ISSN: 0973-7510
E-ISSN: 2581-690X
The study was conducted to observe the bacteriological profile of diabetic foot ulcers in patients at tertiary care hospital. The incidence and pattern of antibiotic resistance of MRSA was studied. A cross-sectional study was carried out in the tertiary care hospital from September 2017 to December 2021. Of a total of 313 DFU cases analysed, 304 cases were infected ulcers accounting for 97.13% of cases and the remaining 09 cases (2.87%) were noninfected. Of the isolates cultured from the infected ulcers, 261 (53.81%) were gram-negative organisms and 224 (46.18%) were gram-positive. Of the gram-positive organisms, Methicillin resistant Staphylococcus aureus (66.9%) was most common, followed by Methicillin sensitive Staphylococcus aureus (31.3%) and Streptococcus pyogenes (1.8%). Of the gram-negative organisms, Pseudomonas aeruginosa 67 (25.7%) was predominant, followed by Escherichia coli 53 (20.3%), Klebsiella species (19.2%), Acinetobacter species 50 (19.2%). The drug resistance pattern of MRSA was, Ofloxacin 82.6%, ciprofloxacin (89.3%), levofloxacin (91.3%), cefoxitin (100%) with Benzyl penicillin showing highest resistance of 100%. There was a significant rise of MRSA infections in DFUs.
Bacterial Isolates, MRSA, Diabetic Foot Ulcer
Diabetes is most common epidemic metabolic disease, causing a great deal of loss in human civilization in terms of physical, psychological, and financial aspects. The most serious pathophysiological consequences of diabetes is diabetic foot ulcer (DFU). The most important cause of chronic DFU is bacterial infection. Multidrug-resistance is demonstrated by bacterial species or their biofilms, which worsens DFU and ultimately necessitates amputation of the affected part.
India is one of the most affected countries, with over 77 million people living with diabetes; by 2045, that figure is expected to increase by 35.7 million. In India, 8.9% of people have diabetes, and the disease is thought to be the cause of one million diabetes-related deaths annually. According to estimates by Singh and Shankhdhar et al., around 25% of Indian diabetic patients will develop DFU.1 Among those with diabetes, infections are the most frequent issue. Due to a compromised vascular supply, these people are more vulnerable to foot infections.2
Foot wounds are currently the most prevalent diabetes-related hospitalization cause. These wounds are becoming more and more common in patients with diabetes.3 Diabetes increases a person’s lifetime risk of foot ulcers by 12-25%, which poses a serious threat to public health.4,5
Many studies done on diabetics with non-healing ulcers, the risk of amputation is almost forty times higher in diabetics with non-healing ulcers than that of trauma patients.6,7
A co-occurrence of DFI and foot ischemia increases the risk of amputation even further.8 Doctors should avoid prescribing antibiotic therapy unnecessarily, undue broad spectrum or for extended time, as it may leads to drug-related side effects, increase the economic cost and antimicrobial-resistance.9
Staphylococcus aureus is main causative bacteria of diabetic foot infections, and in one study, MRSA (methicillin-resistant Staphylococcus aureus) accounted for 23.7% of these infections.10
Infections that have the potential to be fatal are frequently contracted in healthcare and community settings by Staphylococcus aureus.2 It is commonly known that Staphylococcal species develop antibiotic-resistance rapidly and that they follow a distinct epidemiological pattern.
When treating Staphylococcus aureus infections, penicillin was initially the recommended medication. Emergence of penicillin resistance in Staphylococcus aureus is a result of plasmid-borne genetic elements that produce β-lactamases. The best medicine for treating infections due to penicillin-resistant Staphylococcus aureus is then semisynthetic penicillin, also known as methicillin and oxacillin.11
One of the main problems with global health care is antimicrobial-resistance, which makes it more difficult to treat and prevent diseases and outbreaks by bacteria that are spreading quickly. Although the fact that antibiotics have been available for almost 70 years, the growing emergence of antibiotic-resistance, particularly to methicillin and vancomycin, made the therapy difficult for physicians. In about 10-32% of patients with DFIs, antibiotic-resistant organisms-particularly MRSA-are commonly detected, which ultimately raises the risk of treatment failure.10
This was a cross-sectional study done in the Microbiology Department at Krishna Hospital Karad which is tertiary care centre, between September 2017 and December 2021. The approval of the Institution Human Ethics committee was obtained.
