ISSN: 0973-7510
E-ISSN: 2581-690X
The great attention of scientific researches about vancomycin- resistant Enterococcus faecalis due to the great healthy problems in the world. Evaluation of E. faecalis virulence factors showed that all isolates (100%) were hemolysin, protease and aggregation substance producer, 30% were a gelatinase producer, and 40.7% were a lipase producer. The results of the tube method showed that all E. faecalis isolates (100%) were slime layer and biofilm producer but the amount of adherent layer were different among the isolates ranged from strong to moderate and weak. Antibiotics susceptibility test for 20 isolates was done against 11 antibiotics, eighteen E. faecalis isolates were multi drug resistance. Seventeen E. faecalis isolates were determined as Vancomycin-Resistant by the minimum inhibitory concentrations [MICs] using agar dilution method. The virulence factor Enterococcal Surface Protein (esp) which is chromosomal was amplified by Polymerase chain reaction (PCR) technique in a monoplex pattern, results of this investigation showed that 20 (100%) E. faecalis isolates gave the amplicon size 933 base pair for the esp gene. The genetic determinants of Vancomycin-Resistant vanA and vanB genes were amplified using monoplex and multiplex PCR techniques in order to identify vancomycin resistant (van+) and sensitive (lacking van) among (13) E. faecalis. The vanA, vanB genes were detected in 11 and 4 E. faecalis isolates, respectively. The results of monoplex and multiplex PCR revealed that the molecular weight of vanA and vanB genes were 550 and nearly 600 bp, respectively. The results revealed that the vanA and vanB amplicons have a genetic variation in their molecular weight during the electrophoresis of PCR product.
Enterococcos faecalis, Van genes, Antibiotic Resisrant, Virulence Factors, esp gene.
The Enterococci are a diverse and versatile group of bacteria with several intrinsic characteristics that allow them to survive and grow under a variety of conditions (Tellis and Muralidharan, 2012). Among the Enterococcal species described Enterococcus faecalis represented one of the most important causes of nosocomial infections, since it is responsible for 80 – 90% of human Enterococcal infections (Jett and Gilmore, 1990; Jones et al., 2004; Fernandes & Dhanashree, 2013).
The general interest for Enterococci and treatment of Enterococcal infections has increased due to the appearance of antibiotic multi resistant strains (Dahle´n et al., 2012). One of the major reasons why these organisms have survived in the hospital environment is their intrinsic resistance to several commonly used antibiotics and, perhaps more important, their ability to acquire resistance to all currently available antibiotics, either by mutation or by receipt of foreign genetic material through the transfer of plasmids and transposons. Most Enterococci are tolerant to the bactericidal activity of antibiotics; bactericidal synergy between antibiotics is needed to treat most serious Enterococcal infections such as endocarditis and meningitis (Cetinkaya et al., 2000). There are at least three major reasons for the emergence of multidrug-resistant Enterococci: (i) baseline intrinsic resistance to several antimicrobial agents, (ii) acquired resistance via mobility of the resistance genes on plasmids and transposons, and chromosomal exchange, and (iii) the transferability of resistance (Mundy et al., 2000). Resistance to last-line drugs, such as vancomycin is common, and Enterococci are now disseminating this resistance to methicillin-resistant Staphylococcus aureus (MRSA) (Palmer and Gilmore, 2010).Among vancomycin-resistance phenotypes in Enterococci, VanA and VanB possess highest clinical importance (Sharifi et al., 2012). Glycopeptide resistance in Enterococci is mediated predominantly by mobile gene clusters that confer resistance to vancomycin and teicoplanin (Howden et al., 2013).
One hundred thirty – five clinical specimens were collected from urine, blood, teeth root canal and burns of patients’ suffering from urinary tract infection, bacteremia, endodontic infections and burns infections, respectively [Table 1].
Table (1):
Number and Nomenclature of bacterial isolates
specimens |
No. of specimens |
E. faecalis isolate symbol |
|---|---|---|
Urine |
15 |
U |
Blood |
60 |
B |
Root canal |
50 |
R |
Burns |
10 |
W.i |
The collected specimens were streaked on Pfizer Selective Enterococcus; and incubated at 37°C for 24 hr. VITEK system was employed for E. faecalis isolates confirmation.
Determination of minimum inhibitory concentrations [MICs] of Vancomycin
The MICs were determined to all E. faecalis isolates for the Vancomycin antibiotic. This test was achieved according to Morello et al. [12].
The MIC values were based on break point recommended by CLSI [13], for the estimation of response. For Vancomycin, 1-4µg was sensitive, more than 4µg isolate was considered as intermediate resistant and e” 32µg considered as resistant.
Detection of some Virulence factors phenotypically
Haemolysis activity was determined by streaking of pure isolates on blood agar plates, The Egg-Yolk agar and Skim milk agar were inoculated with the E. faecalis isolates for detection lipase and protease production; respectively. Gelatin liquefaction (Harley & Prescott, 2002) activity was determined by streaking of pure isolates on gelatin nutrient medium. Aggregation substance production (Creti et al., 2004) was determined by inoculate a pure E. faecalis isolates in tubes contained 5 ml of Trypticase soy broth with incubation 24 hr. at 37ºC with 5% CO2. Detection of the bacterial ability to produce biofilm was done according to Christensen et al., 1982.
