ISSN: 0973-7510
E-ISSN: 2581-690X
Streptomyces is a genus recognized for combating many plant pathogens, including those affecting post-harvest fruits. This study aimed to identify the Colletotrichum fungus responsible for anthracnose in tomatoes and evaluate the antifungal efficacy of Streptomyces murinus NARZ. The Colletotrichum isolate was identified as C. scovillei C3 through morphological characteristics, ITS region sequencing, and phylogenetic analysis. Culture filtrates (CF) of S. murinus NARZ at concentrations from 0% to 50% (in 10% intervals) were tested for antifungal activity against C. scovillei C3. The Percentage Inhibition of Radial Growth, calculated using colony diameters, showed that a 30.40% CF concentration (EC50) inhibited 50% of C. scovillei C3 growth on PDA plates. The CF exhibited heat stability, with PIRG values ranging from 62.93% to 65.35% across temperatures of 30 °C to 90 °C. In vivo trials involved treating tomatoes with CF using pre-treatment (spraying 24 hours before inoculation) or post-treatment (spraying 24 hours after inoculation). After seven days, pre-treated tomatoes with 50% CF showed a disease incidence of 83.33%, while all other treatments had a 100% incidence. Lesion diameters in the 50% CF treatment were significantly smaller (p < 0.05) compared to lower CF concentrations and showed results similar to the Chlorothalonil treatment. Pre-treatment was more effective than post-treatment, with lesion diameters of 5.40 mm and 8.73 mm, respectively. PCR analysis confirmed that S. murinus NARZ produced antifungal compounds via PKS-I, PKS-II, and NRPS gene clusters. These findings suggest that S. murinus NARZ could be an effective alternative to chemical fungicides for managing tomato anthracnose caused by C. scovillei.
Antifungal Activities, Anthracnose, C. scovillei, Streptomyces murinus, Tomato Fruit
Tomato (Solanum lycopersicum L.) is an important crop cultivated in many countries, including Vietnam. It is rich in various antioxidant compounds such as carotenoids, particularly lycopene, ascorbic acid, vitamin E, and phenolic compounds, especially flavonoids.1 However, tomatoes are susceptible to various diseases, including severe fungal infections that threaten global tomato production, of which anthracnose is the most notable.2
Anthracnose, caused by Colletotrichum species, is a significant disease affecting tomatoes worldwide, leading to post-harvest fruit decay and severely reducing marketability.3 Numerous Colletotrichum species are responsible for tomato anthracnose, including C. gloeosporioides,4 C. gossypii,5 C. acutatum,6 C. truncatum,7 C. scovillei,8 C. coccodes,9 and C. nymphaeae,10 are emerging pathogens causing significant annual yield losses.
Infections by Colletotrichum species in tomato fruit transition from quiescent to active infection as the fruit ripens, resulting in anthracnose in mature tomatoes. When the fruit ripens, Colletotrichum spp. penetrate the pericarp, causing circular depressions that evolve into necrotrophic lesions,11 displaying a dark, sunken, and water-soaked appearance.7 Orange conidial masses appeared on the lesions, sometimes in concentric rings, and black acervuli formed just beneath the fruit skin.6 The anthracnose infection shortens the shelf life of tomatoes12 and reduces crop yields, resulting in a 10-30% loss of total crop production.13 These losses diminish farmers’ income and disrupt food supply chains, putting additional pressure on global agricultural markets.2
Chemical pesticides remain the most common method for controlling post-harvest diseases in fruits and vegetables. However, their improper use threatens human health, fosters pathogen resistance, and contributes to environmental pollution.14-16 In response, biological control using antagonistic microorganisms such as bacteria, yeasts, molds, and actinomycetes has gained traction due to growing concerns about sustainability.2 Biocontrol agents (BCAs) offer an eco-friendly alternative, with Streptomyces standing out for its ability to produce fungicides, antibiotics, and hydrolytic enzymes that inhibit phytopathogens. Unlike synthetic pesticides, Streptomyces suppresses plant diseases naturally, enhances immunity, promotes growth, and improves soil health. With stricter pesticide regulations and a shift toward sustainable agriculture, microbial biocontrol provides a safer, more effective solution.17
Several studies have utilized biological compounds derived from Streptomyces to inhibit pathogenic fungi and extend the shelf life of post-harvest fruits and vegetables.18,19 Streptomyces sp. A1022 inhibited the growth of C. gloeosporioides, which causes anthracnose in pepper and cherry tomatoes.16 S. tuirus AR26 was effective against infection and growth of C. scovillei, C. truncatum, and F. oxysporum on chili fruits.20 S. ahygroscopicus was used for the post-harvest preservation of wax apples and guavas, enhancing their shelf life by delaying quality decline and reducing decay incidences.21 S. murinus JKTJ-3 significantly inhibited Pythium aphanidermatum, which causes damping-off in watermelon, and exhibited broad antifungal activity against various fungi, including Rhizoctonia solani, Colletotrichum gloeosporioides, Botrytis cinerea, Stagonosporopsis cucurbitacearum, Verticillium dahlia, Phomopsis vexans, Fusarium oxysporum f.sp. hiveum, Leptosphaeria biglobosa, Phomopsis asparagi, Fusarium solani, Colletotrichum capsic, and Sclerotiorum.19 More recently, S. distatochromogenes XT34 effectively controlled banana anthracnose caused by C. musae and maintained fruit quality during storage.22 Metabolites from S. murinus THV12 inhibited effectively against Candida albicans,23 while S. murinus SPL-1 showed effectiveness against Trichomonas vaginali.24 The antifungal effects of Streptomyces spp. are believed to result from the presence of biosynthetic gene clusters (BGCs), which are responsible for antibiotics production, such as non-ribosomal peptide synthetase (NRPS), polyketide synthase (PKS), and other ketide synthases.25
Previously, we have demonstrated that Streptomyces murinus NARZ is a potential biological control agent against Penicillium digitatum and Penicillium italicum on oranges,26
C. truncatum causing anthracnose on dragon fruits.27 This study aimed to isolate and identify the fungus causing anthracnose in tomatoes and evaluate the effects of culture filtrate from S. murinus NARZ in controlling C. scovillei growth on PDA plates and anthracnose disease on tomatoes. Furthermore, the putative genes encoding for its antibiotic production were also investigated.
Materials
Tomatoes with typical anthracnose lesions were collected from local markets in Hue City, Vietnam. The fruits were contained in plastic bags with detailed information (collected date, places) and transferred to the laboratory for Colletotrichum sp. isolation.
Fresh, healthy tomatoes were purchased from markets in Hue City, Vietnam, and tomatoes were purchased at pink ripeness (equivalent to 2/3 ripeness) at the ripening stage and transferred to the laboratory.
