statistically significant with a p-value of 0.002.
Table (1):
Incidence of Bacteremia according to age group of the patients
| Age group | Number of case investigated | Bacteremia | |
|---|---|---|---|
| Number | Percentage | ||
| 0-1 month | 778 | 22 | 2.8% |
| 1 month-1 year | 307 | 26 | 8.5% |
| 1-12 years | 847 | 17 | 2.0% |
| Above 12 years | 1497 | 108 | 7.2% |
| Total | 3429 | 173 | 5% |
Table 2 presents the gender distribution of suspected bacteremia cases, showing the total number of cases examined and the number of positive blood cultures for males and females. The data reveals a slightly greater percentage of culture-positive bacteremia cases in males (5.1%) compared to females (4.9%), however, it was not statistically significant as the p-value was 0.70.
Table (2):
Gender distribution among Bacteremia cases
Gender |
Total number of cases investigated |
Positive blood cultures |
|---|---|---|
Male |
1884 |
96 (5.1%) |
Female |
1545 |
77 (4.9%) |
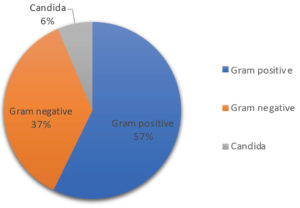
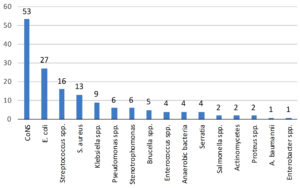
Moreover, Figure 1 illustrates the distribution of blood culture isolates, indicating that 57% of the isolates were Gram-positive bacteria and 37% were Gram-negative bacteria. Candida species were identified in the remaining 6% of the febrile patients. Figure 2 demonstrates the distribution of bacterial isolates obtained from positive blood cultures, with a total of 155 samples. CoNS had the highest number of cases, with a total of 53. This was followed by E. coli, Streptococcus species, S. aureus, and Klebsiella species, with 27, 16, 13, and 9 cases, respectively. The number of cases for Pseudomonas and Stenotrophomonas was 6 for each, followed by Brucella species with a count of 5.
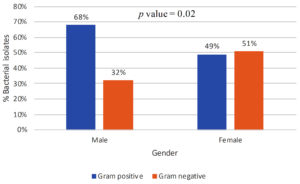
Figure 3. Percent isolation of gram-positive and gram-negative bacterial isolates in relation to the gender of the patients
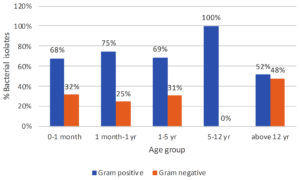
Figure 4. Percent isolation of gram-positive and gram-negative bacterial isolates in relation to the age of the patients
Figure 3 depicts the prevalence of bacteremia about gender and Gram stain characteristics. In the male population, 68% of bacteremia cases were attributed to Gram-positive bacteria, while 32% were attributed to Gram-negative bacteria. In contrast, among females, the distribution of bacteremia was 51% Gram-negative and 49% Gram-positive. The p-value was found to be 0.02, which shows this finding to be statistically significant. Figure 4 shows the prevalence of bacteremia in different age groups by Gram stain distribution. In the age group of 0-1 month, the data shows that 68% of bacteremia cases were attributed to Gram-positive bacteria, while 32% were attributed to Gram-negative bacteria. Moving to the age group of 1 month to 1 year, the prevalence of Gram-positive bacteremia increased to 75%, with Gram-negative bacteremia accounting for 25% of cases. For the age group of 1-5 years, the distribution of bacteremia was 69% Gram-positive and 31% Gram-negative. Notably, in the 5-12-year age group, all cases of bacteremia were attributed to Gram-positive bacteria, with no reported cases of Gram-negative bacteremia. Finally, in the age group above 12 years, the prevalence of Gram-positive bacteremia decreased to 52%, while the prevalence of Gram-negative bacteremia increased to 48%. These findings showed that Gram-positive bacterial isolates were more abundant in younger age groups as compared to adults, however, the results were not statistically significant (p-value = 0.07).
