ISSN: 0973-7510
E-ISSN: 2581-690X
Mangrove forests are ecosystems with highly diverse microorganisms. We aimed to obtain bacterial isolates from mangrove water forests and determine their bacterial diversity and potential for decoloring dye waste. Bacteria were isolated from three sites in the Belawan mangrove forest of North Sumatra. Bacteria were isolated on nutrient agar media. Twenty-nine bacterial strains were isolated, purified, and molecularly identified at the species level. The isolates belonged to 8 genera consisting of 15 species: Aeromonas caviae, Aeromonas salmonicida, Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Enterobacter cloacae, Enterobacter kobei, Klebsiella pneumoniae, Myroides profundi, Providencia huaxiensis, Pseudomonas balearica, Pseudomonas hydrolytica, Pseudomonas khazarica, Shigella flexneri, and Shigella sonnei. Decolorization activity screening was performed in a solid mineral salt medium (MSM) containing 25%, 50%, 75%, and 100% dye waste. Based on colony diameter, the following five out of twenty-nine isolates showed a higher growth response: S. flexneri, B. cereus, A. salmonicida, K. pneumoniae, and E. cloacae. The waste decolorization ability of these five species was quantitatively determined in liquid MSM containing 50% dye waste. All isolates decolorized >15% dye after 15 days, and A. salmonicida and E. cloacae exhibited relatively better decolorization activity than that of the other three strains.
Bacterial Diversity, Belawan, Decolorization, Dye Waste, Mangrove Forest
Mangrove forests exist between the land and sea; therefore, they are often categorized as coastal, tidal, or mangrove forests.1 Indonesia’s mangrove forests cover approximately 3.3 million hectares, representing over 22% of the world’s total mangrove area.2 One of these mangrove forests is situated along the coast of Medan Belawan District, North Sumatra. These ecosystems are characterized by muddy substrates rich in organic matter, which serve as a vital nutrient source for various organisms.3 The productivity and extent of mangrove ecosystems are significantly shaped by environmental factors, including pH levels, dissolved oxygen, temperature, salinity, sediment thickness, organic matter content in sediments, and patterns of land use.4,5
Mangrove aquatic ecosystems support various living organisms with necessary nutrients, maintaining biodiversity, especially for microorganisms. Role of microbial in mangrove ecosystem has been studied around the globe. In addition to the diversity of bacteria, the mangrove ecosystem also holds the potential in terms of applying bacteria to waste bioremediation. Bacteria recycle nutrients, produce and consume gases that affect the global climate, and produce a wide variety of compounds, such as enzymes capable of decomposing pollutants and waste.6
Dyes have become a prominent topic in bioremediation research due to their widespread use in industries such as food, paper, cosmetics, and textiles, including batik production.7,8 Among industrial pollutants, wastewater from textile dyeing is one of the most significant sources of environmental contamination. The batik industry primarily relies on synthetic dyes, which are complex aromatic compounds that are resistant to decomposition.9,10 Additionally, the liquid waste generated in this process is typically non-biodegradable and often contains carcinogenic substances. Due to its persistent nature, dye waste can become toxic in aquatic ecosystems. If discharged untreated, it not only deteriorates the visual quality of water bodies but also poses a serious threat to aquatic organisms.11 Biological decolorization offers an effective approach to removing dye pollution, as it is more environmentally friendly and cost-effective compared to conventional physical and chemical treatment methods.12
The Belawan mangrove waters originate from the Deli River, which flows through Medan City and its surrounding areas. These waters are contaminated with heavy metals such as Pb, Cd, and Zn.13,14 Additionally, oil pollution has been reported in the Belawan waters.15 The region is in close proximity to industrial facilities, ports, and residential areas, contributing to environmental pollution, particularly in the mangrove forest ecosystem. Research on the diversity of bacteria in polluted mangrove waters and their potential for decolorizing waste substances remains limited. This study explores the diversity of culturable bacteria from the Belawan mangrove forest and their potential application in dye waste decolorization.
Sampling sites
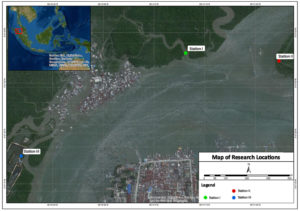
Sampling was conducted in a mangrove forest near the Deli River Estuary in Belawan, North Sumatra, Indonesia. The Deli River is important in Medan; upstream, it passes through residential areas and is close to the estuary through the industrial areas. Water sampling was conducted mangrove ecosystem at three stations: Station I (3°79’33.18’’N, 98°68’50.78’’E), Station II (3°79’28.84’’N, 98°69’01.95’’E), and Station III (3°78’76.50’’N, 98°67’60.74’’E) (Figure 1). Station I is at the edge of the mangrove forest with many shipwrecks that receives flow from a small river. Station II is in an area that only has mangroves but receives direct input from river water. Station III is in a mangrove area close to residential areas. The physicochemical water conditions at the sampling sites were measured, including water pH, temperature, salinity, turbidity, total dissolved solids (TDS), dissolved oxygen (DO), biological oxygen demand (BOD), chemical oxygen demand (COD), and nitrate and phosphate contents.
Figure 1. Map of research locations in mangrove forest waters, Medan Belawan District, Medan City, North Sumatra Province (A: Station I; B: Station II, and C: Station III)
Materials
A global positioning system (GPS), thermometer, pH meter, refractometer, bacterial incubator, shaking incubator, microscope, thermocycler, electrophoresis tools, and ultraviolet (UV) transilluminator were used in this study. The materials used were as follows: Belawan mangrove forest water samples, liquid dye waste obtained from local batik industry, nutrient agar (NA), nutrient broth (NB), mineral salt medium (MSM) containing 1% NaCl, glucose, Gram dyes, agarose, Tris-acetate-EDTA 1X (TAE), GoTaq green mastermix (Promega), nuclease free water (NFW), primers [27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3’)], and 1 kb DNA markers.