Streptomyces murinus NARZ strain (PQ680203) was deposited at the Faculty of Engineering and Technology Laboratory, University of Agriculture and Forestry, Hue University, Vietnam.
Methods
Isolation and identification of Colletotrichum sp. causing anthracnose on tomatoes
Colletotrichum sp. was isolated from anthracnose lesions on tomatoes according to the method of Shahriar et al.7 The fruits were surface disinfected as follows: rinsed under running tap water, surface disinfected with 70% ethanol for 3 min, soaked in 1% sodium hypochlorite for 3 min, and finally washed three times with sterilized distilled water. From the edge of the developing lesion, 2 mm2 tissues were cut using a sterilized knife, and each piece was placed onto the center of Petri plates containing PDA (Potato dextrose agar) medium. These plates were incubated at 25 ± 2 °C for seven days. The resulting fungi were purified by transferring hyphal tips on new PDA plates. Pure colonies were preliminarily examined for Colletotrichum species based on their colony and spore morphological characteristics described in previous studies.28-30
The suspected Colletotrichum sp. was sequenced based on the ITS region by the DNA sequencing company (Can Tho, Vietnam). The resulting sequence was compared with the databases on NCBI (National Center for Biotechnology Information) using BLASTn to determine its taxonomy based on similarity. A phylogenetic tree was built based on resulted sequences and reference strains deposited on GenBank using MEGA1131 to identify species classification. The identified strain was artificially re-inoculated into healthy tomatoes to test its pathogenicity. Furthermore, the fungus was reisolated from the developing lesion to test its morphologies compared to the inoculated strain.32
Culture filtrate (CF) preparation
S. murinus NARZ was cultured in ISP4 medium (International Streptomyces Project 4) on a shaker with 180 rpm at a temperature of 28 ± 2 °C for seven days. The culture was centrifuged at 10,000 rpm for 10 minutes at 4 °C. The supernatant was collected and passed through a 0.2 µm syringe Millipore filter. The obtained cell-free filtrate was used for antifungal examinations.33
Antifungal effect of CF from S. murinus NARZ against C. scovillei C3 growth
The CF was added to the PDA plates at 10%, 20%, 30%, 40% and 50% (v/v) concentrations. Plates without CF addition and plates containing nystatin (100 µg/mL) were negative and positive controls, respectively. From seven-day C. scovillei C3 plates, 2 mm2 pieces were removed from the colony edge using a sterilized knife and placed onto the center of the prepared plates. These plates were incubated at 28 ± 2 °C, and colony diameters were recorded for seven days. Each concentration was repeated thrice. The inhibitory effect of the CF against C. scovillei C3 growth was assessed based on the Percentage Inhibition of Radial Growth (PIRG%). The PIRG was calculated as PIRG = (R1 – R2) * 100/R1 whereas R1 and R2 were the diameter of the colonies in the control and tested plates, respectively (mm).34 The concentration inhibited 50% of the growth of C. scovillei C3 (EC50 value) calculated based on the logarithmic regression equation generated from the PIRGs and CF concentrations graph.35
Heat stability of CF from Streptomyces murinus NARZ against C. scovillei
The CF of S. murinus NARZ was heated at temperature ranges of 30 °C, 50 °C, 70 °C, and 90 °C for 30 minutes and then separately added to the PDA medium at a concentration of 50%. A 2 mm2 piece of seven-day-old C. scovillei C3 was placed at the center of the plates. The fungal diameters were recorded after 7 days incubated at 28 ± 2 °C. The PIRG (%) was calculated using the above equation. Plates without CF addition were controls.33 Each heat treatment was repeated three times.
C. scovillei C3 spore suspension preparation and artificial inoculation
The spore suspension was prepared from plates containing C. scovillei C3 after seven days incubated at 28 ± 2 °C on PDA medium plates. These plates were flooded with sterilized distilled water, and the colony’s surface was scratched using an inoculating loop. The spore suspension was collected through a sterilized four-layer cheesecloth. Spore concentration was adjusted to 105 spore/µL using a hemocytometer for artificial inoculation.36
Healthy tomatoes were surface cleaned under running tap water; then they were dipped in ethanol 70% for 10 min and then in sodium hypochlorite 2% for 3 min. Finally, they were washed with sterilized distilled water three times and dried using sterilized tissue. On the equator of each fruit, a wound (2 x 2 x 2 mm in width, length, and depth) was created using a sterilized knife. Each tomato was artificially inoculated with C. scovillei by pipetting 10 µL prepared spore suspension onto the wounds.37
Effect of the CF on anthracnose development in tomatoes
Two treatment methods were conducted to evaluate the effect of the CF collected from S. murinus NARZ against anthracnose caused by C. scovillei C3 on tomatoes. The first treatment was tomato fruits, which were inoculated with 10 µL C. scovillei C3 prepared spore suspension for 24 hours and then sprayed with the CF using an aerosol sprayer (post-treatment). The second was tomato fruits were sprayed with the CF for 24 hours and inoculated with 10 µL C. scovillei C3 spore suspension afterward (pre-treatment). The CF concentrations in both treatments were 10%, 20%, 30%, 40%, and 50% (v/v). The two treatments were conducted in parallel and repeated three times for each. Sterilized distilled water and Chlorothalonil 0.026% were used as negative and positive controls, respectively. The control fruits were treated in the same way as described.38
All treated fruits were individually placed in plastic boxes (173 x 118 x 70 mm) disinfected with ethanol 70% and containing moistened sterilized tissue for seven days at 28 ± 2 °C. Disease incidence and lesion diameters were recorded every two days. Disease incidence was calculated using the formula DI = number of infected wounds * 100/total number of wounds. The PIRGs (%) were calculated using the formula PIRG = (R1 – R2) * 100/R1, whereas R1 and R2 were the diameter of lesions in the control and tested fruits, respectively (mm).38
Determination of genes encoding for antibiotics production of S. murinus NARZ
Degenerate primers were designed to recognize conserved regions in PKS-I ketosynthase (KS), NRPS adenylation (AD), PKS-II ketosynthase alpha (KSa), and ketosynthase beta (KSb) that are responsible for antibiotic biosynthetic of S. murinus NARZ strains (Table 1).