The antimicrobial susceptibility of Gram-positive bacterial isolates is shown in Table 3. Our data shows that S. aureus exhibits varying levels of resistance to different antibiotics. Penicillin was found 100% resistant, while oxacillin, ciprofloxacin, levofloxacin, clindamycin, and erythromycin exhibited resistance ranging from 23.1% to 46.2%. Least resistance was noted in gentamicin and co-trimoxazole (7.7%), whereas no resistance was seen against tetracycline, linezolid, and vancomycin in S. aureus isolates. Among these cases, MRSA accounted for 38% (n = 5), and methicillin-sensitive Staphylococcus aureus (MSSA) accounted for 62% (n = 8). In CoNS, the highest resistance was noted against penicillin similar to S. aureus, i.e. 97.8%, followed by oxacillin (67.4%). Additionally, Methicillin-resistant Staphylococcus epidermidis (MRSE) constituted 75% (n = 18) and Methicillin-sensitive Staphylococcus epidermidis (MSSE) constituted 25% (n = 6), totaling 45% (n = 24) of CoNS isolates. In Enterococcus and Streptococcus species, the percent resistance against erythromycin was reported to be highest (100% and 66.7%, respectively). However, no resistance was found against vancomycin and linezolid among any of the Gram-positive isolates in our hospital settings.
Table (3):
Antimicrobial resistance of gram-positive bacterial isolates (no. (%))
Antibiotics |
S. aureus |
CoNS |
Enterococcus spp. |
Streptococcus spp. |
|---|---|---|---|---|
Penicillin |
13 (100%) |
45 (97.8%) |
1 (25%) |
7 (46.7%) |
Oxacillin |
5 (38.5%) |
31 (67.4%) |
NA |
NA |
Ciprofloxacin |
3 (23.1%) |
14 (30.4%) |
1 (25%) |
NA |
Levofloxacin |
4 (30.8%) |
14 (30.4%) |
1 (25%) |
1 (6.7%) |
Erythromycin |
6 (46.2%) |
28 (60.9%) |
4 (100%) |
10 (66.7%) |
Clindamycin |
3 (23.1%) |
14 (30.4%) |
NA |
3 (20%) |
Co-trimoxazole |
1 (7.7%) |
7 (15.2%) |
1 (25%) |
2 (13.3%) |
Gentamicin |
1 (7.7%) |
13 (28.3%) |
NA |
NA |
Tetracycline |
0 |
12 (26.1%) |
1 (25%) |
NA |
Vancomycin |
0 |
0 |
0 |
0 |
Linezolid |
0 |
0 |
0 |
0 |
Ampicillin |
NA |
NA |
1 (25%) |
7 (46.7%) |
Ceftriaxone |
NA |
NA |
NA |
3 (20%) |
Cefotaxime |
NA |
NA |
NA |
3 (20%) |
CoNS: Coagulase-negative Staphylococci, NA: Not applicable, S. aureus: Staphylococcus aureus
Table 4 displays the percentage of resistance exhibited by Gram-negative bacterial isolates. The remarkable resistance of E. coli to ampicillin, which stands at 55.6%, is worth mentioning. The susceptibility of E. coli to cephalosporin antibiotics varies between 14.8% and 51.9%. Within the group of quinolones, ciprofloxacin exhibited the highest level of resistance at 22.2%, while levofloxacin and ofloxacin had equal rates of resistance at 18.5% each. Resistance to co-trimoxazole was observed in 22.2% of the isolated E. coli strains. The lowest levels of resistance were observed in gentamicin (3.7%) and piperaillin/tazobactam (7.4%). Every strain of E. coli exhibited complete susceptibility to carbapenems and amikacin. In our hospital setting, among bacteremia cases, the occurrence of E. coli that produces ESBL is 22%.