Total 313 diabetic patients with foot ulcer who were admitted to the Krishna Hospital, Karad were the study subjects. Informed consent from each patient was taken. The subjects’ clinical and sociodemographic information was collected using a structured questionnaire. From patients with diabetic foot ulcers samples were taken aseptically as wound swabs or pus samples for culture and sensitivity testing and processed as per the CLSI guidelines.
Microscopy
The heat fixed smears were stained by Gram stain technique and examined under oil immersion objective of the light microscope. The smears were examined for the presence of pus cells, Gram-positive and Gram-negative organisms. The size, shape, arrangement of bacteria was noted.
Culture of aerobic bacteria
The specimen was culture inoculated onto appropriate culture media viz., blood agar and MacConkey agar plate. The inoculated media was incubated at 37 °C overnight. Bacterial isolates identification done as per standard microbiological methods.
Isolation and identification of Staphylococcus aureus
Conventional method for identification of Staphylococcus aureus
A single well-isolated colony of Staphylococcus aureus was selected based on Gram stain reaction and further identification of the bacterial isolate was done by catalase, coagulase, mannitol fermentation and VP test. Standard bacteriological techniques were used to identify Staphylococcus aureus.
Antimicrobial susceptibility testing
As per the Clinical and Laboratory Standard Institute (CLSI 27th ed. supplement M100 2017) guidelines antimicrobial susceptibility testing of methicillin-resistant Staphylococcus aureus was done on Mueller Hinton agar plate using Kirby-Bauer disc diffusion method.12
The antibiotic discs Benzyl penicillin (10 units), Levofloxacin (5 µg), Cotrimoxazole (25 µg), Vancomycin (30 µg), Gentamicin (10 µg), Ciprofloxacin (5 µg), Ofloxacin (1 µg), Cefoxitin (30 µg), Erythromycin (15 µg), Clindamycin (2 µg), Linezolid (30 µg), Tigecycline (15 µg), Netilmicin (30 µg), and Nitrofurantoin (300 µg) were used to evaluate the antibiotic susceptibility pattern of the MRSA isolates. Staphylococcus aureus ATCC 25923 was used as control.
Detection of methicillin-resistant Staphylococcus
Standard disk diffusion procedure
Staphylococcus species were tested for Methicillin-resistance using 30 µg Cefoxitin discs by Kirby-Bauer Disc Diffusion method. Direct colony suspension of test and control organisms was prepared to obtain 0.5 McFarland standard turbidity and inoculated onto Mueller-Hinton agar plates. Plates were incubated at 33-35 °C; ambient air for 16-18 hours. MRSA: The diameter of zone of inhibition was measured and interpreted according to National Committee for Clinical Laboratory Standards guidelines (CLSI 27th ed. supplement M100 2017).12 ATCC Staphylococcus aureus 25923 (cefoxitin zone 23-29 mm) was used as control strain for the test.
Table 1 shows that, out of 313 cases of DFU, culture was positive in 304 cases and 09 cases (2.87%) were sterile. Among total 485 bacterial isolates obtained from culture positive cases, 184 (58.78%) cases yielded monomicrobial organism, i.e., single organism isolate (37.9%) and 120 (38.33%) cases were polymicrobial (62.1%). Among the polymicrobial organisms, 74 cases yielded two organisms, 33 cases three organism, 12 cases four organisms, with a single case of six organisms.
Table (1):
Total number and percentage of bacterial isolates
| Parameters | Number of cases n (%) | Number of Organisms |
|---|---|---|
| Culture positive cases | 304 (97.13%) | |
| Culture negative cases | 09 (2.87%) | |
| Pattern of organisms isolated from culture positive cases | ||
| Monomicrobial organism | 184 (58.78%) | 184 (37.9%) |
| Polymicrobial organisms | 120 (38.33%) | 38.33% |
| Two organisms | 74 | 148 (30.51%) |
| Three organisms | 33 | 99 (20.41%) |
| Four organisms | 12 | 48 (0.41%) |
| Six organisms | 01 | 06 (0.20%) |
| Total | 313 | 485 |
Table 2 shows the percentage of different organisms isolated from DFUs. Of the culture positive isolates gram-negative organisms were most common isolates 261 (53.81%), then gram-positive 224 (46.18%).