Table (1):
Primer sequences used to amplify putative genes emcpdomg for antibiotic production
| Genes | Length (bp) | Primers | Primer sequences (5’ to 3’) |
|---|---|---|---|
| Type I polyketide synthase β-ketoacyl synthase (KS) domain fragments | 670 | KS-F | CCS CAG SAG CGC STS YTS CTS GA |
| KS-R | GTS CCS GTS CCG TGS GYS TCS A | ||
| Type II polyketide synthases KSα and KSβ domain fragments | 800-900 | KSα | TSG RCT ACR TCA ACG GSC ACG G |
| KSβ | TAC SAG TCS WTC GCC TGG TTC | ||
| Type II polyketide synthases KSα domain fragments | 613 | KS1-F | TSG CST GCT TGG AYG CSA TC |
| KS1-R | TGG AAN CCG CCG AAB CCT CT | ||
| NRPS Adenylation domain (AD) fragments | 700 | A3F | GCS TAC SYS ATS TAC ACS TCS GG |
| A7R | SAS GTC VCC SGT SCG GTA S |
The 25 µL reaction mixture for PCR contained 12.5 µL Master Mix (2X) (Bioline, England), 2 µL forward primer (10 µM), 2 µL reverse primer (10 µM), 7.5 µL DEPC water, and 1 µL 140 ng DNA template. Reaction mixtures were prepared for each DNA sample. The primer pairs KS-F and KS-R were used to amplify the genes that synthesize the polyketide synthase type I b-ketoacyl synthase (KS) region fragments. The PCR cycle consisted of an initial denaturation at 95 °C for 15 min, one cycle of 95 °C for 1 min, 65 °C for 1 min, and 72 °C for 1 min, followed by 35 cycles of 95 °C for 1 min, 62 °C for 1 min, and 72 °C for 1 min, with a final extension of 10 min at 72 °C. The PKS-II ketoacyl synthase alpha (KSa) and ketoacyl synthase beta (KSb) regions were amplified using two sets of primers: KSa and KSb and KS1-F and KS1-R. The PCR cycle was as follows: initial denaturation at 95 °C for 5 min; 40 cycles of 95 °C for 1 min, 58 °C for 1 min, and 72 °C for 2 min; and a final extension for 10 min at 72 °C. Meanwhile, to amplify AD gene fragments, A3F and A7R primer set was used. PCR conditions included an initial denaturation at 95 °C for 5 min, followed by 40 cycles at 95 °C for 30 s, 59 °C for 1.5 min, and 72 °C for 1 min, with a final extension for 10 min at 72 °C. PCR amplicons were analyzed using a 2% (w/v) agarose gel to determine the presence of the target genes.25
Statistical analysis
All experiments were performed in triplicates. Statistical analysis was performed using the SPSS statistics program 20 (IBM, USA) based on average values of three replicates. One-way ANOVA (Duncan’s Multiple Range Test) was used to evaluate the differences in the PIRG% and lesion diameters at various concentrations of CF. The paired-sample t-test was used to compare the efficiency of the two treatments of CF on anthracnose lesion diameters in tomatoes.
Fungal isolation and identification
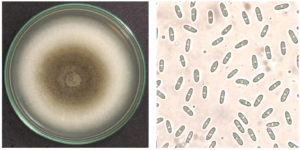
An isolate named C3 was purified from tomato fruits, which showed anthracnose symptoms, and its morphologies were examined. The mycelium was grayish-white to grayish-green, creating concentric circles. The mycelium was thick and fluffy, with abundant small orange and slimy spores. The back of the plate was light orange, and black spots formed on it. The diameter of the colony after 7 days was 47 mm. The spores (40 x magnification) were transparent and cylindrical, with blunt ends and straight shapes, showing oil droplets inside (Figure 1).
Our observation was similar to the previous description of Colletotrichum species. According to the description of Damm et al. an isolate with cottony hyphae, pale white to pale gray, with an olive gray center, and cylindrical transparent spores with a slightly pointed tip and a rounded tip belonged to the acutatum complex.29 Furthermore, Kanto et al. also described the colony on PDA of C. scovillei as gray or pale gray to pale orange, sometimes with black spots on the back. However, traditional taxonomy based on morphological characteristics is unreliable because Colletotrichum species are significantly affected by cultural conditions such as light, temperature, etc.39 Therefore, molecular analysis based on the ITS region of isolate C3 was conducted to identify its taxonomy. The BLASTn result exhibited isolate C3 was 99.11% similar to C. scovillei C212 (MK327142.1). Furthermore, phylogenetic analysis indicated that isolate C3 had the nearest relationship with C. scovillei LD242 (LC488852.1) (Figure 2).
Figure 2. A phylogenetic tree was built using ITS region sequences of isolate C3 and reference sequences downloaded from GenBank
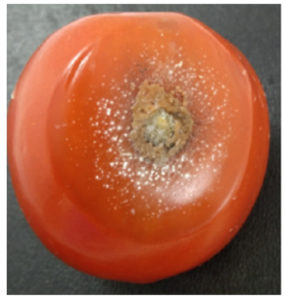
Therefore, it can be concluded that isolate C3 was C. scovillei and named C. scovillei C3. C. scovillei C3 was re-infected in tomato fruits to test its pathogenicity. The inoculated tomatoes showed typical symptoms of anthracnose. The lesions were circular, slightly sunken, and waterlogged, with orange sclerotia at the inoculated site, white mycelium formed around them and spread over the entire fruit surface. These symptoms were similar to the anthracnose lesions on the initially infected tomatoes (Figure 3). Furthermore, fungi re-isolated from the lesion had identical morphological characteristics with C. scovillei C3.
Effect of the CF against the growth of C. scovillei C3
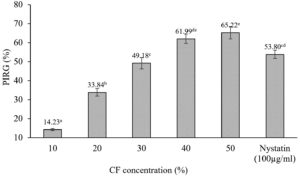
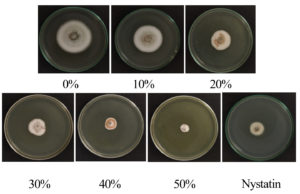
Colony diameters of C. scovillei C3 were significantly smaller in the plates containing the CF of S. murinus NARZ than those of control plates without CF addition. The PIRGs (%) increased as the CF concentration was raised (Figure 4), gradually rising from 14.23% to 65.52%. The concentration inhibited 50% of the growth of C. scovillei C3 (EC50 value) was 30.40%, calculated based on a regression equation y = 33.102; ln(x) – 63.022 (R2 = 0.9907). Nystatin (100 µg/mL) showed 53.8% PIRG against C. scovillei C3, lower than the efficiency of 40% CF, with 61.99% PIRG. The presence of CF not only reduced the colony diameters of C. scovillei C3 but also altered its morphology. The colonies grew slower, and mycelia shrank, forming an orange layer in the center (Figure 5).