Water sampling was performed using sterile bottles, stored in a cool box, and transported to the laboratory. Bacterial isolation was performed following a standard procedure: sample (1 mL) was diluted × 1000 using sterile distilled water containing 1% NaCl. The suspensions were evenly inoculated onto a NA plate containing 1% NaCl and incubated at room temperature (28 ± 2 °C) for 24 hr. Independent colonies with distinct appearance were then transferred onto fresh NA plates to obtain pure cultures.
Pure isolate characterization
Pure isolates with different morphotypes were macroscopically observed, including characteristics such as the shape, color, consistency (slimy or non-slimy), elevation, and edge of the bacterial colonies. Microscopic observations were performed to observe Gram staining and cell morphology.
Molecular identification
Species identification was performed based on 16S rRNA gene sequences. Bacterial DNA was isolated through freeze-thaw processes. An Eppendorf tube (1.5 mL) was filled with 100 µL distilled water under aseptic conditions, and pure culture bacteria were inoculated into the tube. The suspensions were frozen at -10 °C for 10 min, then thawed at 90 °C for 10 min. The cycle was repeated five times to determine the cell breakdown efficiency. The isolated DNA was used for gene amplification by polymerase chain reaction (PCR) using the 16S rRNA bacterial primers 27F and 1492R.16
The PCR reaction mixture was prepared to a total volume of 25 µL, containing the following components: 12.5 µL GoTaq green master mix (Promega), 1 µL forward primer, 1 µL reverse primer, 8.5 µL NFW, and 2 µL DNA template. The thermocycler was programmed and run at 94 °C (2 min), 90 °C denaturation (30 sec), 60 °C annealing (30 sec), 72 °C elongation (1 min), and 72 °C post-PCR (5 min). This process was repeated for 40 cycles. The amplified DNA samples were then visualized by electrophoresis using 1% agarose (1 g agarose in 100 mL TAE 1X with 1 µl EtBr), with a 1 kb marker and DNA samples loaded into the wells. Electrophoresis was performed at 80 V and 400 mA for 60 min and then detected using a transilluminator.
Bioinformatics
Bioinformatic analysis was performed after analyzing the 16S rRNA sequences of commercially amplified DNA. Sequence data were compared with data in GenBank from the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) program, and a phylogenetic tree was created using MEGA XI Construct/Test Neighbor-Joining Tree software and the Kimura 2-parameter model method, with a bootstrap value of 1000.
Decolorization potential test
Potential screening was performed on solid MSM containing dye waste at concentrations of 25%, 50%, 75%, and 100%. Media containing 100% dye waste meaning that MSM components was dissolved in liquid dye instead distilled water. The MSM plates were divided into several parts and the bacterial isolates were inoculated onto the surface using a sterile toothpick. The plates were incubated for 4 days at 27 °C, and the diameters of the bacterial colonies were measured. A potential bacterial test for the dye waste decolorization process was performed for five isolates selected from the screening results on liquid MSM media. The selected bacterial isolates were inoculated into 5 mL NB medium and incubated at 37 °C with aeration in a shaker incubator at 150 rpm for 16 hr. The suspension was transferred to 100 mL of the test medium containing 50% dye waste with the addition of 1% glucose and no glucose in a 250 mL Erlenmeyer with three replicates. MSM containing 50% dye waste without bacterial inoculation was used as the control. The culture was incubated at 37 °C for 15 days with aeration in a shaker incubator at 150 rpm, and color change was monitored. Observations were made every five days during the incubation period. Aliquots (3 mL) of the culture medium were collected at specified time intervals and centrifuged at 7000 rpm for 20 min.17 Decolorization was observed by measuring the absorbance of the culture supernatant using a spectrophotometer at the maximum wavelength of the dye waste, which is 365 nm.18
The percentage decolorization was calculated as follows:
% Decolorization = I – F / I × 100
Where: I = initial absorbance and F = absorbance of decolorization medium
Physico-chemical factors of Belawan mangrove forest waters
The physicochemical factors of the water were measured at each sampling location. The physicochemical factors were relatively similar among the study sites, as shown in Table 1.
Table (1):
Physicochemical data of Belawan mangrove waters at the three sampling stations
Parameter |
Unit |
Station I |
Station II |
Station III |
|---|---|---|---|---|
Water pH |
– |
6.8 |
6.8 |
7.0 |
Water temperature |
oC |
28.5 |
29.8 |
30.0 |
Salinity |
% |
1.5 |
1.3 |
1.0 |
Turbidity |
NTU |
1.53 |
1.85 |
2 |
TDS |
mg/L |
3.57 |
3.83 |
8.14 |
DO |
mg/L |
7.11 |
7.58 |
8.17 |
BOD |
mg/L |
0.48 |
0.42 |
0.23 |
COD |
mg/L |
4.1 |
5.6 |
3.8 |
Nitrate |
mg/L |
5.5 |
4 |
2.2 |
Phosphate |
mg/L |
0.35 |
0.48 |
0.38 |
TDS, total dissolved solids; DO, dissolved oxygen; BOD, biological oxygen demand; COD, chemical oxygen demand
The pH of the water at the sampling locations at stations I and II was 6.8, while that at station III was 7.0. According to Mustapha and Halimoon,19 the optimum pH for bacterial growth is 6.5-7.5, which indicates that the sampling location was the correct pH for obtaining neutrophilic bacterial isolates. The optimum pH for the hydrocarbon biodegradation process ranges between 6.0-8.0, which indicates that the bacteria in the Belawan mangrove waters are environmentally adapted to support the hydrocarbon degradation process. The water temperature at the three stations was 28.5, 29.8, and 30.0 °C, which is normal. At this temperature, the thriving bacteria were mesophilic. The highest salinity was observed at station I (1.5%), which was likely due to the closer proximity to the high seas than the other sites. The highest turbidity was at station III (2 NTU), with a TDS value of 8.14 mg/L, which could be attributable to the close proximity to settlements (contaminated by household waste).