- DNA Extraction from E. faecalis isolates by using Wizard® Genomic DNA Purification Kit.
- Plasmid DNA extraction from E. faecalis by using AccuPrep® Plasmid Mini Extraction Kit.
- Estimating of DNA concentration and purity: The DNA concentration and purity was determined by using Nano drop instrument from ACT gene.
- Polymerase Chain Reaction (PCR) Technique: PCR assay was performed in a monoplex and multiplex (for vanA and vanB) patterns in order to amplify genes under study for Vancomycin-Resistant E. facalis. The primers listed in table (2) were selected for this study; these primers were provided in lyophilized form.
Table (2):
The primers and their sequences used in conventional PCR
| Gene | Primer name | Sequence 5ˋ→3ˋ | Size (bp) | References |
|---|---|---|---|---|
| vanA | VAF | GGGAAAACGACAATTGC | 732 | Satak et al., 1997; Depardieu et al., 2004; Fatholahzadeh et al., 2006; Biendo et al., 2010; Tellis and Muralidharan, 2012. |
| vanA | VAR | GTACAATGCGGCCGTCGTTA | 732 | |
| vanB | VBF | ATGGGAAGCCGATAGTC | 635 | Dutka-Malen et al., 1995; Jayaratne and Rutherford, 1999; Biendo et al., 2010; Tellis and Muralidharan, 2012. |
| vanB | VBR | GATTTCGTTCCTCGACC | 635 | |
| Esp | EF | TTGCTAATGCTAGTCCACGACC | 933 | Eaton and Gasson, 2001; Creti et al., 2004; Jankoska et al., 2008. |
| Esp | ER | GCGTCAACACTTGCATTGCCGAA | 933 |
The extracted DNA, primers and PCR premix, were thawed at 4ºC, PCR mixture was set up in a total volume of 25 µl included: 5 µl PCR premix, 2.5 µl of each primer (forward and reverse), 5 µl of DNA template and the rest volume was completed with sterile deionized distilled water, the negative control contained all material except template DNA, so instead that deionized distilled was added. The PCR reaction tubes were mixed briefly and placed into thermocycler PCR instrument where DNA was amplified according to the PCR program as indicating in Tables 3, 4, 5.
Table (3):
Program was processed in PCR amplification of vanA gene according to Fatholahzadeh et al., 2006
| Stage | Temperature | Time | |
|---|---|---|---|
| Initial denaturation | 94 ºC | 5 min. | 30 cycles |
| Denaturation | 94 ºC | 45 sec. | |
| Annealing | 54 ºC | 45 sec. | |
| Extension | 72 ºC | 45 sec. | |
| Final Extension | 72 ºC | 5 min. | |
Table (4):
Program was processed in PCR amplification of vanB gene according to Dutka-Malen et al., 1995
| Stage | Temperature | Time | |
|---|---|---|---|
| Initial denaturation | 94 ºC | 2 min. | 30 cycles |
| Denaturation | 94 ºC | 1 min. | |
| Annealing | 54 ºC | 1 min. | |
| Extension | 72 ºC | 1 min. | |
| Final Extension | 72 ºC | 10 min. | |
Table (5):
Program was processed in PCR amplification of esp gene according to Creti et al., 2004
| Stage | Temperature | Time | |
|---|---|---|---|
| Initial denaturation | 95 ºC | 5 min. | 30 cycles |
| Denaturation | 95 ºC | 1 min. | |
| Annealing | 63 ºC | 1 min. | |
| Extension | 72 ºC | 1 min. | |
| Final Extension | 72 ºC | 10 min. | |
Determination of PCR Specificity. The PCR products were analyzed and the result confirmed by using 1.5 % agarose gel electrophoresis, by dissolving 1.5 gm.
Isolation and Identification of Vancomycin-Resistant E. faecalis
Twenty isolates of the genus Enterococcus were isolated from 135 clinical specimens have the ability to grow on Pfizer selective Enterococcus. The highest numbers of isolates were distributed among urine specimens and the lowest one was observed among wound infection specimens, VITEK were employed to confirm the presence of E. faecalis isolates, regarding samples’ type and isolated Enterococcus spp., there were a 20 isolates identified, they were demonstrated in table 7.
Table (6):
Program was processed in Multiplex PCR amplification of vanA and vanB genes
| Stage | Temperature | Time | |
|---|---|---|---|
| Initial denaturation | 94 ºC | 5 min | 30 Cycles |
| Denaturation | 94 ºC | 1 min | |
| Annealing | 54 ºC | 1 min | |
| Extension | 72 ºC | 1 min | |
| Final Extension | 72 ºC | 10 min | |
Table (7):
Numbers and percentages of E. faecalis isolates from clinical specimens
Sample type |
No. of samples |
E. faecalis |
% of E. faecalis from each source |
|---|---|---|---|
Urine |
15 |
7 |
46.6% |
Blood |
60 |
5 |
8.3% |
Root canal |
50 |
7 |
14% |
Burns |
10 |
1 |
10% |
Total |
135 |
20 |
78.9% |
E. faecalis isolates from urine in this study was 46.6%, Alebouyeh et al. [19] showed that the percentage of E. faecalis isolates from urine was 75%.