Figure 5. Colony morphologies of C. scovillei C3 on PDA medium with various concentrations of CF from S. murinus NARZ and controls after seven days
Heat stability of the CF against C. scovillei C3
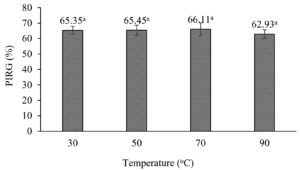
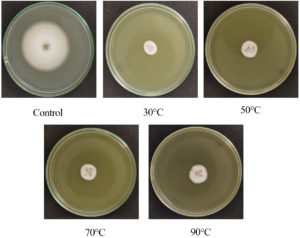
The efficiency of the CF from S. murinus NARZ against C. scovillei C3 remained highly stable when subjected to heat treatment, with PIRGs fluctuating between 62.93% and 65.35%. After being treated at 90 °C for 15 minutes, the CF’s efficiency against C. scovillei C3 was 62.93%, showing a 2.45% decrease compared to the 30 °C treatment. However, statistical analysis revealed no significant differences in PIRGs across all treatments (p > 0.05) (Figures 6 and 7).
Figure 6. Effect of heat treatment on antifungal activities of CF from S. murinus NARZ against C. scovillei C3
Figure 7. Colony diameters of C. scovillei C3 after 7 days on PDA medium with 50% CF treated at various temperatures
Effect of the CF from S. murinus NARZ against anthracnose disease caused by C. scovillei C3 on tomatoes
Table 2 indicates the positive efficacy of the CF from S. murinus NARZ against anthracnose disease caused by C. scovillei C3 on tomatoes under two treatments. However, the two treatments differed in disease incidences and lesion diameters (Tables 2 and 3).
Table (2):
Anthracnose disease incidence (%) in tomatoes under the two treatments
| CF (%) | Incubation time (days) | |||||
|---|---|---|---|---|---|---|
| Pre-treatment | Post-treatment | |||||
| 3 | 5 | 7 | 3 | 5 | 7 | |
| 0 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 10 | 66.67 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 20 | – | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 30 | – | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 40 | – | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 50 | – | 66.67 | 83.33 | 100.00 | 100.00 | 100.00 |
| Chlorothalonil (0.026%) | – | 83.33 | 100.00 | 100.00 | 100.00 | 100.00 |
(-): no lesions.
Table (3):
Effect of CF from S. murinus NARZ on anthracnose lesion diameter (mm) on tomatoes
| CFS (%) | Incubation time (days) | |||||
|---|---|---|---|---|---|---|
| 3 | 5 | 7 | ||||
| Post-treatment | Pre-treatment | Post-treatment | Pre-treatment | Post-treatment | Pre-treatment | |
| 0 (Control) | 12.23 ± 0.68d | 11.53 ± 0.48b | 14.95 ± 3.92c | 16.50 ± 1.42d | 22.47 ± 1.91d | 23.87 ± 2.61e |
| 10 | 10.17 ± 2.47*d | 4.13 ± 2.80a | 13.73 ± 2.11c | 11.63 ± 0.76c | 19.13 ± 1.52*d | 17.80 ± 0.62d |
| 20 | 6.33 ± 1.53c | – | 10.17 ± 1.42bc | 9.37 ± 2.05bc | 16.77 ± 1.28c | 15.20 ± 1.39d |
| 30 | 5.34 ± 0.58bc | – | 8.50 ± 2.14ab | 8.07 ± 1.12ab | 11.03 ± 1.68b | 10.40 ± 0.82c |
| 40 | 4.67 ± 0.78bc | – | 6.90 ± 2.20*a | 5.50 ± 0.87ab | 9.80 ± 0.69b | 8.57 ± 1.60bc |
| 50 | 3.40 ± 0.53b | – | 6.27 ± 3.84*a | 3.73 ± 0.32a | 8.73 ± 1.79*a | 5.40 ± 0.36a |
| Chlorothalonil (0.026%) | – | – | 5.10 ± 1.45a | 4.80 ± 1.69a | 7.63 ± 1.09a | 6.39 ± 1.25ab |
(-): no lesions.
In the same rows, the (*) following the SD values indicate a statistically significant difference at the significance level of p ≤ 0.05. In the same columns, different letters following the SD values indicate a statistically significant difference at the significance level of p ≤ 0.05.
After three days of incubation, all post-treated tomatoes exhibited anthracnose symptoms, resulting in a 100% disease incidence. In contrast, pre-treated tomatoes sprayed with over 20% CF before artificial inoculation showed no signs of anthracnose. Only the tomatoes treated with 10% CF exhibited a disease incidence of 66.67%. After five days, lesions developed at all inoculated wounds on tomatoes treated with 20%, 30%, and 40% CF, while those treated with 50% CF and Chlorothalonil (0.026%) showed disease incidences of 66.67% and 83.33%, respectively. Notably, after seven days, tomatoes treated with 50% CF had a disease incidence of 83.33%, while all other treatments reached 100%.
The CF treatments helped reduce tomato lesion diameters in both methods (Table 3). After three days of incubation, the lesion diameters reduced from 12.23 mm to 3.40 mm with 0% (control) to 50% CF in the post-treatment method. Lesions on tomatoes treated with 10% CF in pre-treatment were significantly larger than those in the post-treated tomatoes, 10.17 mm compared to 4.13 mm. After five days of treatment, there were no differences in lesion diameters in tomatoes treated with 10%, 20%, and 30% CF between the two treatments (p ≤ 0.05). However, at 40% and 50% CF, lesions in pre-treated tomatoes were smaller than those of post-treated tomatoes. A significant difference in lesion diameters between the two treatments was observed only at a 50% CF concentration after seven days, with lesions measuring 5.40 mm in the pre-treatment and 8.73 mm in the post-treatment tomatoes (p ≤ 0.05). Furthermore, the growth of lesion diameters in tomatoes treated with higher CF concentrations was slower than those treated with lower concentrations during incubation time. At 10% CF in the post-treatment method, lesion diameters increased from 10.17 mm after three days to 19.13 mm after seven days, while tomatoes treated with 50% CF showed a slower increase from 3.40 mm after three days to 7.63 mm after seven days. Chlorothalonil (0.026%) demonstrated equivalent efficiency to 50% CF in preventing the progression of anthracnose.
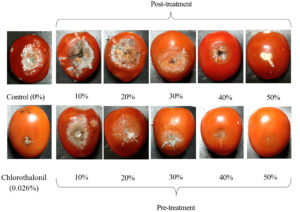
Disease symptoms of post-treatment tomatoes were more severe than those in the pre-treatment at the same concentration of CF (Figure 8). The fruits were sunken, covered by the mycelium of C. scovillei C3 and orange conidia, with large cracks in fruits treated with 10%, 20%, and 30% CF concentrations and negative controls. Fruits treated with 50% CF had white mycelium with small lesions.