According to Ikhtiar et al.20 the higher the DO value, the better the water quality, whereas the higher the BOD value, the lower the water quality. Based on the regulations of the State Minister of Environment No. 03 of 2010, the maximum allowable levels of BOD and COD is 50 mg/L and 100 mg/L, respectively. The highest DO, BOD, and COD values were at Station III (8.17 mg/L), Station I (0.48 mg/L), and Station II (5.6 mg/L), respectively.
According to Patty et al.21 water P and N contents are crucial for organism growth and are indicators of water quality. If their thresholds are exceeded, it can be deadly to several types of marine life. The main sources of P and N in the water is plants or organism decomposition; however, it can also originate from industrial waste containing organic compounds. The highest P and N contents were at Station II (0.48 mg/L) and Station I (5.5 mg/L), respectively. The P and N levels at the three stations were high based on the Decree of the Minister of the Environment, which stipulates that the quality standard for P and N concentrations are 0.015 ml/L and 0.008 mg/L, respectively, indicating that the mangrove waters of Belawan are polluted.
Culturable bacterial isolates
Bacteria were isolated and purified using NA, a general medium for bacterial cultivation. A total of 29 different morphotype isolates were successfully purified from 3 sampling sites. Using Gram staining, macroscopic colony observations included color, shape, edge, elevation, surface, and consistency. The results showed that the number of purified isolates from each station was relatively similar: ten isolates from stations I and III and nine isolates from station II. The macro-and micro characteristics of the isolates are listed in Table 2.
Table (2):
Macro-and microscopic characteristics of bacteria isolated from the waters of the Belawan mangrove forest
| Isolate code | Colony characteristics | Cell shape/Gram stain | |||||
|---|---|---|---|---|---|---|---|
| Color | Shape | Edge | Elevation | Surface | Consistency | ||
| Station I | |||||||
| A1 | Pale yellow | Circular | Entire | Convex | Smooth | Butyrous | Bacillus (-) |
| A2 | Milky White | Circular | Entire | Convex | Smooth | Butyrous | Bacillus (-) |
| A3 | Clear White | Circular | Entire | Convex | Smooth | Butyrous | Coccobacillus (-) |
| A4 | Clear White | Circular | Entire | Convex | Smooth | Butyrous | Coccobacillus (-) |
| A5 | Clear White | Circular | Entire | Convex | Smooth | Butyrous | Coccobacillus (-) |
| A6 | Yellowish White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| A7 | Milky White | Circular | Entire | Raised | Smooth | Butyrous | Bacillus (-) |
| A8 | White | Circular | Undulate | Convex | Smooth | Butyrous | Bacillus (-) |
| A9 | Yellowish White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| A10 | Milky White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| Station II | |||||||
| B1 | Yellowish White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| B2 | Yellowish White | Irregular | Entire | Umbonate | Rough | Butyrous | Bacillus (-) |
| B3 | Yellowish White | Irregular | Entire | Umbonate | Rough | Butyrous | Bacillus (-) |
| B4 | Milky White | Circular | Entire | Convex | Rough | Mucoid | Bacillus (-) |
| B5 | Milky White | Irregular | Undulate | Convex | Rough | Mucoid | Bacillus (-) |
| B6 | Milky White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| B7 | White | Circular | Entire | Umbonate | Smooth | Butyrous | Bacillus (+) |
| B8 | Milky White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| B9 | Milky White | Irregular | Entire | Convex | Smooth | Butyrous | Bacillus (+) |
| Station III | |||||||
| C2 | Milky White | Circular | Entire | Raised | Rough | Butyrous | Bacillus (+) |
| C3 | White | Circular | Entire | Umbonate | Smooth | Butyrous | Bacillus (+) |
| C4 | Milky White | Irregular | Undulate | Umbonate | Smooth | Mucoid | Bacillus (-) |
| C5 | Milky White | Irregular | Entire | Convex | Rough | Butyrous | Bacillus (+) |
| C6 | Milky White | Circular | Entire | Convex | Smooth | Butyrous | Bacillus (-) |
| C8 | Milky White | Irregular | Entire | Convex | Rough | Butyrous | Bacillus (+) |
| C9 | Milky White | Irregular | Entire | Umbonate | Rough | Butyrous | Bacillus (+) |
| C10 | Milky White | Circular | Entire | Raised | Smooth | Butyrous | Bacillus (-) |
| C11 | Milky White | Irregular | Entire | Convex | Rough | Butyrous | Bacillus (+) |
| C12 | Milky White | Irregular | Entire | Convex | Rough | Butyrous | Bacillus (+) |
(+) = Gram-positive; (-) = Gram-negative
The isolate colors were pale yellow (n = 1), white (n = 3), clear white (n = 3), yellowish white (n = 5), and milky white (n = 17). Based on the colony shape, 13 isolates were circular and 16 were irregular. The edges of the colonies on each isolate included three isolates with undulated edges and 27 isolates with entire edges. There were 3 isolates with elevated elevation, 6 umbonate, and 20 convex. Nineteen isolates had smooth surfaces and 10 isolates had rough surfaces. Three isolates exhibited a mucoid (slimy) consistency and the others were butyrous (not slimy/creamy). Fourteen isolates were Gram-negative and 15 were Gram-positive. Twenty-six isolates were bacillus-shaped cells and three coccobacilli.
Molecular identification
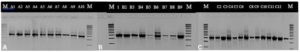
16S rRNA gene amplification using primers 27F and 1492R, visualized by electrophoresis, were observed on a 1% agarose gel using a 1 kb marker. All the bacterial isolates showed DNA bands measuring approximately 1500 bp in length (Figure 2).
Figure 2. PCR product of 16S rRNA gene with 27F and 1492R primers of bacterial isolates from Belawan mangrove visualized in electrophoresis gel with 1 kb marker.
Note = A: Isolate of station I; B: Isolate of station II; C: Isolate of station III; M: Marker
The sequence results were then compared with data from GenBank in the National Center for Biotechnology Information (NCBI) database (Table 3).
Table (3):
Results of BLAST analysis of bacterial isolates from the Belawan mangrove waters.