For blood specimens, the percentage of E. faecalis isolates were 8.3%, this result was compatible with the results by Tellis and Muralidharan [2]; they showed that percentage of E. faecalis isolated from blood was 18%. While other studies reported by Al-Jarousha et al. [21], AL-khafaji et al. [22] and Mira et al. [23] revealed that the percentage of E. faecalis isolated from blood were 3%, 0% and 55.05%, respectively.
The percentage of E. faecalis isolated from root canal specimens was 14%, this result was higher than the result of local study obtained by Mahmoudpour et al. [24], who showed the percentage of E. faecalis isolated from root canal was 10%, another studiy by Zoletti et al. [25] and Preethee et al. [26], showed that the percentage of E. faecalis isolated from root canal was 80% and 46.87%, respectively. The percentage of E. faecalis isolated from wounds was 10%, this result was compatible with the result of Giacometti et al. [27] and Al-Jarousha et al. [21], who indicated that the percentage of E. faecalis isolated from wounds was 5.6%, 1.9%, respectively.
The differences between isolation percentages may be related to the number of specimens, the differences in the source of isolates, hospitals included in each study, their geographical regions and differences in the identification methods.
Determination of Vancomycin susceptibility
Vancomycin susceptibility was determined by the minimum inhibitory concentration [MIC] for all E. faecalis isolates, according to Clinical and Laboratory Standards Institute [CLSI], if the MIC d” 4 ¼g / ml then the isolate is sensitive, MIC 8–16 ¼g/mL the isolate have intermediate resistance and if the MIC e” 32 ¼g/ml the isolate is resistant to vancomycin.
The MICs result of vancomycin for E. faecalis isolates were indicted in the table 8.
Table (8):
The Minimum Inhibitory Concentrations [MICs] of Vancomycin for E. faecalis isolates
Id |
isolates |
specimen |
MIC[μg/ml] |
susceptibility |
|---|---|---|---|---|
1 |
1U |
urine |
32 |
R |
2 |
2U |
urine |
4 |
S |
3 |
3U |
urine |
64 |
R |
4 |
4U |
urine |
32 |
R |
5 |
5U |
urine |
64 |
R |
6 |
6U |
urine |
64 |
R |
7 |
7U |
urine |
32 |
R |
8 |
3B |
Blood |
64 |
R |
9 |
4B |
Blood |
4 |
S |
10 |
6B |
Blood |
16 |
IR |
11 |
7B |
Blood |
128 |
R |
12 |
8B |
Blood |
16 |
IR |
13 |
1w.i |
Wound |
4 |
S |
14 |
1R |
Root canal |
16 |
IR |
15 |
2R |
Root canal |
32 |
R |
16 |
3R |
Root canal |
64 |
R |
17 |
4R |
Root canal |
128 |
R |
18 |
5R |
Root canal |
32 |
R |
19 |
6R |
Root canal |
64 |
R |
20 |
7R |
Root canal |
16 |
IR |
S: Sensitive, IR: Intermediate Resistant, R: Resistant.
Results of Vancomycin sensitivity test obtained by this study showed that from 20 isolates, 13 isolates [65%] were resistant to Vancomycin, 4 isolates [20%] were intermediate resistant and 3 isolates [15%] were sensitive.
In a study reported by Fatholahzadeh et al. [28] stated that 38% of E. facalis isolates were resistant to vancomycin. While Camargo et al. [29] demostrated that 20.8% of E. faecalis isolates were resistant to vancomycin and 79.1% of isolates were sensitive. The results of this study was close to the results of Chabuck et al. [30], Al-jmor [20] and Praharaj et al.[31], who showed that the percentages of vancomycin resistant were 71.43%, 50% and 90.6%, respectively.
The multidrug resistance (MDR) bacteria was defined as resistance to 3 or more types of antibiotics (Al-Jarousha et al. 2008), the emergence of multidrug-resistant (MDR) pathogens seriously threatens this classes of lifesaving drugs (Queenan and Bush, 2007), therefore; This study was showed the percentage of multidrug resistant E. faecalis isolates was 90% as shown in table (9). This result was adjacent to the results reported by Al-Jarousha et al. (2008) and Chabuck et al. (2011) were showed the percentage of MDR is 66.6% and 100%, respectively.
Table (9):
Multi-drug resistant pattern of E. faecalis isolates
Number of Antibiotic types were resistant by isolates |
Number of isolates |
Percentage of isolates |
|---|---|---|
2 or less |
2 |
10% |
3 |
3 |
15% MDR |
4 |
2 |
10% MDR |
5 or more |
13 |
65% MDR |
The unique E. faecalis (isolated from blood) was resistant to (Penicillin G, Ceftazidime, Gentamycin, Ciprofloxacin, Rifampin, Amoxiclav and Imipenem), therefore; this isolate was considered as Extended-resistant E. faecalis (Diekema et al., 1999; Queenan and Bush, 2007; Papp-Wallace et al., 2011).
Resistance to multiple classes of antibiotics is common in Enterococci as had seen in this study, this finding was alarming as infection due to MDR Enterococcal isolates are difficult to treat. Enterococci showed a great resistance to most of the commonly used antibiotics in hospitals where development of antibiotic resistance is often related to the overuse, and misuse of the antibiotics prescribed (Saifi et al., 2008).