Figure 8. Anthracnose symptoms caused by C. scovillei C3 on tomatoes treated with CF from S. murinus NARZ at various concentrations in two treatments after seven days
Detection of genes encoding for antibiotic biosynthesis of S. murinus NARZ
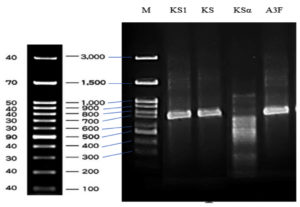
Polyketide synthase type I (PKS-I), polyketide synthase type II (PKS-II), and non-ribosomal peptide synthase (NRPS) genes were identified through PCR analysis. The size of fragments varied from 700 to 850 bp (Figure 9), reaching the expected length when using their respective primer sets. The PKS-I gene was amplified using the KS-F and KS-R primer pair, with 700 to 800 bp length. The PKS-II gene was amplified using the KS1-F and KS1-R primer pair, yielding an approximate 800 bp fragment. The NRPS gene was amplified with the A3F and A7R primer pair, producing a fragment of approximately 800-850 bp. The KSa and KSb primer pairs did not amplify any PKS-II gene fragments, suggesting either the absence of these PKS-II genes or the low specificity of the primers used. The presence of the polyketide synthase type I (PKS-I), polyketide synthase type II (PKS-II), and non-ribosomal peptide synthase (NRPS) genes contributed to explain the antibiotic biosynthesis of S. murinus NARZ.
Figure 9. Agarose gel electrophoresis of PCR amplicons of type II polyketide synthases KSα domain fragments (Lane KS1), type I polyketide synthase β-ketoacyl synthase (KS) domain fragments (Lane KS), type II polyketide synthases KSα and KSβ domain fragments (Lane KSα), and non-ribosomal peptide synthase (NRPS) genes (Lane A3F)
It is essential to understand pathogen-causing diseases to have a proper controlling strategy. Therefore, the isolation of pathogen-causing anthracnose on tomatoes was necessary. Many Colletotrichum species have been recorded as causative agents in tomatoes, including C. C. gloeosporioides,4 C. gossypii,5 C. acutatum,6 C. truncatum,7 C. scovillei,8 C. coccodes,9 and C. nymphaeae.10 However, anthracnose caused by C. scovillei on tomatoes has been rarely documented. This is the first report on C. scovillei causing anthracnose on tomatoes in Vietnam.
During the disease cycle, C. scovillei spores adhere to the surface and produce germ tubes (appressorium), specialized cells that invade host cells. After invading the epidermal cells, the fungus develops anthracnose lesions with typical sunken symptoms and pinkish-green acervuli containing large spores, essential for widespread infection. Polycyclic infection of the fungus contributes to significant disease during the growing season, causing severe economic losses.40
The antifungal activities of S. murinus species have been reported in previous studies, showing good effectiveness against P. tabacinum, which causes root rot disease in White Lupine,41 Pythium, causing damping-off disease on watermelon,19 Candida albicans,23 and Trichomonas vaginali.24 However, this study provides new insights into the antifungal activity of S. murinus NARZ, highlighting its beneficial effects in controlling tomato anthracnose caused by C. scovillei C3.
In our study, CF from S. murinus NARZ significantly reduced the growth of C. scovillei C3 in the PDA medium, with PIRG reaching up to 65.22% at 50% CF. CF of S. katrae had 95.00% inhibitory effect on the growth of C. musae mycelium on a PDA medium.42 Meanwhile, S. tuirus showed antagonistic activity against chili rot pathogens, C. scovillei, C. truncatum, and F. oxysporum.20 The authors explained the inhibitory activity was often associated with disrupted mycelial supra-molecular organization. Culture extracts of Streptomyces spp. induced mycelial shriveling, exhibited fungal cell wall lytic activity, and inhibited spore growth.1,43,44
The antifungal properties of S. murinus have been attributed to the production of antibiotics such as Actinomycin X (Act-X), Actinomycin D (Act-D), Dactinomycin, Pentamycin, and other antifungal compounds.19,23,24,45-47
Although the use of Act-D in medicine is widely known, its use in agriculture is rarely documented. 35 Act-D synthesized by Streptomyces spp. showed effective in controlling plant pathogens and inhibiting the growth of Fusarium spp. causing wilt disease,48 B. cinerea, which causes gray mold in tomatoes.35 Additionally, Act-D produced by Streptomyces sp. Tc022 exhibited antifungal activity against C. musae with a minimum inhibitory concentration of 10 mg/ml.49 Studies explored the mechanisms by which Actinomycin exerts its effects, such as damaging fungal cell membranes through a cleavage mechanism50 and generating reactive oxygen species that harm fungal membranes.51 Pentamycin, a polyene antifungal antibiotic produced by Streptomyces species, disrupts fungal cell membranes by binding to ergosterol, leading to cell death.24
On the other hand, the antifungal mechanism of Streptomyces spp. is believed to involve the production of cell-wall-degrading enzymes, such as β-1,3-glucanase and chitinase, which break down the cell walls of phytopathogenic fungi composed of chitins and glucans. Another secondary compound involved in this process is siderophores. S. globisporus F8 and S. praecox R7 have been shown to enhance lipoxygenase and phenylalanine ammonia-lyase activity in tomatoes, potentially triggering jasmonic acid and phenylpropanoid signaling pathways, thus activating a defense response in tomatoes against R. solani.52
Interestingly, our findings showed that the inhibitory effect of the culture filtrate (CF) from S. murinus NARZ against C. scovillei C3 was stable within a temperature range of 30-90 °C, the PIRGs remaining from 62.93% to 65.35%. Other studies also explained the inhibition of fungal growth by Streptomyces culture filtrate was associated with the presence of thermostable antifungal compounds rather than hydrolytic enzymes.53,54 For example, the CF from the millet broth of S. deccanensis QY-3 demonstrated exceptional stability under thermal (20 °C-100 °C) and pH (2-10) conditions, effectively damaging the hyphae and inhibiting the germination of C. gloeosporioides.42 Similarly, Wonglom et al. showed that the culture fluid of S. angustmyceticus NR8-2 displayed antifungal activity against Colletotrichum spp. and Curvularia lunata, maintaining stable antifungal properties even when exposed to temperatures of 28 °C, 50 °C, and 100 °C.55 Our results suggested that the composition of the CF from S. murinus NARZ contained antibiotic compounds, which play a key role in antifungal activities, rather than enzymes that break down fungal cell membranes or other heat-sensitive compounds.