Table (4):
Antimicrobial resistance of gram-negative bacterial isolates (no. (%))
Antibiotics |
EC |
KLEB |
PSEU |
AB |
ENTB |
PRO |
STEN |
SER |
SAL |
|---|---|---|---|---|---|---|---|---|---|
Ampicillin |
15 (55.6%) |
IR |
IR |
IR |
NA |
1 (50%) |
NA |
NA |
2 (100%) |
Ampicillin Sulbactam |
NA |
NA |
IR |
0 |
NA |
NA |
NA |
NA |
NA |
Amoxicillin/ Clavulanic acid |
11 (40.7%) |
4 (44.4%) |
IR |
IR |
1 (100%) |
1 (50%) |
NA |
4 (100%) |
NA |
Cefalotin |
14 (51.9%) |
4 (44.4%) |
NA |
NA |
1 (100%) |
1 (50%) |
NA |
4 (100%) |
NA |
Cefuroxime |
9 (33.3%) |
4 (44.4%) |
NA |
NA |
1 (100%) |
1 (50%) |
NA |
4 (100%) |
NA |
Cefixime |
9 (33.3%) |
4 (44.4%) |
NA |
NA |
1 (100%) |
1 (50%) |
NA |
3 (75%) |
NA |
Cefpodoxime |
9 (33.3%) |
4 (44.4%) |
NA |
NA |
1 (100%) |
1 (50%) |
NA |
3 (75%) |
1 (50%) |
Ceftriaxone |
6 (22.2%) |
4 (44.4%) |
IR |
0 |
0 |
1 (50%) |
NA |
0 |
1 (50%) |
Ceftazidime |
NA |
NA |
0 |
1 (100%) |
NA |
NA |
1 (16.7%) |
NA |
NA |
Cefepime |
4 (14.8%) |
0 |
0 |
1 (100%) |
0 |
1 (50%) |
NA |
0 |
NA |
Imipenem |
0 |
1 (11.1%) |
3 (75%) |
0 |
0 |
1 (50%) |
3 (50%) |
0 |
NA |
Meropenem |
0 |
0 |
3 (75%) |
0 |
0 |
0 |
3 (50%) |
0 |
NA |
Piperacillin/ Tazobactam |
2 (7.4%) |
0 |
3 (75%) |
0 |
0 |
1 (50%) |
NA |
NA |
NA |
Co-trimoxazole |
6 (22.2%) |
0 |
IR |
1 (100%) |
0 |
1 (50%) |
0 |
0 |
1 (50%) |
Gentamicin |
1 (3.7%) |
0 |
0 |
0 |
0 |
0 |
NA |
0 |
NA |
Amikacin |
0 |
0 |
0 |
NA |
0 |
0 |
NA |
0 |
NA |
Ciprofloxacin |
6 (22.2%) |
1 (11.1%) |
3 (75%) |
0 |
0 |
1 (50%) |
NA |
0 |
0 |
Ofloxacin |
5 (18.5%) |
1 (11.1%) |
3 (75%) |
0 |
0 |
1 (50%) |
NA |
0 |
0 |
Levofloxacin |
5 (18.5%) |
1 (11.1%) |
3 (75%) |
0 |
0 |
1 (50%) |
0 |
0 |
0 |
Tigycycline |
NA |
NA |
NA |
0 |
NA |
NA |
NA |
NA |
NA |
Colistin |
NA |
NA |
NA |
0 |
NA |
NA |
NA |
NA |
NA |
Aztreonam |
NA |
NA |
0 |
NA |
NA |
NA |
NA |
NA |
NA |
Minocycline |
NA |
NA |
NA |
NA |
NA |
NA |
4 (66.7%) |
NA |
NA |
Azithromycin |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1 (50%) |
AB: Acinetobacter baumannii, EC: Escherichia coli, ENTB: Enterobacter species, IR: Intrinsic resistant, KLEB: Klebsiella species, NA: Not applicable, PSEU: Pseudomonas species, PRO: Proteus species, STEN: Stenotrophomonas maltophilia, SER: Serratia marcescens, SAL: Salmonella species.