The BLAST analysis conducted on bacteria isolated from the Belawan mangrove waters revealed the presence of 8 distinct genera, which collectively represent a total of 15 different bacterial species
| Isolate code | Genbank Data Similarities | AC number (Accession) | Similarity (%) |
|---|---|---|---|
| Station I | |||
| A1 | Myroides profundi strain D25 | NR_044435.1 | 98.39 |
| A2 | Enterobacter kobei strain JCM 8580 | NR_113321.1 | 87.83 |
| A3 | Shigella sonnei strain CECT 4887 | NR_104826.1 | 99.57 |
| A4 | Shigella flexneri strain AATC 29903 | NR_026331.1 | 98.27 |
| A5 | Shigella flexneri strain AATC 29903 | NR_026331.1 | 98.42 |
| A6 | Bacillus anthracis strain SBSI | NR_118536.1 | 81.14 |
| A7 | Pseudomonas hydrolytica strain DSWYDI | NR_170428.1 | 97.80 |
| A8 | Providencia huaxiensis strain WCHPr00369 | NR_174258.1 | 98.68 |
| A9 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 98.41 |
| A10 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 97.33 |
| Station II | |||
| B1 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 98.59 |
| B2 | Aeromonas caviae strain CECT 4221 | NR_104824.1 | 97.95 |
| B3 | Aeromonas caviae strain CECT 4221 | NR_104824.1 | 98.99 |
| B4 | Aeromonas salmonicida | NR_118547.1 | 97.95 |
| B5 | Pseudomonas balearica strain SP1402 | NR_025972.1 | 99.71 |
| B6 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 98.18 |
| B7 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 97.41 |
| B8 | Bacillus cereus strain IAM 12605 | NR_113266.1 | 98.45 |
| B9 | Bacillus mycoides strain 273 | NR_036880.1 | 99.50 |
| Station III | |||
| C2 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 98.43 |
| C3 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 97.96 |
| C4 | Klebsiella pneumoniae subsp. Rhinoscleromatis strain R-70 | NR_037084.1 | 96.16 |
| C5 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 96.00 |
| C6 | Enterobacter cloacae strain ATCC 13047 | NR_118568.1 | 98.74 |
| C8 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 93.18 |
| C9 | Aeromonas caviae strain CECT 4221 | NR_104824.1 | 98.68 |
| C10 | Pseudomonas khazarica strain TBZ2 | NR_169334.1 | 99.28 |
| C11 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 96.25 |
| C12 | Bacillus cereus strain IAM 12605 | NR_115526.1 | 93.91 |
Based on the results of the Basic Local Alignment Search Tool (BLAST) analysis of bacterial isolates from the waters of the Belawan mangrove forest, eight species were isolated from station I: Myroides profundi, Enterobacter kobei, Shigella sonnei, Shigella flexneri (two isolates), Bacillus anthracis, Pseudomonas hydrolytica, Providencia huaxiensis, and Bacillus cereus. At station II, five species were detected: Bacillus cereus, Aeromonas caviae (two isolates), Aeromonas salmonicida, Pseudomonas balearica, Bacillus cereus (four isolates), and Bacillus mycoides. Five species were isolated from station III, namely Bacillus cereus (six isolates), Klebsiella pneumoniae, Enterobacter cloacae, Aeromonas caviae, and Pseudomonas khazarica.
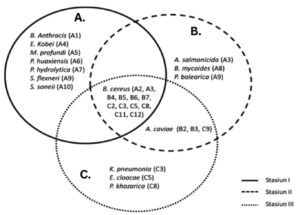
Various bacterial genera have been isolated from mangrove waters, as reported in previous studies, including Bacillus, Nitrococcus, Planococcus, Lactobacillus, Pseudomonas, Micrococcus, Vibrio, Listeria, and Paenibacillus.22-25 These findings highlight the bacterial diversity in mangrove water ecosystems, which aligns with the results of our study. A diagram illustrating the bacterial species isolated from the Belawan mangrove forest at three sampling locations is presented in Figure 3.
Figure 3. Diagram of the presence of bacterial species (A: isolates from station I; B: isolates from station II and C: isolates from station III) from Belawan mangrove forest waters at three stations
Seven species were found only at station I, namely B. anthracis, E. kobei, M. profundi, P. huaxiensis, P. hydrolytica, S. flexneri, and S. sonnei. All the species found at Station I were bacteria found in clinical samples from humans or animals. This could be due to the location of Station I, which is drained by a small river that may carry microorganisms from the mainland. In addition to clinical samples, all species can tolerate the temperature and salinity conditions present at Station I.26 Furthermore, Zhu et al.27 reported that apart from clinical specimens, the Enterobacter group can grow at temperatures of 15-42 °C. The optimal temperature of S. sonnei is 37 °C, pH 5.0-6.0, and salinity 1-4%.28 Mallick et al.29 stated that the optimal temperature of S. flexneri is 37 °C. The optimal temperature for the growth of P. huaxiensis is 35-37 °C and pH 6.0-7.0.30 P. hydrolytica grows optimally at 37 °C, pH 7.0, and has a salinity tolerance of more than 4%. The environmental conditions at Station I supported the growth of the bacterial species present there.
The species A. salmonicida, B. mycoides, and P. balearica were found at station II, which was surrounded by mangroves and received direct input from the river flow. These three species were able to live at station II, which was 29 °C and 1.3% salinity. P. balearica was first isolated on the Balearic Islands, which can grow at temperatures up to 45 °C and salinity up to 8.5%,31 which is supported by Nejad et al.32 who stated that P. balearica is a salt-tolerant bacterium. Station III was located close to the residential areas. Three isolates were found at Station III: K. pneumoniae, E. cloacae, and P. khazarica, which may have been influenced by the presence of waste generated by human and animal activities. Several researchers have reported that these three isolates can be isolated from several samples related to human and animal life. Klebsiella can be isolated from clinical samples (the urine and blood of patients with urinary tract infections and bacteremia), surface water,33 and sediment and seawater samples.34 E. cloacae can be found in diseased rice seedlings,35 activated sludge from municipal wastewater treatment plants,36 and sediment and seawater samples.34 P. khazarica was first isolated from the Khazar Sea, and decomposes polycyclic aromatic hydrocarbons. This strain is able to grow without NaCl, tolerates up to 8.5% NaCl, and can grow at pH 3.0-10.0 (optimal pH 6.0-7.0) at a temperature of 10-45 °C (optimally 30 °C).