The application of CF obtained from S. murinus NARZ indicated the positive efficacy of preventing anthracnose disease caused by C. scovillei C3 in tomatoes. Pre-treatment tomatoes showed lower disease incidence than post-treatment tomatoes, with lesions measuring 5.40 mm in pre-treatment and 8.73 mm in post-treatment after seven days. At 50% concentration, CF was as effective as Chlorothalonil (100 µg/mL). These results were consistent with the report of Lian et al. Tomato samples 24 h pre-treated with 10 µL cell suspension of S. pratensis LMM15 (106 cells/mL) before artificial infection (pre-treatment) showed 76.64% antifungal efficacy against gray mold caused by B. cinerea. Meanwhile, procymidone pre-treated fruits had higher efficacy, reaching 83.18%. Compared with 24 h post-treated fruits, there were no differences in antifungal efficacy between cell suspension of S. pratensis LMM15 and Procymidone treatments, with 49.32% and 50.26%, respectively.36 Thus, it can be concluded that fruits treated with antifungal compounds before the pathogen infection were more effective and efficient than treatments applied after the pathogen infection.
The antifungal mechanisms of Streptomyces spp. in inhibiting fungal growth on fruit include the suppression of mycelial growth and spore production, disruption of fungal cell walls and membranes, and interference with mitochondrial function.56 Spore germination is key in the early stages of fungal penetration and disease progression in fruit.43 Prevention of spore adhesion results in fewer spore germination and forming an adsorbent layer on the plant surface, leading to a lower disease incidence. Furthermore, the enhanced inhibition observed with pre-treatment fruits can also be linked to the protease production of Streptomyces spp., which reduces sporulation, adhesion, and the formation of adsorbent layers. Protease preparations from S. phaeourepureus Expro 138 effectively reduced sporulation and prevented anthracnose disease in tomato plants.57 Moreover, treatment with Streptomyces spp. can activate plant defense responses. Variations in antifungal activity may be influenced by factors such as the growth conditions of the microorganisms (e.g., carbon sources, nutrients, pH) and the inoculation method.43
Biosynthetic gene clusters (BGCs) are groups of genes responsible for producing bioactive compounds, such as antibacterial and antifungal agents.24 Streptomyces species have demonstrated that their genomes possess abundant biosynthetic gene clusters (BGCs).25 In our study, the presence of the polyketide synthase type I (PKS-I), polyketide synthase type II (PKS-II), and non-ribosomal peptide synthase (NRPS) genes in S. murinus NARZ contributed to explaining its antifungal action.
The S. murinus SPC1 strain, isolated from fox tail palm seeds, showed a potent antifungal effect against Fusarium oxysporum f. sp. palmarum, Ganoderma zonatum, and Thielaviopsis paradoxa. Genome analysis of S. murinus SPC1 revealed that this strain possesses a complete secondary metabolite cluster, 37 secondary metabolite BGCs, involved in producing the antifungal compound pentamycin.58 Interestingly, a genome comparison study compared the genome of seven different S. murinus strains to the genome of S. murinus SPC1 revealed the presence of pentamycin-producing BGCs in all tested S. murinus strains.24
Antifungal compounds, including Actinomycin D, pentamycin, desferrioxamine E, and cinnabaramide A, were identified in the secondary metabolites of the S. murinus THV12 strain using MS/MS analysis. The S. murinus THV12 genome analysis revealed that the strain harbored 47 secondary metabolite biosynthetic gene clusters. Type 1 PKS gene clusters encode pentamycin production, while NRPS gene clusters are associated with actinomycin biosynthesis. Both type 1 PKS and NRPS gene clusters are responsible for cinnabaramide A synthesis, and siderophore gene clusters are related to desferrioxamin E secretion.23
The study by Tenebro et al. on the analysis of 19 Streptomyces species from marine sediments in the Philippines revealed variability among species regarding the presence of antibiotic biosynthesis genes. The results showed that all 19 Streptomyces species contained the NRPS gene based on amplifying the adenylation (AD) domain. Still, there were differences in the PKS-I and PKS-II genes across species. Most species contained the PKS-I gene, with only the Streptomyces sp. DSD3025 strain lacking PKS-I. Strains closely related to S. mutabilis did not have the conserved KS domain when amplified with the KS-F and KS-R primers. Diversity among species was particularly evident in the PKS-II domain. Only four species, S. parvulus, S. griseorubens, S. carpaticus, and S. xiamenensis, contained the KSa and KSb domains of PKS-II, with a fragment size of 500-600 bp. Strains, such as S. kunmingenesis, S. sedi, and Streptomyces sp. DSD3025, lacked all PKS-II domains in their genome. The variability in PKS genes reflects differences in metabolite production, explaining species’ observed variation in antibiotic activity.25
Based on the above analysis, we supposed that the S. murinus NARZ strain produced antifungal compounds, including pentamycin, actinomycin D, and other metabolites, through PKS-I, PKS-II, and NRPS gene clusters in its genome.
This study demonstrated that S. murinus NARZ effectively inhibited C. scovillei C3, the pathogen causing anthracnose in tomatoes. Its culture filtrate was highly thermostable, making it suitable for practical use. The application of CF on tomatoes infected with C. scovillei C3 suggests that antifungal treatments should be applied shortly after harvest to prevent pathogen invasion. This is the first study to explore the biocontrol potential of S. murinus isolated from soil against C. scovillei, which was first recorded as a pathogen responsible for anthracnose on tomatoes in Vietnam.
ACKNOWLEDGMENTS
The authors would like to thank Prof. Dr. Markus Kalkum of the City of Hope, California, USA, for his assistance with this manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HTN and TTTN designed the experiments. TLL, TDHN, and TTTT carried out the experiments and performed data analysis. HTN wrote the manuscript. TTTN revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This research was funded by the Hue University of Agriculture and Forestry, Hue University, under Project NCM.׀HNL.2021-04. This research was also partly funded by the International Foundation for Science, under Project number I1-F-6276-1.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Li J, Liu F, Wu Y, et al. Evaluation of nutritional composition, biochemical, and quality attributes of different varieties of tomato (Solanum lycopersicum L.). J Food Compos Anal. 2024;132(106384).
Crossref - Sopialena S, Subiono T, Rosyidin AU, Tantiani D. Control of antracnose disease in tomato (Solanum lycopersicum) using endophytic fungi. KnE Life Sciences. 2022;7(3):393-408.
Crossref - Saini TJ, Gupta SG, Anandalakshmi R. Detection of tomato anthracnose caused by Colletotrichum truncatum in India. Australas Plant Dis Notes. 2017;12(1):1-3.