The highest percentage of resistance was observed in amoxicillin/clavulanic acid, cefalothin, cefuroxime, cefixime, and cefpodoxime among Klebsiella and Enterobacter species isolated from blood cultures. For Klebsiella species, the resistance rate was 44.4%, while for Enterobacter species, it was 100%. Quinolones and imipenem had a lower resistance rate, with 11.1% for Klebsiella species and 0% for Enterobacter species. No resistance was observed against cefepime, meropenem, piperacillin/tazobactam, co-trimoxazole, amikacin, and gentamicin. In our circumstances, the proportion of Klebsiella that produces extended-spectrum beta-lactamase (ESBL) within the Enterobacteriaceae family among bacteremia cases is 44%.
Proteus species had a resistance rate of 50% to all beta-lactam antibiotics tested, except meropenem (which had a resistance rate of 0%), co-trimoxazole, and quinolones. ESBL-producing Proteus species accounted for 50%. However, amikacin and gentamicin exhibited a complete susceptibility of 100%. Salmonella species exhibited complete resistance (100%) to ampicillin, and lower resistance rates were seen for ceftriaxone, cefpodoxime, co-trimoxazole, and azithromycin (50%).
Additionally, 75% of Pseudomonas species exhibited carbapenemase production, rendering them resistant to imipenem and meropenem. Furthermore, the percentage of resistance to quinolones was similarly 75%. No resistance was seen against anti-pseudomonas cephalosporins, aztreonam, and the tested aminoglycosides. During the study period, a single strain of Acinetobacter baumannii was isolated in blood culture specimens. This strain was determined to be completely resistant (100%) to ceftazidime, cefepime, and co-trimoxazole while being susceptible to all other antibiotics tested. Stenotrophomonas maltophilia demonstrated the highest resistance to minocycline (66.7%) followed by carbapenems and ceftazidime (50% and 16.7% respectively). All isolates exhibited susceptibility to co-trimoxazole and levofloxacin, with no resistance observed against these antibiotics. All Serratia marcesens isolates exhibited complete resistance (100%) to amoxicillin/clavulanic acid, cefalotin, and cefuroxime. However, 75% resistance was seen against cefixime and cefpodoxime, while all other tested drugs remained susceptible to it.
This study demonstrated the prevalence of bacteremia with the age and gender of the suspected patients including antimicrobial susceptibility patterns of the isolated bacteria. The incidence of bloodstream infections in our study was 5%. This incidence rate is by another study conducted in Alkharj military hospital where bacteremia cases were found to be 7.53%.21 The prevalence of bacteremia in different age groups and gender aligns with findings from various studies. For instance, Kolesnichenko et al. reported on the prevalence of bacteremia in febrile neonates and infants, which is relevant to the observed high occurrence of bacteremia in the 1-month to 1-year age group.22 Male predominance of culture-positive bacteremia was also in parallel with Uslan et al. who reported 341 male bacteremia cases and 309 female bacteremia cases over a study period of three years.23
The distribution of blood culture isolates indicates that 57% of the isolates were Gram-positive bacteria, 37% were Gram-negative bacteria, and the remaining 6% were Candida species. This distribution aligns with findings from a study, which reported that Gram-positive and Gram-negative bacteria constituted 69% and 31% of the culture isolates, respectively.24 These findings are also supported by a study done on 200 blood culture samples that also reported Gram-positive bacterial predominance in all the age groups of suspected bacteremia cases.25 Ahmed et al. also reported the same predominance of Gram-positive bacteremia cases.21 Furthermore, the study by Yao and Fu revealed that 59.9% of positive blood culture samples had Gram-negative bacteria, and 39.3% had gram-positive bacteria,26 in contrast to the distribution observed in our study. The most probable reason could be a selection of the study population. Additionally, the distribution of bacterial isolates obtained from positive blood cultures for CoNS had the highest number of cases, followed by E. coli, Streptococcus species, S. aureus, and Klebsiella species, among others. These findings are consistent with the literature, as CoNS are frequently isolated from blood cultures and are an important cause of nosocomial bloodstream infections, especially in catheterized patients.21,27