Table (2):
Distribution of isolated organisms
| Microorganisms isolated | Frequency |
|---|---|
| Total number of microorganisms | 485 |
| Aerobes | |
| Gram-positive organisms | 224 (46.18%) |
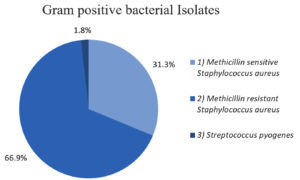
| 1. Methicillin sensitive S. aureus | 70 (31.3%) |
| 2. Methicillin resistant S. aureus | 150 (66.9%) |
| 3. Streptococcus pyrogens | 4 (1.8%) |
| Gram-negative organisms | 261 (53.81%) |
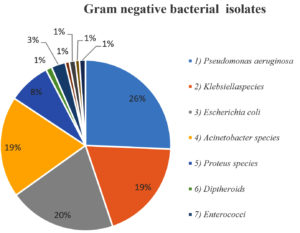
| 1. Pseudomonas aeruginosa | 67 (25.7%) |
| 2. Klebsiella species | 50 (19.2%) |
| 3. Escherichia coli | 53 (20.3%) |
| 4. Acinetobacter species | 50 (19.2%) |
| 5. Proteus species | 21 (8.0%) |
| 6. Diptheriods species | 03 (1.1%) |
| 7. Enterococci species | 7 (2.7%) |
| 8. Citrobacter species | 2 (0.8%) |
| 9. Morganella morgani | 03 (1.1%) |
| 10. Aeromonas hydrophila | 02 (0.8%) |
| 11. Serratia marcescens, Providencia rettgeri, Providencia stuartti | 03 (1%) |
Figure 1 shows that, of the total 224 gram-positive bacteria, Staphylococcus aureus was the most common gram-positive organism 220 (98.2%) isolated. Methicillin-resistant Staphylococcus aureus accounted for the majority of gram-positive organisms 150 (66.9%), followed by Methicillin-sensitive Staphylococcus aureus (31.3%) and Streptococcus pyogenes (1.8%).
Among gram-negative aerobes, Pseudomonas aeruginosa 67 (25.7%) and Escherichia coli 53 (220%) were the most common isolates (Table 2).
Figure 2 shows the distribution of Gram-negative organisms isolated from Diabetic foot ulcer patients in our study.
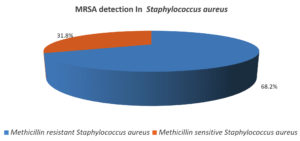
Table 3 shows that, of the total Staphylococcus aureus isolates, MRSA was (150) followed by MSSA (70).
Table (3):
MRSA detection in Staphylococcus aureus
Gram-positive organisms |
No. of isolates |
|---|---|
MRSA |
150 (68.2%) |
MSSA |
70 (31.8%) |
Total |
220 |
Figure 3 shows of the Staphylococcus aureus isolated 68.2% were Methicillin resistant 31.8% were Methicillin sensitive.
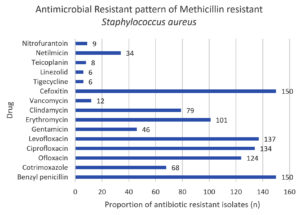
In our study, Table 4 shows that, MRSA isolates showed maximum level of resistance to benzyl penicillin and cefoxitin (100%) followed by levofloxacin (91.3%), ciprofloxacin (89.3%), and oxacillin (82.6%).