Iraq is one of the developing countries where all types of antibiotics are sold over the counter, an attitude that encourages self-medication.
From this study results, it can be conclude that the resistance to multiple classes of antibiotics is common to E. faecalis, these findings were alarming the infections due the MDR and extended -resistant E. facalis isolates and difficult to treat (Al-Jmor , 2012).
Detection the Virulence factors of E. faecalis
Hemolysin production was detecting by culturing the isolates on blood agar base supplemented with 5% blood, and hemolytic activity was observed as clear zone (²- hemolysis) surrounding the bacterial colonies. All the bacterial isolates showed ²-hemolysis (Figure 1). This result was similar to the result reported by Al-Jmor (2012) that showed 100% of E. faecalis isolates have the ability to produce hemolysin. Jankoska et al. (2008) reported that 50% of isolates produce hemolysin. In local studies reported by AL-Khafaji et al. (2010) and Mousa (2012) that showed the percentage of hemolysin production by E. faecalis isolates was 15.1%, 40%. Tellis and Muralidharan (2012) showed the percentage of E. faecalis isolates which have ability to produce hemolysin is 44%.
Lipase production was detected on egg-yolk agar plates, the hydrolyzed clear zone around the colonies were considered a positive result. The results of this study showed that 25% of the E. faecalis isolates were lipase producer (Figure 1).
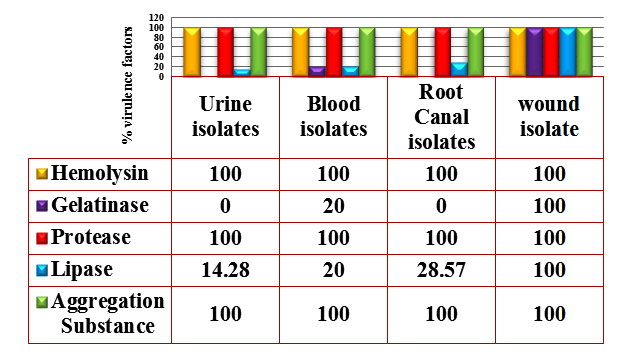
Fig. 1. Percentages of Virulence factors produced by E. faecalis isolates
AL-Khafaji et al. (2010) showed that the percentage of lipase producer was 3%; while Al-Jmor (2012) has reported 2.94% of isolates were lipase producer, whereas; the result of this study was similar to the findings of Elsner et al. (2000), Dworniczek et al. (2005) were 35% and 33%, respectively.
The biological role of lipase in infection might be considered the most important step in bacterial infection, the most prominent role of microorganism extracellular lipases may be the digestion of host cellular lipids for nutrient acquisition, which results in sticking to host tissue and neighboring cell (Furumura et al., 2006).
Protease production was detected on skim milk agar plates; the hydrolyzed clear zones around the colonies were indicating a positive result. The results showed 100% of isolates were protease producers (Figure 1). This result was close to Furumura et al. (2006) who demonstrated 75% of E. faecalis isolates was protease producer. Al-Jmor (2012) was reported a result similar to this study, showing that 100% of E. faecalis isolates was protease producing.
Gelatinase is extracellular metalloprotease secreted by E. faecalis, hydrolyzes gelatin, collagen, and casein, and has been implicated as a virulence factor in animal models. The ability of this enzyme to hydrolyze collagens and certain bioactive peptides suggests its participation in the initiation and propagation of inflammatory processes involving E. faecalis (Furumura et al., 2006).
Gelatinase production was detected on nutrient gelatin, liquefaction of gelatin after placed nutrient gelatin in refrigerator considered a positive result.
The results of this study showed that 10% of E. faecalis isolates were gelatinase producer (Figure 1). This result was in agreement to the result of Al-Jmor (2012) that represented the percentage of gelatinase producer was 11.76%, whereas Vergis et al. (2002) showed that 64% of E. faecalis isolated from patients with bacteremia, produced gelatinase, while Jankoska et al. (2008) was reported that 68% of E. faecalis isolates were gelatinase producer.
Aggregation substance (AS) of E. faecalis is a pheromone inducible surface protein encoded by pheromone plasmids that facilitates conjugative exchange, by mediating strong binding between cells and formation of cell aggregates (clumps). AS also contributes to pathogenicity by enhancing cell adhesion and internalization as well as favoring intracellular survival within macrophages (Glimore, 2002).
Aggregation substance was detected on trypticase soy broth, the presence of adherent dense cell growth in the bottom of the tube was considered a positive result (Figure 2).
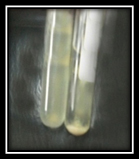
Fig. 2. Cell aggregation of E. faecalis strains in Trypticase soy broth (A) Negative AS, (B) Positive AS
The results showed that all tested isolates (100%) have produced a slime layer by this method but in different levels of adherent layer which ranged from weak to moderate and strong (Figure 3). The result of this study was similar with the result reported by Al-Jmor (2012), the result of this study was higher than the result reported by Necidova et al., (2009) and Oli et al., (2012) who found that 28% and 85% of E. faecalis isolates were produce slime layer, respectively.