The antimicrobial susceptibility of Gram-positive bacterial isolates, particularly S. aureus, has been a subject of interest due to varying levels of resistance to different antibiotics and multidrug-resistant strains reported globally. Except for one isolate of CoNS, all Staphylococci showed 100% resistance to penicillin which was consistent with the previous findings by Vasudeva et al. However, this penicillin resistance was reported up to 90% by Zhu et al.28 The most likely reasoning could be the different geographical practices to prescribe beta-lactam antibiotics resulting in the acquisition of resistant bugs. Furthermore, resistance rates of Staphylococci to clindamycin, tetracycline, gentamycin, co-trimoxazole, and quinolones were less than 50%. Therefore, these antibiotics could be the effective therapeutic choice for patients having bacteremia due to Staphylococci, to lessen the overwhelming use of vancomycin and linezolid. All Staphylococci isolates were 100% susceptible to vancomycin and linezolid and these results were in line with other reported studies from Asian countries.27-29 Moreover, vancomycin-resistant Enterococci (VRE) from bloodstream infections had been reported in many previous studies,28,30 however, in contrast to the above-mentioned study, we did not find any VRE in our clinical settings. Furthermore, linezolid susceptibility was also found 100% in all isolated Enterococcus species, whereas erythromycin resistance was 100%. In Streptococcus species penicillin/ampicillin resistance was 46.7% in our setup in contrast to Vasudeva et al., who reported 100% susceptibility.27 The most acceptable reason is the inclusion of all the isolated species of Streptococci in our study, whereas Vasudeva et al., reported penicillin susceptibility in Streptococcus pneumoniae only. However, Zhu et al, reported 30% Streptococci resistant to penicillin.28 Furthermore, our findings for vancomycin and linezolid susceptibilities in Streptococci were in parallel with previous studies that also showed 100% susceptibilities against these antibiotics.28
The prevalence of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, particularly in E. coli and Klebsiella species, has been well-documented in various hospital settings from all over the world including Saudi Arabia.31-33 These bacteria exhibit resistance to a range of antibiotics, including ampicillin, cephalosporins, and quinolones, with varying levels of resistance observed across different antibiotics.31,32 Notably, ESBL-producing E. coli and Klebsiella species strains have shown complete susceptibility to carbapenems and amikacin and this finding is in parallel with Kateregga et al.31 The emergence of ESBL-producing Enterobacteriaceae has raised concerns due to their resistance to nearly all antibiotics, necessitating the need for effective antimicrobial strategies.34
The resistance of Pseudomonas species to carbapenem and quinolones has been a growing concern. Studies have shown that a significant percentage of Pseudomonas species exhibit carbapenemase production, rendering them resistant to imipenem and meropenem.35,36 In our study, we found 75% carbapenemase-producing Pseudomonas strains from blood cultures. Additionally, the prevalence of resistance to quinolones among Pseudomonas species has been reported to be as high as 75%. This finding is supported by Tilahun et al. who found 54.3% quinolone resistance in Pseudomonas species.37 However, it is noteworthy that no resistance has been observed against anti-pseudomonas cephalosporins, aztreonam, and the tested aminoglycosides.
However, this study has some limitations that should be considered. Initially, data collection relied on medical records from the VIDA system, entered by clinicians and health workers, rather than interviewing patients and evaluating clinically. Our study was limited to a single hospital, which may have biased our findings due to the small sample size. Ultimately, a considerable amount of drug susceptibility data could not be examined due to the lack of established criteria. Furthermore, not all samples were tested identically for antimicrobial sensitivity, resulting in a lack of resistance comparisons among the isolates in the current study. Further research and surveillance are needed to monitor and address the emergence of antibiotic-resistant strains.