The species A. caviae was detected at stations II and III. Based on the physical and chemical factors at stations I and III, which were not significantly different, the adjacent location points allowed the presence of the same species at these two locations. Although it receives direct inputs from river flows carrying various compounds and wastes, this species can still grow. This is supported by Shamim et al.37 who reported that A. caviae isolated from estuarine surfaces in India showed resistance to lead and tolerance to several heavy metals, including Zn, Cd, Cu, and Hg.
B. cereus was isolated from all three stations and dominated the water in the Belawan mangrove forest. This may be because B. cereus naturally thrives on many substrates. Many studies have isolated this species from various locations, one of which is Hemalatha and Banu38 who isolated B. cereus from six different samples, including soil, wastewater, air, freshwater, seawater, and milk. Based on the results of this study, the waters of the Belawan mangrove forest were predominantly inhabited by Bacillus species. This finding is consistent with the research conducted by Alhelaify et al.,39 who studied bacterial diversity in mangrove environments in Saudi Arabia and reported that Bacillus spp. accounted for up to 50% of the isolated bacteria. Bacillus spp. play a crucial role in the decomposition of mangrove litter, contributing to nutrient cycling and ecosystem sustainability.
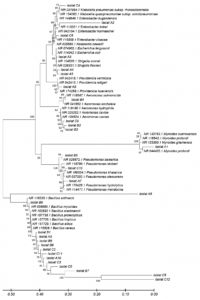
The bacterial DNA sequences obtained from the Belawan mangrove waters were then used to create a phylogenetic tree to determine their level of kinship and genetic evolutionary distance using the MEGA X software (Figure 4). Based on the phylogenetic tree, the bacterial species from the waters of the Belawan mangrove forest had very high genetic variation. Each end of the phylogenetic tree represents a unique species in all isolates. Bacillus cereus, a species that dominates the waters of the Belawan mangrove forest, has very high genetic variation. Water P content is not directly related to bacteria but is related to the level of water fertility. Excessive nutrients in the ecosystem can be caused by high N and P contents from the decomposition of organic matter or human activities that produce waste, which can cause eutrophication. The high N and P contents at the three stations, due to exposure to toxic compounds and waste carried by the river, allowed this species to mutate and survive environmental changes. The high genetic variation and the results obtained were then tested for their potential to decolorize dye waste.
Figure 4. The phylogenetic tree by bacteria from the waters of the Belawan mangrove forest was reconstructed using MEGA XI software using the Kimura-2-parameter method and a bootstrap value of 1000×
Decolorization potential
Screening potential for decolorization activity of isolates was performed in MSM with liquid dye waste as a carbon source using concentrations of 25-100%. Bacterial growth was measured based on the diameter of the colony after four days of incubation. Relative growth responses are shown in Table 4.
Table (4):
Colony growth diameter scale of bacterial isolates from Belawan mangrove waters on solid mineral salt medium (MSM) at different concentrations of dye waste (25%, 50%, 75%, and 100%)
Isolates code |
Species |
25% |
50% |
75% |
100% |
|---|---|---|---|---|---|
A1 |
Myroides profundi strain A1 |
+++ |
+++ |
+++ |
++ |
A2 |
Enterobacter kobei strain A2 |
+++ |
+++ |
+++ |
++ |
A3 |
Shigella sonnei strain A3 |
+++ |
+++ |
++ |
++ |
A4 |
Shigella flexneri strain A4 |
+++ |
+++ |
+++ |
+++ |
A5 |
Shigella flexneri strain A5 |
++ |
++ |
– |
– |
A6 |
Bacillus anthracis strain A6 |
+++ |
+++ |
++ |
++ |
A7 |
Pseudomonas hydrolytica strain A7 |
++ |
++ |
++ |
+ |
A8 |
Providencia huaxiensis strain A8 |
++ |
+ |
+ |
+ |
A9 |
Bacillus cereus strain A9 |
+ |
+ |
+ |
– |
A10 |
Bacillus cereus strain A10 |
+++ |
+++ |
+++ |
+++ |
B1 |
Bacillus cereus strain B1 |
++ |
++ |
++ |
+ |
B2 |
Aeromonas caviae strain B2 |
+ |
++ |
++ |
+ |
B3 |
Aeromonas caviae strain B3 |
+ |
++ |
++ |
+ |
B4 |
Aeromonas salmonicida strain B4 |
+++ |
+++ |
+++ |
+++ |
B5 |
Pseudomonas balearica strain B5 |
+++ |
+++ |
+++ |
++ |
B6 |
Bacillus cereus strain B6 |
++ |
++ |
++ |
+ |
B7 |
Bacillus cereus strain B7 |
++ |
++ |
+ |
+ |
B8 |
Bacillus cereus strain B8 |
+ |
+ |
+ |
+ |
B9 |
Bacillus mycoides strain B9 |
+ |
+ |
+ |
+ |
C2 |
Bacillus cereus strain C2 |
+ |
+ |
++ |
+ |
C3 |
Bacillus cereus strain C3 |
++ |
– |
+ |
– |
C4 |
Klebsiella pneumoniae strain C4 |
+++ |
+++ |
+++ |
+++ |
C5 |
Bacillus cereus strain C5 |
+ |
+ |
+ |
+ |
C6 |
Enterobacter cloacae strain C6 |
+++ |
+++ |
+++ |
+++ |
C8 |
Bacillus cereus strain C8 |
+++ |
+++ |
++ |
++ |
C9 |
Aeromonas caviae strain C9 |
++ |
+ |
+ |
– |
C10 |
Pseudomonas khazarica strain C10 |
+ |
– |
+ |
– |
C11 |
Bacillus cereus strain C11 |
++ |
++ |
++ |
+ |
C12 |
Bacillus cereus strain C12 |
+ |
+ |
– |
– |
All bacterial isolates grew on solid MSM with a concentration of 25% dye waste; however, not all isolates grew at concentrations of 50%, 75%, and 100%. Eleven isolates grew very well on media with 25% and 50% dye waste concentrations, eight isolates grew on media with 75% dye waste concentration, and only five isolates grew on media with up to 100% dye waste concentration.