Crossref - Barad S, Sela N, Dubey AK, et al. Differential gene expression in tomato fruit and Colletotrichum gloeosporioides during colonization of the RNAi-SlPH tomato line with reduced fruit acidity and higher pH. BMC Genomics. 2017;18(1):579.
Crossref - Nawaz HH, Anam U, He Q, Liu WB, Miao W. First report of anthracnose caused by Colletotrichum gossypii on tomato in Hainan, China. Plant Dis. 2018;103(1):161.
Crossref - Zivkovic S, Stojanovic S, Ivanovic Z, et al. Morphological and molecular identification of Colletotrichum acutatum from tomato fruit. Pesticidi i fitomedicina. 2010;25(3):231-239.
Crossref - Shahriar SA, Husna A, Paul TT, et al. Colletotrichum truncatum causing anthracnose of tomato (Solanum lycopersicum L.) in Malaysia. Microorganisms. 2023;11(1):226.
Crossref - Thao LD, Choi H, Choi Y, Mageswari A, Lee D, Hong SB. Re-identification of Colletotrichum acutatum species complex in Korea and their host plants. Plant Pathol J. 2023;39(4):384-396.
Crossref - Hassine M, Aydi-Ben-Abdallah R, Jabnoun-Khireddine H, Daami-Remadi M. Soil-borne and compost-borne Penicillium sp. and Gliocladium spp. as potential microbial biocontrol agents for the suppression of anthracnose-induced decay on tomato fruits. Egypt J Biol Pest Control. 2022;32(1):20.
Crossref - Chechi A, Stahlecker J, Zhang M, Luo CX, Schnabel G. First Report of Colletotrichum fioriniae and C. nymphaeae causing anthracnose on cherry tomatoes in South Carolina. Plant Dis. 2019;103(5):1042.
Crossref - Fabian ML, Zhang C, Sun J, et al. Steroidal glycoalkaloids contribute to anthracnose resistance in Solanum lycopersicum. J Exp Bot. 2023;74(12):3700-3713.
Crossref - Wani AH. An overview of the fungal rot of tomato. Mycopath. 2011;9(1):33-38.
- Wakene DM, Sharew T. A comprehensive review of tomato post-harvest losses: Understanding impacts and contributing factors in Ethiopia. Asian Science Bulletin. 2024;2(4):525-535.
Crossref - Taha NA, Elsharkawy MM, Shoughy AA, El-Kazzaz MK, Khedr AA. Biological control of postharvest tomato fruit rots using Bacillus spp. and Pseudomonas spp. Egypt J Biol Pest Control. 2023;33(1):106.
Crossref - Sonowal S, Konwar AN, Hazarika SN, Gurumayum S, Borah JC, Thakur D. Unveiling the biocontrol potential of Streptomyces sp. OR02 against Rhizoctonia solani in tomato fruit. Physiol Mol Plant Pathol. 2024;134:102425.
Crossref - Kim HJ, Lee EJ, Park SH, Lee HS, Chung N. Biological control of anthracnose (Colletotrichum gloeosporioides) in pepper and cherry tomato by Streptomyces sp. A1022. J Agric Sci. 2014;6(2):54.
Crossref - Khan S, Srivastava S, Karnwal A, Malik T. Streptomyces as a promising biological control agents for plant pathogens. Front Microbiol. 2023;14:1285543.
Crossref - Evangelista-Martinez Z, Contreras-Leal EA, Corona-Pedraza LF, Gastelum-Martinez E. Biocontrol potential of Streptomyces sp. CACIS-1.5CA against phytopathogenic fungi causing postharvest fruit diseases. Egypt J Biol Pest Control. 2020;30(117).
Crossref - Ge M, Cai X, Wang D, et al. Efficacy of Streptomyces murinus JKTJ-3 in suppression of Pythium damping-off of watermelon. Microorganisms. 2023;11(6):1360.
Crossref - Renuka R, Prabakar K, Anandham R, et al. Exploring the potentiality of native actinobacteria to combat the chilli fruit rot pathogens under post-harvest pathosystem. Life. 2023;13(2):426.
Crossref - Bai JL, Wang HH, Zhang JM, et al. Postharvest quality maintenance of wax apple and guava fruits by use of a fermented broth of an e-poly-L-lysine-producing Streptomyces strain. PLoS One. 2022;17(3):e0265457.
Crossref - Zeng W, Feng J, Wei Y, et al. Biocontrol efficiency and potential mechanism of Streptomyces distatochromogenes XT34 against postharvest anthracnose caused by Colletotrichum musae on banana fruit. Postharvest Biol Technol. 2024;212:112899.
Crossref - Das V, Chatterjee NS, Pushpakaran PU, Lalitha K V., Joseph TC. Exploration of natural product repository by combined genomics and metabolomics profiling of mangrove-derived Streptomyces murinus THV12 strain. Fermentation. 2023;9(6):576.
Crossref - Kurani D, Korisapati V, Mathew S, et al. Comparative genomics across Streptomyces murinus strains to study conservation of biosynthetic gene cluster producing study conservation of biosynthetic gene cluster producing antifungal compound, pentamycin. NSU Undergraduate Student Journal. 2024;2024(4):1-8.
- Tenebro CP, Trono DJVL, Vicera CVB, et al. Multiple strain analysis of Streptomyces species from Philippine marine sediments reveals intraspecies heterogeneity in antibiotic activities. Sci Rep. 2021;11(1):1-31.
Crossref - Nguyen TTT, Nguyen HT, Le TL, Nguyen TDH. Antifungal efficacy of Streptomyces murinus against postharvest pathogens Penicillium digitatum and Penicillium italicum in oranges. Asian J Agric Biol. 2025;(1):2024088.
Crossref - Nguyen TTT, Nguyen HT, Nguyen TDH, Le TL. Evaluation antifungal ability of Streptomyces sp. against Collectotrichum causing anthracnose on dragon fruit. Vietnam Journal of Agricultural Sciences. 2022;20(12):1591-1598.
- Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012;73:115-180.
Crossref - Damm U, Cannon PF, Woudenberg JHC, Crous PW. The Colletotrichum acutatum species complex. Stud Mycol. 2012;73(1):37-113.
Crossref - Zakaria L. Diversity of Colletotrichum species associated with anthracnose disease in tropical fruit crops – A review. Agriculture (Switzerland). 2021;11(4):297.
Crossref - Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022-3027.
Crossref - Ain N, Zainudin IM, Sattar MM. Characterization and pathological diversity of Colletotrichum species associated with anthracnose disease on mango in Peninsular Malaysia. Asian J Agric Biol. 2019;Special Issue:261-270.