Table (4):
Antibiotic resistance pattern of MRSA (n = 150)
Drug |
Methicillin-resistant S. aureus n (%) |
|---|---|
Benzyl penicillin |
150 (100) |
Cotrimoxazole |
68 (45.3) |
Ofloxacin |
124 (82.7) |
Ciprofloxacin |
135 (90) |
Levofloxacin |
137 (91.3) |
Gentamicin |
47 (31.3) |
Erythromycin |
102 (68) |
Clindamycin |
79 (52.7) |
Vancomycin |
12 (8) |
Cefoxitin |
150 (100) |
Tigecycline |
6 (4) |
Linezolid |
6 (4) |
Teicoplanin |
8 (5.3) |
Netilmicin |
34 (22.7) |
Nitrofurantoin |
10 (6.7) |
By 2045, there will likely be 35.7 million people with diabetes worldwide, up from the current predicted 77 million.1 A hospital stay is necessary for the majority of problems resulting from diabetic foot infections, which are common. Majority of the study subjects were over fifty years of age and maximum number 174 (55%) of them were in the age group 50-70 years. Of the total patients, 244 were males and 69 females. Diabetic foot ulcers showed a male predominance, (78%) in present study which is similar to the research by Shah. et al.13 Of the 485 bacterial isolates obtained, majority were gram-negative isolates, which are more prevalent isolates found in many prior studies conducted in India.11
The studies from the western country shows predominance of Gram-positive organisms in diabetic foot infection.14,15 In our study, gram-negative organisms were more commonly isolated than gram-positive, which is in accordance with findings by Umadevi et al.16 and Mohanasoundaram et al.17 As per the previous studies, Pseudomonas aeruginosa accounted for the majority of gram-negative organisms (53.81%), with a prevalence of 25.7%.18 Also, a study by Shah et al.13 reported Pseudomonas aeruginosa and Escherichia coli as most predominant among gram-negative organisms which is similar to our study. South Indian research indicates that monobacterial infections predominate over polymicrobial infections,19 similar results were detected in our study. A predominance of monomicrobial infections i.e. (77.3%) were observed in study by Hassan et al., while polymicrobial infections were found in 22.7%.3 In our study, 58.78% of cases were monomicrobial infections which is comparable to results in the study done by Shah et al.13
The frequency of MRSA, MSSA, and Streptococcus pyogenes among gram-positive organisms was 66.9%, 31.3%, and 1.8%, respectively. The percentage of MRSA cases in India varies from 6.9% to 81%.20 A few studies have shown as high as 64% incidence of MRSA.21 A study by Shah et al.13 showed 77.8% of Methicillin-resistant Staphylococcus aureus.
In line with previous research by Vidhani et al.22 stating that the MRSA isolates were 100% resistance to penicillin, similar observation was seen in present study, demonstrating resistance 100% to benzyl penicillin; the antibiotic with the highest level of resistance (Figure 4).
Drug resistance and inadequate antibiotic treatment have become increasingly serious issues over time. Considering that India has one of the greatest populations of diabetics, it is very important to evaluate the Methicillin-resistant Staphylococcus aureus in diabetic foot ulcers at the earliest. New resistance keeps emerging and developing at new places in spite of more research and discovery. Consequently, there is still a great need for new antibiotics, particularly those directed against MRSA.
In conclusion, empirical antibiotics are a common treatment for diabetic foot ulcers. The choice of empirical treatment is frequently dictated by the severity of the wound and the antimicrobial susceptibility pattern. Prevalence of MRSA in DFI cases was 68.2% in our study. Managing DFI presents numerous hurdles due to the intricate pathophysiology of MRSA. The results of this study showed a higher rate of antimicrobial-resistance to frequently prescribed medicines: benzyl penicillin (100%), cefoxitin (100%), levofloxacin (91.3%), Ciprofloxacin 89.3% and Ofloxacin 82.6%. Clinicians are being compelled to reconsider treatment options and minimize problems as soon as possible due to the increasing resistance to multiple antimicrobials and safe usage of anti-MRSA drugs now in use. For rapid MRSA identification, the use of fast diagnostic tools such as polymerase chain reaction for detection of MecA gene, PVL gene, Coa gene, Pulse field gel electrophoresis, and Multilocus sequence typing can be used.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Krishna Institute of Medical Sciences, Karad, India, vide reference number KIMSDU/IEC/02/2017.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Kale DS, Karande GS, Datkhile KD. Diabetic foot Ulcer in India: Aetiological Trends and Bacterial Diversity. Indian J Endocrinol Metab. 2023;27(2):107-114
- Mahjoob MO, Abdalla AE, Elawad HE, Abdulla AA , Altayeb SO, Abdalla UH. The Incidence of Methicillin Resistance Staphylococcus aureus in Diabetic Septic Foot. J Trop Dis. 2019;7:333.