Each isolate showed a different colony diameter growth scale, which could be due to the ability of each isolate to use the dye waste contained in the test medium as the only carbon source. However, an increase in the concentration of dye waste also affected colony growth. The higher the concentration of dye waste, the lower was the growth of bacterial cells in the test medium. This could be because the dye contained in the waste used is a synthetic compound that is toxic to bacteria; thus, bacterial isolates with a high colony diameter scale are considered better for using dye waste in their growth as the only carbon source. Therefore, isolates with the highest colony diameter scale and able to grow up to 100% concentration of dye waste were selected and tested for their decolorization ability on liquid MSM media, they were A4 (S. flexneri), A10 (B. cereus), B4 (A. salmonicida), C4 (K. pneumoniae), and C6 (E. cloacae).
Decolorization activity
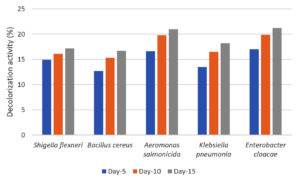
The decolorization activity of the five potential isolates from the screening process was evaluated in liquid MSM with dye waste as the carbon source. Qualitatively, decolorization activity was observed by reducing the color intensity of the culture medium during cultivation. Qualitatively, the rate of decolorization of dye waste was evaluated by measuring the decrease in the absorbance of the culture media at 365 nm. The decolorization activity profile of the five isolates after cultivation for 15 days is shown in Figure 5.
Figure 5. Percentage of decolorization of dye waste by selected isolates (S. flexneri, A. salmonicida, E. cloacae, B. cereus, and K. pneumoniae) on liquid mineral salt medium (MSM)
Five species exhibited the ability to reduce absorbance values in media containing 50% dye waste. Decolorization ranged between 12-21%, and the color reduction profile was similar during cultivation among the isolates. No decrease of absorbance value was detected for control until 15 days. A. salmonicida and E. cloacae exhibited slightly better decolorization than the other three isolates. Even at low decolorization, compared with our previously reported cases for fungi, the results of this study clearly indicate that the bacteria used the carbon available in the liquid waste. The decolorization rate was determined by several factors, of which the concentration and toxicity of the dyes are among the most important. Different decolorization activities have been reported for bacteria and fungi using various dyes. For instance, Moosvi et al.40 reported that the decolorization of azo dyes by bacteria with the addition of glucose showed a maximum decolorization of 94%, whereas without glucose it was only 40%, this could be due to glucose metabolism resulting in the production of reduced nucleotides (NADH and FADH) causing an increase in decolorization efficiency. Seyedi et al.41 also reported that in the decolorization of dyes by bacteria of the genus Halomonas, the highest percentage of decolorization was achieved with the addition of supplements (glucose, peptone, and yeast extract). The percentage decolorization by the strain requires a certain glucose concentration. Decolorization increased with an increase in glucose concentration up to 10%, followed by a decrease with an increase in glucose concentration. Low glucose concentrations (1-5%) did not support the growth of bacteria in the test medium, and at higher concentrations
(>10%), bacteria preferred to use glucose.
The bacterium A. salmonicida induces degradation and decolorization. A. salmonicida has five enzymes that have the potential to degrade chitin,42 which are resistant to lauric acid, dyes, and aromatic amines. Furthermore, these enzymes have shown technical feasibility for regenerating biomaterials during the decolorization of wastewater.43 Wanyonyi et al.44 reported that B. cereus, which produces proteases, has a decolorization efficiency of up to 98% within 24 hrs with a dye concentration of 1.0 × 10-5 M. The lactase enzyme produced by K. pneumoniae also exhibited high decolorization and oxidation efficiencies for azo and malachite green dyes under acidic and neutral conditions.45 Chantarasiri46 isolated K. pneumoniae and Enterobacter sp. from mangroves and found that they showed strong decolorization efficiencies within 72 hrs of incubation with 0.01% reactive dye, with decolorization percentages of 20% and 92%, respectively. E. cloacae effectively decolorizes azo dyes and anthraquinones with different structures, the decolorization rate increases with increasing temperature from 20 °C to 30 °C .47 The potential of the S. flexneri strain for the decolorization of dye waste has not yet been reported; however, Said et al.48 reported that Shigella was able to use polycyclic aromatic hydrocarbons (PAHs) containing up to four rings as the sole carbon source.
Conclusively, this study isolated 29 bacterial species from the waters of the Belawan mangrove forest. Based on molecular identification, the isolates consisted of eight genera of bacteria from 15 different species: A. caviae (three isolates), A. salmonicida, B. anthracis, B. cereus (12 isolates), B. mycoides, E. cloacae, E. kobei, K. pneumonia. The bacterial species Profundi, P. huaxiensis, P. balearica, P. hydrolytica, P. khazarica, S. flexneri (two isolates), and S. sonnei. Based on the screening test for potential dye waste in solid MSM, five different bacterial species with the best growth potential were obtained: Shigella flexneri, Bacillus cereus, Aeromonas salmonicida, Klebsiella pneumoniae, and Enterobacter cloacae. The percentage of dye waste decolorized for 15 days with the addition of 1% glucose showed a better decolorization ability than that without glucose. The best decolorization ability was shown by Aeromonas salmonicida, with a percentage value of decolorization on two types of media: 36.2% with glucose and 25.0% without glucose.
In this study, we successfully isolated and identified twenty-nine bacterial strains from the mangrove water forests of Belawan, North Sumatra, belonging to eight genera and fifteen species. Notable species included A. salmonicida, B. cereus, and K. pneumoniae. Screening for dye waste decolorization revealed that five isolates S. flexneri, B. cereus, A. salmonicida, K. pneumoniae, and E. cloacae exhibited enhanced growth in the presence of dye waste. All five species demonstrated significant decolorization abilities, with A. salmonicida and E. cloacae showing superior decolorization activity after 15 days. These findings highlight the potential of specific mangrove-derived bacterial isolates for bioremediation applications in dye waste management.