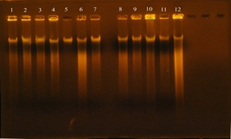
Fig. 3. DNA bands extracted from 20 E. faecalis isolates
The results in figure (1) , showed that all E. faecalis isolates (100%) were AS producer. The result of this study was similar to Paoletti et al. (2007) was showed that 100% of E. faecalis isolates were AS producer, While Tomita and Ike (2008) were reported (33%) of E. faecalis isolates have the ability to produce AS. Alebouyeh et al. (2005) was showed 23.3% of E. faecalis isolates were AS producer.
A positive correlation between the presence of Esp and the ability of an Enterococcal strain to form biofilms in vitro has been reported. None of the esp-deficient isolates tested in that study were capable of forming biofilms. However, it was also observed that insertional inactivation of esp did not cause a loss of the biofilm phenotype in every mutant tested (Toledo-Arana et al., 2001).
The involvement of Enterococcal surface protein in biofilm formation in the presence of a higher glucose concentration has been reported by Tendolkar et al., (2004). Two E. faecalis esp-positive strains and produce significantly more bio volume and thickness of biofilm than their controls, esp-negative strains. In the same study, the presence of 0.5% glucose in the growth medium influenced the biofilm production by E. faecalis (Tendolkar et al., 2006).
Di Rosa et al., (2006) have explained that neither esp seemed to be required for biofilm formation: both E. faecalis and did not show a correlation between the presence of esp and biofilm formation. The presence of esp and biofilm together was only found in strains from clinical settings, suggesting that there exists a synergy between this factor which serves as an advantage for the process of infection.
DNA and plasmid Extraction from E. faecalis isolates
The concentration results of DNA were ranged between 100-1800 ng/ml, while the purity of Genomic DNA was ranged between 1.8-1.9 in wavelength 260/280. This indicates the method was efficient in extraction and purification of DNA as shown in figure 4. The purpose of DNA extraction is for detection the virulence factor Enterococcal surface protein esp gene.
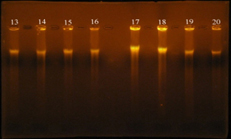
Fig. 4. DNA bands extracted from 20 E. faecalis isolates
Gel electrophoresis: agarose (0.8%), TBE buffer (1x), 70 volt for 1 hr., stained with ethidium bromide. 1: DNA extracted from the isolate 3B; 2: DNA extracted from the isolate 7B; 3: DNA extracted from the isolate 6U; 4: DNA extracted from the isolate 6B; 5: DNA extracted from the isolate 4B; 6: DNA extracted from the isolate 1R; 7: DNA extracted from the isolate 8B; 8: DNA extracted from the isolate 5R; 9: DNA extracted from the isolate 1U; 10: DNA extracted from the isolate 7U; 11: DNA extracted from the isolate 1W.i; 12: DNA extracted from the isolate 2U; 13: DNA extracted from the isolate 4R; 14: DNA extracted from the isolate 4U; 15: DNA extracted from the isolate 3U; 16: DNA extracted from the isolate 2R; 17: DNA extracted from the isolate 7R; 18: DNA extracted from the isolate 5U; 19: DNA extracted from the isolate 6R; 20: DNA extracted from the isolate 3R.
The plasmid DNA was extracted from 13 isolates of E. faecalis using Accuprep ® PCR plasmid mini extraction kit, the isolates were (1w.i) isolate from wound infection was sensitive to vancomycin (MIC 4µg/ml), (1R) isolate from root canal was intermediate resistant to vancomycin (MIC 16 µg/ml), (1U, 7U, 5R) isolates from Urine and Root canal were resistant to vancomycin (MIC 32 µg/ml), (3B, 5U, 3U, 6U, 3R, 6R) isolates from Blood, Urine and Root canal, respectively (MIC 64 µg/ml) and (4R , 7B) isolated from root canal and blood (MIC 128 µg/ml). The result of agarose gel electrophoresis (figure 5), showed that all thirteen isolates have one large (Mega) plasmid, this result was similar to study reported by Kandela (2006) showed that E. faecalis isolates contain one large (Mega) plasmid. Flannagan et al. (2003) were showed that Vancomycin-resistant E. faecalis was found to contain two plasmids, designated pAM830 (45 kb) and pAM831 (95 kb). The pMG1-like plasmids were include the pHT plasmids (pHT² (65.9 kbp), pHT² (63.7 kbp), and pHT² (66.5 kbp) are highly conjugative pheromone-independent (Tomita and Ike, 2005).
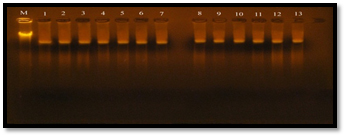
Fig. 5. Plasmid DNA bands extracted from 13 E. faecalis isolates
Enterococcal plasmids have been classified into 3 groups by a mixture of these methods; the Inc 18 group of plasmids (rep group 1, represented by pIP501 and partially rep family 2, represented by pRE25), the rolling circle replication plasmids (mostly rep family 4 and 6, represented by pMBB1 and pS86, respectively) and the pheromone responsive plasmids of E. faecalis (rep family 8 and 9, represented by pAM373 and pCF10) (Rosvoll, 2012).