The data presented in this study aligns with findings from relevant literature, providing valuable insights into the prevalence of bacteremia, distribution of blood culture isolates, antimicrobial susceptibility patterns, and resistance of specific bacterial species. The study highlights the importance of understanding the demographics and antibiotic resistance profiles of bacteremia cases to inform effective treatment strategies.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
FK conceptualized and designed the study. FK, AE and CP performed data collection. FK and AM performed analysis and results interpretation. FK and OTK wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
The study was approved by the Research Committee of Dr. Sulaiman Al Habib Medical Group Research Center, Riyadh, Kingdom of Saudi Arabia (IRB study number RC23.11.18).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Karakonstantis S, Kritsotakis EI. Systematic review and meta-analysis of the proportion and associated mortality of polymicrobial (vs monomicrobial) pulmonary and bloodstream infections by Acinetobacter baumannii complex. Infection. 2021;49(6):1149-1161.
Crossref - Li Z, Zhuang H, Wang G, Wang H, Dong Y. Prevalence, predictors, and mortality of bloodstream infections due to methicillin-resistant Staphylococcus aureus in patients with malignancy: systemic review and meta-analysis. BMC Infect Dis. 2021;21(1):74.
Crossref - Hattori H, Maeda M, Nagatomo Y, et al. Epidemiology and risk factors for mortality in bloodstream infections: A single-center retrospective study in Japan. Am J Infect Control. 2018;46(12):e75-e79.
Crossref - Huson MAM, Stolp SM, van der Poll T, Grobusch MP. Community-acquired bacterial bloodstream infections in HIV-infected patients: a systematic review. Clin Infect Dis. 2014;58(1):79-92.
Crossref - Ramphal R. Changes in the Etiology of Bacteremia in Febrile Neutropenic Patients and the Susceptibilities of the Currently Isolated Pathogens. Clin Infect Dis. 2004;39(Suppl 1):S25-31.
Crossref - Ostrosky-Zeichner L, Sable C, Sobel J, et al. Multicenter retrospective development and validation of a clinical prediction rule for nosocomial invasive candidiasis in the intensive care setting. Eur J Clin Microbiol Infect Dis. 2007;26(4):271-276.
Crossref - Ramanan P, Deziel PJ, Wengenack NL. Gordonia Bacteremia. J Clin Microbiol. 2013;51(10):3443-3447.
Crossref - Bassetti M, Righi E. Multidrug-resistant bacteria: what is the threat? Hematology. 2013;2013(1):428-432.
Crossref - Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock, 2012. Intensive Care Med. 2013;39(2):165-228.
Crossref - Fleischmann-Struzek C, Goldfarb DM, Schlattmann P, Schlapbach LJ, Reinhart K, Kissoon N. The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir Med. 2018;6(3):223-230.
Crossref - Gonzalez MD, Chao T, Pettengill MA. Modern Blood Culture: Management Decisions and Method Options. Clin Lab Med. 2020;40(4):379-392.
Crossref - Banerjee R, Humphries R. Rapid Antimicrobial Susceptibility Testing Methods for Blood Cultures and Their Clinical Impact. Front Med. 2021;8:635831.
Crossref - Watson RS, Carcillo JA, Linde-Zwirble WT, Clermont G, Lidicker J, Derek G. The Epidemiology of Severe Sepsis in Children in the United States. Am J Respir Crit Care Med. 2003;167(5):695-701.
Crossref - Williams MD, Braun LA, Cooper LM, et al. Hospitalized cancer patients with severe sepsis: analysis of incidence, mortality, and associated costs of care. Crit Care. 2004;8(5):R291.
Crossref - Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5(1):4-11.
Crossref - Giacobbe DR, Del Bono V, Trecarichi EM, et al. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect. 2015;21(12):1106.e1-1106.e8.
Crossref - Bandy A, Almaeen AH. Pathogenic spectrum of blood stream infections and resistance pattern in Gram-negative bacteria from Aljouf region of Saudi Arabia. PLOS ONE. 2020;15(6):e0233704.
Crossref - World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2016-2017. World Health Organization; 2017. https://iris.who.int/handle/10665/259744. Accessed November 7, 2023.
- Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200-211.
Crossref - Banawas SS, Alobaidi AS, Dawoud TM, et al. Prevalence of Multidrug-Resistant Bacteria in Healthcare-Associated Bloodstream Infections at Hospitals in Riyadh, Saudi Arabia. Pathogens. 2023;12(9):1075.