ACKNOWLEDGMENTS
The authors would like to thank Universitas Sumatera Utara for its support under the International Research Collaboration Program.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by Universitas Sumatera Utara under the scheme of International Collaboration of World Class University (WCU) research of 2021 and USU EQUITY Research Program funded by LPDP of 2023.
DATA AVAILABILITY
All data generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Saenger P. Mangrove Ecology Silviculture and Conservation. Springer. Australia. 2002.
Crossref - Alongi DM, Murdiyarso D, Fourqurean JW, et al. Indonesia’s blue carbon: a globally significant and vulnerable sink for seagrass and mangrove carbon. Wetlands Ecol Manag. 2016;24: 3-13.
Crossref - Palit K, Rath S, Chatterjee S, Das S. Microbial diversity and ecological interactions of microorganisms in the mangrove ecosystem: Threats, vulnerability, and adaptations. ESPR. 2022;29(22):32467-32512.
Crossref - Holguin G, Vazquez P, Bashan Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils. 2001;33:265-278.
Crossref - Sakho I, Mesnage V, Copard Y, et al. A cross-section analysis of sedimentary organic matter in a mangrove ecosystem under dry climate conditions: The Somone estuary, Senegal. J. Afr. Earth Sci. 2015;101:220-231.
Crossref - Thatoi H, Behera BC, Mishra RR, Dutta SK. Biodiversity and Biotechnological potential of Microorganisms from Mangrove Ecosystems: A Review. Ann. Microbiol. 2012;63:1-19.
Crossref - Hasanpour M, Hatami M. Photocatalytic performance of aerogels for organic dyes removal from wastewaters: Review study. J Mol Liq 2020;309:113094.
Crossref - Brazesh B, Mousavi SM, Zarei M, Ghaedi M, Bahrani S, Hashemi SA. Biosorption. In Interface Sci. Elsevier. 2021;33;587-628.
Crossref - Mehra S, Singh M, Chadha P. Adverse impact of textile dyes on the aquatic environment as well as on human beings. Toxicol. Int. 2021;28(2):165-176.
Crossref - Zakaria N, Rohani R, Wan Mohtar WHM, Purwadi R, Sumampouw GA, Indarto A. Batik effluent treatment and decolorization—a review. Water. 2023;15(7): 1339.
Crossref - Alsukaibi AKD. Various approaches for the detoxification of toxic dyes in wastewater. Processes. 2022;10(10): 1968.
Crossref - Hayat H, Mahmood Q, Pervez A, Bhatti ZA, Baig SA. Comparative decolorization of dyes in textile wastewater using biological and chemical treatment. Sep Purif Technol. 2015;154: 149-153.
Crossref - Sinaga MP, Simanullang AF. Analysis And Prevention of Pb Metal Content Suspended In Belawan Waters. JGISE. 2021;5(2).
Crossref - Mirandha A, Irvan, Wahyuningsih H. Spatial distribution of water quality in Belawan River, North Sumatra. In IOP Conf. Ser.: Earth Environ. Sci. 2021;713(01): 012011. IOP Publishing.
Crossref - Tarigan LNIS, Qodri N, Rangkuti SLD, Zubir M. Pb (II) and Oil Contamination Analysis of Belawan Sea, Medan City, North Sumatera. IJCST. 2019;2(2): 131-135.
Crossref - Sangi F, Mohammadi A, Afzali N, Mirabolfathy M. Identification of new aflatoxin B1-degrading Bacteria from Iran. Iran J Toxicol. 2018;12(3):39-44.
Crossref - Mane UV, Gurav PN, Desmusk AM, Govincwar SP. Degradation of Textile Dye Reactive Navy-blue Rx (Reactive blue-59) by an isolated Actinomycete Streptomyces krainskii SUK-5. Malaysian J Microbiol. 2008;4(2):1-5.
Crossref - Khehra MS, Saini HS, Sharma DK, Chadha BS, Chimni SS. Decolorization of Various Azo Dyes by Bacterial Consortium. Dyes Pigm. 2005;67(1):55-61.
Crossref - Mustapha MU, Halimoon N. Screening and isolation of heavy metal tolerant bacteria in industrial effluent. Procedia Environ. 2015;30:33-7.
Crossref - Ikhtiar M, Alzad H, Paramita S, Yadi. Microbiological assessment of indoor air of Takalar County hospitalwards in South Sulawesi, Indonesia. Sci J Public Health. 2017;5(3):172-177.
Crossref - Patty JO, Wagey BT, Tilaar FF, Lumingas LJL, Sondakh CFA, Undap S, Sumilat D. Seagrass mapping using Landsat-8 satellite images in Tanjung Merah waters, Bitung City, North Sulawesi. AES Bioflux. 2021;13(1):37-47.
- Widawati S, Suliasih, Sugiharto A, Suyadi, Sudiana IM. Characterization of plant growth promoting bacteria isolated from water in mangrove ecosystem. In IOP Conf. Ser.: Earth Environ. Sci. 2022;976(01): 012039. IOP Publishing.
Crossref - Gomes NCM, Borges LR, Paranhos R, Pinto FN, Mendonça-Hagler LCS, Smalla K. Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol. Ecol. 2008;66(1): 96-109.
Crossref - Ghizelini AM, Mendonça-Hagler LCS, Macrae A. (2012). Microbial diversity in Brazilian mangrove sediments: a mini review. Braz. J. Microbiol. 2012;43(4):1242-1254.
Crossref - Behera BC, Singdevsachan SK, Mishra RR, Dutta SK, Thatoi HN. Diversity, mechanism and biotechnology of phosphate solubilising microorganism in mangrove-a review. Biocatal Agric Biotechnol. 2014;3(2): 97-110.