Crossref - Hassan MA, Tamer TM, Ragch AA, Abou-Zeid AM, Abd El-Zaher EHF, Kenawy E. Insight into multidrug-resistant microorganisms from microbial infected diabetic foot ulcers. Diabetes Metab Syndr. 2019;13(2):1261-1270.
Crossref - Singh N, Armstrong DG, Lipsky BA. Preventing Foot Ulcers in patients with Diabetes. JAMA. 2005;293(2):217-228.
Crossref - urbancic-Rovan V. Causes of foot lesions. Lancet. 2005;366:1675-1676.
Crossref - Otta S, Debata NK, Swain B. Bacteriological profile of diabetic foot ulcers. Chris med J Health Res. 2019;6(1):7-11.
Crossref - Louie TJ, Bartlett JG, Tally FP, Gorbach SL. Aerobic and anaerobic bacteria in diabetic foot ulcers. Ann Intern Med. 1976:85(4):461-463.
Crossref - Skrepnek GH, Armstrong DG, Mills JL. Open bypass and endovascular procedures among diabetic foot ulcer cases in the United States from 2001 to2010. J Vase Surg. 2014;60(5):1255-1265.
Crossref - Lipsky BA, Berendt AR, Cornia PB, et al. 2012 Infectious Diseases Society of America Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis. 2012;54(12):132-173.
Crossref - Vishwanathan V, Pendsey S, Radhakrishnan C, Rege TD, Ahdal J, Jain R. Methicillin resistant Staphylococcus aureus in diabetic foot infection in India: A Growing menace. Int J Low Extrem Wounds. 2019;18(3):236-246.
Crossref - Gandi PS, Someshwaran R, Inteti K. Prevalence of methicillin resistant staphylococcus aureus among patients with diabetic foot ulcers in a tertiary care hospital. Int J Recent Sci Res. 2022;13(03):752-756
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100.Wayne, PA: Clinical and Laboratory Standards Institute. 2017.
- Shah P, Eswarawaka M, Anne D, Reddy MSC, Shah P, Srivastava N. Bacteriological profile of Diabetic foot. Int Surg J. 2021;8(2):704-709.
Crossref - Dang CN, Prasad YDM, Boulton AJM, Jude EB. Methicillin-resistant Staphylococcus aureus in the diabetic foot clinic: a worsening problem. Diabet Med. 2003;20(2):159-161.
Crossref - Lipsky BA, Baker PD, Landon GC, Fernau R. Antibiotic therapy for diabetic foot infections: comparison of two parenteral-to-oral regimens. Clin Infect Dis. 1997;24(4):643-648.
Crossref - Umadevi S, Kumar S, Joseph NM, et al. Microbiological study of diabetic foot infections. Indian J Med Specialities. 2011;2(1):12-7.26.
Crossref - Mohanasoundaram KM. The microbiological profile of diabetic foot infections. J Clin Diag Res. 2011;5:1-3.
- Sughandhi P, Prasanth DA. Bacteriological profile of diabetic foot infections. Int J Innov Res Sci Eng Technol. 2014;3:14688-14692.28.
- Lipsky BA, Pecoraro RE, Wheat LJ. The diabetic foot: soft tissue and bone infection. Infect Dis Clin North Am. 1990; 4(3):409-432.
Crossref - Verma S, Joshi S, Chitnis V, Heemwani N, Chitnis D. Growing problem of methicillin resistant staphylococci–Indian scenario. Indian J Med Sci. 2000;54(12):535-540.
- Banoo S, Shubha DS, Shashidhar V, Venkatesha D. Bacterial and clinical profile of diabetic foot patients. Ann Trop Med Public Health. 2012;5(2):69-73.
- Vidhani S, Mehndiratta PL, Mathur MD. Study of Methicillin resistant S. aureus (MRSA) isolates from high-risk patients. Indian J Med Microbiol. 2001;19(2):87-90.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.