The results obtained by Al-Jmor (2012) as result of plasmid extraction, the vancomycin resistant E. faecalis isolates have two large plasmids with molecular weight higher than (1 Kb) of DNA ladder, while the vancomycin sensitive isolates have one large plasmid.
vanA and vanB genes are carried on Tn- 1546 like transposon on the pHT² plasmid which have molecular weight(63.7 kbp) (Tomita and Ike, 2005, Tomita and Ike, 2008).
Gel electrophoresis: agarose (0.8%), TBE buffer (1x), 60 volt for 2 hr., stained with ethidium bromide. M: chromosomal DNA as ladder; 1: plasmid DNA extracted from the isolate 6R; 2: plasmid DNA extracted from the isolate 1R; 3: plasmid DNA extracted from the isolate 5R; 4: plasmid DNA extracted from the isolate 5U; 5: plasmid DNA extracted from the isolate 4R; 6: plasmid DNA extracted from the isolate 3U; 7: plasmid DNA extracted from the isolate 6U; 8: plasmid DNA extracted from the isolate 7U; 9: plasmid DNA extracted from the isolate 7B; 10: plasmid DNA extracted from the isolate 3B; 11: plasmid DNA extracted from the isolate 2U; 12: plasmid DNA extracted from the isolate 3R; 13: plasmid DNA extracted from the isolate 1w.i.
esp gene amplification by monoplex PCR technique
To detect the virulence factor Enterococcal surface protein esp gene in E. faecalis, Twenty E. faecalis isolates was subjected to PCR technique in a monoplex pattern.
The Positive result of esp gene was confirmed by 1.5% agarose gel electrophoresis stained with ethidium bromide, electrophoresed in 70 volt for 1.5 hrs and photographed under ultraviolet (UV) trans illuminator (figure 3-10). The results of present study showed that esp gene band detected at 933 bp region, all E. faecalis isolates have esp gene as virulence factor.
Figure (6) demonstrated that the twenty (100%) of E. faecalis isolates were obtained from clinical samples found it has esp gene, Shankar et al. (1999) was reported that 26.3% of E. faecalis isolates having the virulence factor ESP. Creti et al. (2004) and Jankoska et al. (2008) were reported that E. faecalis which have the esp gene was represented to 44.6% and 76%, respectively.
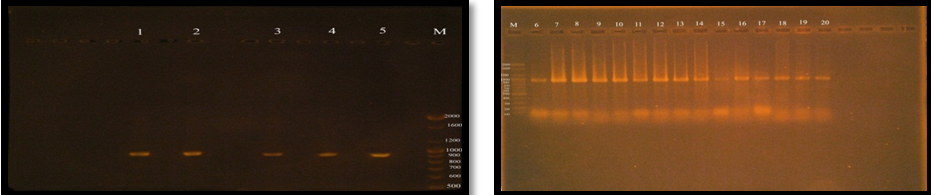
Fig. 6. Gel electrophoresis of amplified PCR product of virulence factor Enterococcal surface protein (esp gene) in monoplex pattern
The results of this study were similar to results reported by; Jankoska et al. (2008) Eaton and Gasson (2001), who showed that the molecular weight of PCR product (esp) bands in gel electrophoresis is 933bp.
The results in this study have stated all E. faecalis isolates were produced esp gene regardless of their susceptibility to vancomycin (resistant or sensitive).
Sharifi et al. (2012) and Comerlato et al. (2013) have stated there is no association between the virulence factor ESP and any other virulence factor or antibiotic resistance.
In this study all E. faecalis isolates which is esp producer have produced biofilm which was ranged between strong to moderate and weak. This explains a strong positive correlation between the presence of esp and biofilm formation, and E. faecalis biofilm productivity dependent on esp expression.
Chuang-Smith et al. (2010) speculated that Esp may mediate the interaction with primary surfaces and participate in the formation of this phenotype. In addition, Heinkens et al. (2007) showed that Esp is involved in biofilm formation.
There are diverging opinions concerning the role of Esp in biofilm production (Garth et al. 2008). Dworniczek et al. (2012) concluded that there is no clear relationship between the expression of esp or gelE and biofilm formation. Indeed, an analysis by Sillanpaa et al. (2010) showed efficient biofilm production in the absence of Esp.
Comerlato et al. (2013) was showed there is no association between the presence of esp and biofilm production, they were assumed that other factors are associated with this phenotype.
vanA, vanB Genes Amplification by Monoplex PCR Technique
The accurate and rapid diagnosis of antibiotic resistance genes in the treatment of E. faecalis infections is extremely important in preventing the spread of infections. PCR-based molecular methods are often preferred for determination of antibiotics resistant genes. Using PCR technique, the genetic determinants of vancomycin resistant vanA and vanB were amplified to identify susceptible (lacking vanA or vanB) and resistant (vanA or vanB) for 13 E. faecalis isolates.
The results of the present study showed that, vanA gene bands were detected at 550 bp regions, Eleven E. faecalis isolates (84.6%) were produced 550 bp band (Figure 7), Four E. faecalis isolates (30.7%) produced vanB gene bands were nearly detected at 600 bp region, (Figure 8).