Crossref - Zhang XY, Zhang YJ, Chen XL, et al. Myroides profundi sp. nov., Isolated from Deep-sea Sediment of the Southern Okinawa Trough. FEMS Microbiol Lett. 2008;287(1):108-112.
Crossref - Zhu B, Lou MM, Xie GL, et al. Enterobacter mori sp. nov., Associated with Bacterial Wilt on Morus alba L. Int J Syst Evol Microbiol. 2011;61(11):2769-2774.
Crossref - Bagamboula CF, Uyttendaele M, Debevere J. Antimicrobial Effect of Species and Herbs on Shigella sonnei and Shigella flexneri. J Food Prot. 2003;66(4):668-673.
Crossref - Mallick B, Mondal P, Dutta M. Morphological, Biological and Genomic Characterization of a Newly Isolated Lytic Phage Sfk20 Infecting Shigella flexneri, Shigella sonnei, and Shigella dysenteriae1. Sci Rep. 2021;11(1):19313.
Crossref - Hu Y, Feng Y, Zhang X, Zong Z. Providencia huaxiensis sp. nov., Recovered from a Human Rectal Swab. Int J Syst Evol Microbiol. 2019;69(9):2638-2643.
Crossref - Bennasar A, Rossello-Mora R, Lalucat J, Moore ERB. 16S rRNA Gene Sequence Analysis Relative to Genomovars of Pseudomonas stutzeri and Proposal of Pseudomonas balearica sp. nov. Int J Syst Evol Microbiol. 1996;46(1):200-205.
Crossref - Nejad YS, Jaafarzadeh N, Ahmadi M, Abtahi M, Ghafari S, Jorfi S. Remediation of Oily Sludge Wastes Using Biosurfactant Produced by Bacterial Isolate Pseudomonas balearica strain Z8. J Environ Health Sci Eng. 2020;18:531-539.
Crossref - Struve C, Krogfelt KA. Pathogenic Potential of Environmental Klebsiella pneumoniae Isolates. Environ Microbiol. 2004;6(6):584-590.
Crossref - Matyar F, Kaya A, Dinחer S. Antibacterial Agents and Heavy Metal Resistance in Gram-negative Bacteria Isolated from Seawater, Shrimp and Sediment in Iskenderun Bay, Turkey. Sci Total Environ. 2008;407(1):279-285.
Crossref - Cao X, Wu L, Wu M, Zhu C, Jin Q, Zhang J. Abscisic acid mediated proline biosynthesis and antioxidant ability in roots of two different rice genotypes under hypoxic stress. BMC Plant biology. 2020;20:1-4.
Crossref - Wang PC, Mori T, Komori K, Sasatsu M, Toda K, Ohtake H. Isolation and Characterization of an Enterobacter cloacae strain that Reduces Hexavalent Chromium under Anaerobic Conditions. Appl Environ Microbiol. 1989;55(7):1665-1669.
Crossref - Shamim K, Naik MM, Pandey A, Dubey SK. Isolation and Identification of Aeromonas caviae strain KS-1 as TBTC- and Lead-resistant Estuarine Bacteria. Environ Monit Assess. 2013;185:5243-5249.
Crossref - Hemalatha S, Banu N. DNA Fingerprinting of Bacillus cereus from Diverse Sources by Restriction Fragment Length Polymorphism Analysis. Adv Biosci Biotechnol. 2010;1(02):136-144.
Crossref - Alhelaify SS, Hozzein WN, Alharbi SA. Diversity and metagenomic sequence analysis of bacterial strains isolated from different Mangrove environments in Saudi Arabia.SAJB. 2022;10(3): 53-61.
Crossref - Moosvi S, Kher X, Madamwar D. Isolation, Characterization and Decolorization of Textile Dyes by a Mixed Bacterial Consortium JW-2. Dyes Pigm. 2007;74(3):723-729.
Crossref - Seyedi ZS, Zahraei Z, Kashi FJ. Decolorization of reactive black 5 and reactive red 152 Azo Dyes by New Faloalkaliphilic Bacteria Isolated from the Textile Wastewater. Curr Microbiol. 2020;77:2084-2092.
Crossref - Pentekhina I, Hattori T, Tran DM, et al. Chitinase System of Aeromonas salmonicida and Characterization of Enzymes Involved in Chitin Degradation. Biosci Biotechnol Biochem. 2020;84(9):1936-1947.
Crossref - Chen G, Huang MH, Chen L, Chen DH. A Batch Decolorization and Dinetic Study of Reactive Black 5 by a Bacterial Strain Enterobacter sp. GY-1. Int. Biodeterior. Biodegrad. 2011;65(6):790-796.
Crossref - Wanyonyi WC, Onyari JM, Shiundu PM, Mulaa FJ. Enzymatic Decolorization of Malachite Green Dye by a Newly Isolated Bacillus cereus strain wwcp1. IOSR J Environ Sci Toxicol Food Technol. 2014;8(12):58-64.
Crossref - Liu Y, Huang L, Guo W, Jia L, Fu Y, Gui S, Lu F. Cloning expression, and characterization of a Thermostable and pH-stable Laccase from Klebsiella pneumoniae and its application to Dye Decolorization. Process Biochem. 2017;53:125-134.
Crossref - Chantarasiri A. Klebsiella and Enterobacter Isolated from Mangrove Wetland Soils in Thailand and Their Application in Biological Decolorization of Textile Reactive Dyes. Int J Environ. Res. 2020;17(20):7531.
Crossref - Chen BY, Shiau TJ, Wei YH, Chen WM. Feasibility Study on Polyhydroxybutyrate Productiun of Dye-decolorizing Bacteria using Dye and Amine-bearing Cultures. J Taiwan Inst Chem. Eng. 2012;43(2):241-245.
Crossref - Said OB, Goni-Urriza MS, Bour M, Dellali M, Aissa P, Duran R. Characterization of Aerobic Polycyclic Aromatic Hydrocarbon degrading Bacteria from Bizerte Lagoon Sediments, Tunisia. J Appl Microbiol. 2008;104(4):987-997.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.