Crossref - Ahmed NJ, Mahmoud S, Menshawy MA. The Prevalence of Bacteremia among Patients Admitted to a Military Hospital in Alkharj. J Pharm Res Int. 2020;32(30):93-99.
Crossref - Biondi EA, Lee B, Ralston SL, et al. Prevalence of Bacteremia and Bacterial Meningitis in Febrile Neonates and Infants in the Second Month of Life: A Systematic Review and Meta-analysis. JAMA Netw Open. 2019;2(3):e190874.
Crossref - Uslan DZ, Crane SJ, Steckelberg JM, et al. Age- and Sex-Associated Trends in Bloodstream Infection: A Population-Based Study in Olmsted County, Minnesota. Arch Intern Med. 2007;167(8):834-839.
Crossref - Dagnew M, Yismaw G, Gizachew M, et al. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC Res Notes. 2013;6(1):283.
Crossref - Kolesnichenko SI, Lavrinenko AV, Akhmaltdinova LL. Bloodstream Infection Etiology among Children and Adults. Int J Microbiol. 2021;2021(1):6657134.
Crossref - Yao JF, Li N, Jiang J. Clinical Characteristics of Bloodstream Infections in Pediatric Acute Leukemia: A Single-center Experience with 231 Patients. Chin Med J. 2017;130(17):2076-2081.
Crossref - Vasudeva N, Nirwan PS, Shrivastava P. Bloodstream infections and antimicrobial sensitivity patterns in a tertiary care hospital of India. Ther Adv Infect Dis. 2016;3(5):119-127.
Crossref - Zhu Q, Yue Y, Zhu L, et al. Epidemiology and microbiology of Gram-positive bloodstream infections in a tertiary-care hospital in Beijing, China: a 6-year retrospective study. Antimicrob Resist Infect Control. 2018;7(1):107.
Crossref - Fayyaz M, Mirza IA, Ikram A, Hussain A, Ghafoor T, Shujat U. Pathogens Causing Blood Stream Infections and their Drug Susceptibility Profile in Immunocompromised Patients. J Coll Physicians Surg Pak. 2013;23(12):848-851.
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266-278.
Crossref - Kateregga JN, Kantume R, Atuhaire C, Lubowa MN, Ndukui JG. Phenotypic expression and prevalence of ESBL-producing Enterobacteriaceae in samples collected from patients in various wards of Mulago Hospital, Uganda. BMC Pharmacol Toxicol. 2015;16:14.
Crossref - Kanamori H, Yano H, Hirakata Y, et al. Molecular Characteristics of Extended-Spectrum Beta-Lactamases and qnr Determinants in Enterobacter Species from Japan. PLOS ONE. 2012;7(6):e37967.
Crossref - Hassan H, Abdalhamid B. Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Ctries. 2014;8(3):282-288.
Crossref - Khademi F, Vaez H, Neyestani Z, Sahebkar A. Prevalence of ESBL-Producing Enterobacter Species Resistant to Carbapenems in Iran: A Systematic Review and Meta-Analysis. Int J Microbiol. 2022;2022(1):8367365.
Crossref - Ogba RC, Nomeh OL, Edemekong CI, et al. Molecular Characterization of Carbapenemase Encoding Genes in Pseudomonas aeruginosa from Tertiary Healthcare in South Eastern Nigeria. Asian J Biochem Genet Mol Biol. 2022;12(4):161-168.
Crossref - Bajpai V, Govindaswamy A, Khurana S, et al. Phenotypic & genotypic profile of antimicrobial resistance in Pseudomonas species in hospitalized patients. Indian J Med Res. 2019;149(2):216-221.
Crossref - Tilahun M, Gedefie A, Bisetegn H, Debash H. Emergence of High Prevalence of Extended-Spectrum Beta-Lactamase and Carbapenemase Producing Acinetobacter Species and Pseudomonas aeruginosa Among Hospitalized Patients at Dessie Comprehensive Specialized Hospital, North-East Ethiopia. Infect Drug Resist. 2022;15:895-911.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.