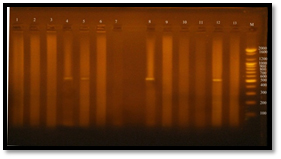
Fig. 7. Gel electrophoresis of amplified PCR product of vanA gene (550 bp) of E. faecalis isolates in monoplex pattern
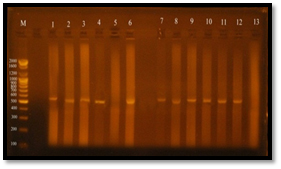
Fig. 8. Gel electrophoresis of amplified PCR product of vanB gene (nearly 600 bp) of E. faecalis isolates in monoplex pattern
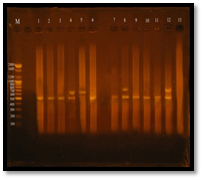
Fig. 9. Gel electrophoresis of amplified PCR product of vanA, vanB genes of E. faecalis isolates in multiplex pattern
Regarding vanA gene bands, the sensitive isolate (1w.i) did not have vanA gene, since it is specific marker for vancomycin resistance.
While the resistant isolates (6R, 1R, 5R, 5U, 3U, 6U, 7U, 7B, 3B, 1U, 3R) have shown vanA gene bands except the resistant isolate (4R) haven’t vanA band (it showed vanB genotype), the molecular weight of vanA bands was 550 bp.
vanA resistance is usually inducible by sub inhibitory concentrations of vancomycin and teicoplanin and it associated with production of a 38-40 kD membrane protein called VanA and encoded by vanA gene (Nicas et al., 1989).
The deduced amino acid sequence of vanA indicates that it is related to the bacterial D-Ala:D-Ala ligases, which mediate the D-alanyl-D-alanine (D-Ala-D-Ala) linkages of the growing cell wall peptidoglycan (Malen et al., 1990). VanA appears to be a D-Ala:D-X ligase of relaxed substrate specificity that can condense D-alanine with other amino acids, fatty acids, or hydroxy acids (Arthur et al., 1992). In resistant isolates, D-Ala-D-Ala is replaced with D-Ala-D-lactate (D-Lac), which cannot bind glycopeptides, The production of D-Ala-D-Lac depends on the cooperative activity of three enzymes, VanA, VanH, and VanX (Arthur et al., 1991). In vancomycin-resistant E. faecalis, the genes encoding these proteins are located on a plasmid and arranged sequentially within an operon. Induction of resistance is mediated by the transcription activator vanR and the membrane sensor vanS, located upstream of the other members of the gene cluster (Arthur et al., 1992; French, 1998).
The results showed by Satak et al. (1997); Depardieu et al. (2004); Fatholahzadeh et al. (2006); Biendo et al. (2010); Tellis and Muralidharan, (2012), they were detected vanA gene in E. faecalis isolates and the molecular weight was 732 bp. While Clark et al. (1993); Kariyama et al. (2000) were reported the molecular weight of vanA from PCR product was 1030 bp. Miele et al. (1995); Patel et al. (1997); Lim et al. (2006); were reported that molecular weight of vanA gene from PCR product was 1029bp, 885 bp, 1114 bp, respectively. These results indicate that vanA gene have variation in molecular weight.
The results vanA in this study revealed that the gene have a genetic variation as showed in their molecular weight during electrophoresis of vancomycin resistant E. faecalis strains.
Figure 3-12shows that the molecular weight of vanB gene bands were near 600 bp region, Vancomycin resistant E. facalis strains and their MIC were (5U and 3R=64 µg/ml, 4R=128 µg/ml and 7U=32 µg/ml).
The vanB operon contains genes encoding a dehydrogenase, a ligase, and a dipeptidase, all of which have a high level of sequence identity (67%–76% identity) with the corresponding deduced proteins of the vanA operon, the function of the additional vanW protein found only in the vanB cluster is unknown (Patel et al., 1998; Dahl et al., 1999)
The regulatory system in class B strains appears insensitive to induction by teicoplanin. Teicoplanin induces the synthesis of VanA-related proteins but does not induce the production of VanB-related proteins. On the other hand, vancomycin induces the synthesis of the resistance proteins of both systems, and in fact, if a teicoplanin-susceptible Enterococcus with the vanB gene cluster is preexposed to vancomycin, the strain then tests teicoplanin resistant as well (Cetinkaya et al., 2000).
Regarding to genetic variation, the results of this study showed that vanB gene bands have molecular weight closely to 600 bp, while the results reported by Satak et al. (1997); Depardieu et al. (2004); Biendo et al. (2010); Tellis and Muralidharan, (2012), they were detected vanB gene in E. faecalis isolates and the molecular weight was 635 bp. While Clark et al. (1993); Kariyama et al. (2000) were reported the molecular weight of vanA gene from PCR product was 433 bp. Miele et al. (1995); Patel et al. (1997) were reported that molecular weight of vanA gene from PCR product was 457bp, 885 bp, respectively.
vanA and vanB genes carried on Tn-1546 transposon that it is ubiquity, this gene could move (transpose) from one plasmid to other in the same or different bacterial strain, this explained by the conjugation experiment. The complete genome sequence of E. faecalis, a vancomycin-resistant clinical isolate, revealed that more than a quarter of the genome consists of probable mobile or foreign DNA. One of the predicted mobile elements is a previously unknown vanB vancomycin-resistance conjugative transposon (Paulsen et al., 2003).