- Jacob J, Rajendran RU, Priya SH, Purushothaman J, Saraswathy Amma DKBN. Enhanced antibacterial metabolite production through the application of statistical methodologies by a Streptomyces nogalater NIIST A30 isolated from Western Ghats forest soil. PLoS One. 2017;12(4):1-21.
Crossref - Rejon-Martinez GA, Rios-Muniz DE, Contreras-Leal EA, Evangelista-Martinez Z. Antagonist activity of Streptomyces sp. y20 against fungi causing diseases in plants and fruits. Trop Subtrop Agroecosystems. 2022;25(2):1-9.
Crossref - Yong D, Li Y, Gong K, et al. Biocontrol of strawberry gray mold caused by Botrytis cinerea with the termite associated Streptomyces sp. sdu1201 and actinomycin D. Front Microbiol. 2022;13:1051730.
Crossref - Tovar-Pedraza JM, Mora-Aguilera JA, Nava-Díaz C, et al. Distribution and pathogenicity of Colletotrichum species associated with mango anthracnose in Mexico. Plant Dis. 2020;104(1):137-146.
Crossref - Lian Q, Zhang J, Gan L, Ma Q, Zong Z, Wang Y. The biocontrol efficacy of Streptomyces pratensis LMM15 on Botrytis cinerea in tomato. Biomed Res Int. 2017;2017:9486794.
Crossref - Montesdeoca-Flores DT, Hernandez-Bolanos E, Leon-Barrios M, et al. Antifungal Activity of Streptomyces spp. Extracts in vitro and on post-harvest tomato fruits against plant pathogenic fungi. Horticulturae. 2023;9(12):1319.
Crossref - Kanto T, Uematsu S, Tsukamoto T, et al. Anthracnose of sweet pepper caused by Colletotrichum scovillei in Japan. J Gen Plant Pathol. 2014;80(1):73-78.
Crossref - Gao YY, Li XX, He LF, Li BX, Mu W, Liu F. Effect of pyrisoxazole on Colletotrichum scovillei infection and anthracnose on chili. Plant Dis. 2020;104(2):551-559.
Crossref - Youssef YA, El-Tarabily KA, Hussein AM. Plectosporium tabacinum root rot disease of white lupine (Lupinus termis Forsk.) and its biological control by Streptomyces species. J Phytopathol. 2001;149(1):29-33.
Crossref - Shu C, Chen Q, Pi L, Zhang D, Panhwar QA, Zhou E. Identification and antifungal activity analysis of two biocontrol antagonists to Colletotrichum musae. J Phytopathol. 2017;165(7-8):554-561.
Crossref - Gu L, Zhang K, Zhang N, Li X, Liu Z. Control of the rubber anthracnose fungus Colletotrichum gloeosporioides using culture filtrate extract from Streptomyces deccanensis QY-3. Antonie Van Leeuwenhoek. 2020;113(11):1573-1585.
Crossref - Li X, Jing T, Zhou D, et al. Biocontrol efficacy and possible mechanism of Streptomyces sp. H4 against postharvest anthracnose caused by Colletotrichum fragariae on strawberry fruit. Postharvest Biol Technol. 2021;175.
Crossref - Frommer, W. Zur Systematik der Actinomycin bildenden Streptomyceten. Archiv. Mikrobiol. 1959; 32:187-206.
Crossref - Piedra JLL, Cabeza JG, Pico AAR, Cornejo MAS. Identification and in vitro antifungal evaluation of Streptomyces sp. of desert soil against Colletotrichum sp. Biotecnologia Aplicada. 2019;36(2). http://elfosscientiae.cigb.edu.cu/Biotecnologia.asp
- Tunvongvinis T, Jaitrong W, Samung Y, Tanasupawat S, Phongsopitanun W. Diversity and antimicrobial activity of the tropical ant-derived actinomycetes isolated from Thailand. AIMS Microbiol. 2024;10(1):68-82.
Crossref - Toumatia O, Yekkour A, Goudjal Y, et al. Antifungal properties of an actinomycin D-producing strain, Streptomyces sp. IA1, isolated from a Saharan soil. J Basic Microbiol. 2015;55(2):221-228.
Crossref - Taechowisan T, Wanbanjob A, Tuntiwachwuttikul P, Taylor WC. Identification of Streptomyces sp. Tc022, an endophyte in Alpinia galanga, and the isolation of actinomycin D. Ann Microbiol. 2006;56(2):113-117.
Crossref - Zeng H, Feng PX, Wan CX. Antifungal effects of actinomycin D on Verticillium dahliae via a membrane-splitting mechanism. Nat Prod Res. 2019;33(12):1751-1755.
Crossref - Gao L, Kumaravel K, Xiong Q, et al. Actinomycins produced by endophyte Streptomyces sp. GLL-9 from navel orange plant exhibit high antimicrobial effect against Xanthomonas citri susp. citri and Penicillium italicum. Pest Manag Sci. 2023;79(11):4679-4693.
Crossref - Ebrahimi-Zarandi M, Bonjar GHS, Riseh RS, El-Shetehy M, Saadoun I, Barka EA. Exploring two Streptomyces species to control Rhizoctonia solani in tomato. Agronomy. 2021;11:1384. 2021;11(7):1384.
Crossref - Prapagdee B, Kuekulvong C, Mongkolsuk S. Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. Int J Biol Sci. 2008;4(5):330-337.
Crossref - Choudhary B, Nagpure A, Gupta RK. Biological control of toxigenic citrus and papaya-rotting fungi by Streptomyces violascens MT7 and its extracellular metabolites. J Basic Microbiol. 2015;55(12):1343-1356.
Crossref - Wonglom P, Suwannarach N, Lumyong S, Shin-ichi I, Matsui K, Sunpapao A. Streptomyces angustmyceticus NR8-2 as a potential microorganism for the biological control of leaf spots of Brassica rapa subsp. pekinensis caused by Colletotrichum sp. and Curvularia lunata. Biological Control. 2019;138:104046.
Crossref - Yuan G, Jia-Qi L, Ming-Jie X, et al. Antifungal volatile organic compounds from Streptomyces setonii WY228 control black spot disease of sweet potato. Appl Environ Microbiol. 2022;88(6):e02317-21.
Crossref - Palaniyandi SA, Yang SH, Suh JW. Extracellular proteases from Streptomyces phaeopurpureus ExPro138 inhibit spore adhesion, germination and appressorium formation in Colletotrichum coccodes. J Appl Microbiol. 2013;115(1):207-217.
Crossref - Dhillon B, Chakrabarti S. Draft genome sequence of Streptomyces murinus strain SPC1 with antimicrobial potential against fungal pathogens of palms. Microbiol Resour Announc. 2023;12(12):e0082623.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.