In addition to transcriptional and translational regulatory mechanisms, many bacteria have evolved additional mechanisms to enable them to respond to changing environments. In these bacteria a genetically divers population is generated by reversible genetic changes known as phase variation, therefore; these isolates to survive in their environment, it must gain a new characters that enable them for the survivor, they are gained these characters with missing some of unimportant sequences from genes that still activated in absence these sequences (Dale and park, 2003).
Detection of vanA, vanB genes using Multiplex PCR Technique
The multiplex PCR assay in our hands is a convenient and rapid method for determining glycopeptide resistance genotypes for Enterococcus spp. in the clinical microbiology laboratory. The assay provides a more specific and rapid alternative to classical phenotypic methods for the detection of low level glycopeptide resistance (MIC range, 4 to 8 mg/ml) (Patel et al. 1997).
Figure 3-13 demonstrated that Twelve (95%) E. faecalis isolates out of thirteen isolates were obtained from clinical samples found to be vancomycin resistant isolates, Eleven E. facalis isolates (84.6%) have vanA gene bands were detected at 550bp regions, Four E. faecalis isolates (30.7%) have vanB gene bands were detected nearly at 600bp region. Three of E. faecalis isolates (5U, 7U and 3R) were having vanA-vanB cluster.
All E. faecalis isolates were showing vanA or vanB genotype in PCR assay, no other genotype were showed because of only the extracted pHT² plasmids were amplified in PCR for detection vanA or vanB genes or both, the other van genes are chromosomally.
The results of MIC showed a perfect similarity with the results of PCR, this reflect the efficiency of MIC method. Vancomycin-sensitive E. faecalis isolates were have MIC of vancomycin 4 µg/ml and they were havn’t any van genes in gel electrophoresis of multiplex PCR product, While the Intermediate-resistant E. faecalis isolates showed vanA genotype and its MIC was 16 µg/ml (Table 3-6). The resistant E. faecalis isolates have shown identical results between MICs and PCR which was varied in its van genes, these isolates showed vanA, vanB or vanAvanB cluster genotypes. While it’s MIC was e”32.
Table (3-6):
Comparison between the MIC values of E. faecalis ioslates and their van genotype
| Id | isolates | MIC | Susceptibility | van |
|---|---|---|---|---|
| (µg/ml) | genotype | |||
| 1 | 1U | 32 | R | vanA |
| 3 | 3U | 64 | R | vanA |
| 5 | 5U | 64 | R | vanA, vanB |
| 6 | 6U | 64 | R | vanA |
| 7 | 7U | 32 | R | vanA, vanB |
| 8 | 3B | 64 | R | vanA |
| 11 | 7B | 128 | R | vanA |
| 13 | 1w.i | 4 | S | ÜÜÜÜÜÜ |
| 14 | 1R | 16 | IR | vanA |
| 16 | 3R | 64 | R | vanA, vanB |
| 17 | 4R | 128 | R | vanB |
| 18 | 5R | 32 | R | vanA |
| 19 | 6R | 64 | R | vanA |
Resistance to glycopeptides is disseminating rapidly and has recently spread to methicillin-resistant S. aureus (Sievert et al. 2002). Rapid and accurate methods are thus essential for the detection of such clinical isolates and the prevention of their transmission. In addition, MIC determination is time-consuming and does not detect GRE with low-level glycopeptide resistance and misidentification of E. faecalis can occur with commercial systems (Willey et al, 1999; Depardieu et al. 2004).One of the limitations of the method proposed, or of other similar assays, could be the sequence variability among van genes that has occasionally been observed (Perichon et al. 1997).
- AL-Jmor, S. A. Detaction of some virulence factors of Vancomycin – resistant Enterococcus faecalis and the effect of Punica granatum and Thuja orientalis extracts on it. M.Sc. Thesis College of Science. Baghdad University, 2012.
- AL-Khafaji, J. K. T., Samaan, S. F. and AL-Saeed, M. S. Virulence factors of Enterococcus faecalis. Med. J. Babylon, 2010; 7(4 -3):579-583.
- Arthur, M., Molinas, C., Bugg T.D.H., Wright G.D., Walsh C.T., and Courvalin, P. Evidence for in vivo incorporation of D-lactate into peptidoglycan precursors of vancomycin-resistant Enterococci. 1992; 36(8):67–69.
- Arthur, M., Molinas, C., Dutka-Malen, S. and Courvalin, P. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxy- carboxylic acid dehydrogenases. Gene, 1991; 103:133–134.
- Biendo, M., Adjid´e C., Castelain, S., Belmekki, M., Rousseau, F., Slama, M., Ganry, O., Schmit, J. L. and Eb, F. Molecular Characterization of Glycopeptide-Resistant Enterococci from Hospitals of the Picardy Region (France). Hindawi Publ. Co. Int. J. Microbiol., 2010; P: 1-8.
- Camargo, I. L. B. C., Zanella, R. C., Gilmore, M. S. and Darini, A. L. C. Virulence factors in vacomycin-resistant and vancomycin- susceptible Enterococcus faecalis from Brazil. Brazilian J. Microbiol., 2008; 39:273-278.
- Cetinkaya, Y., Falk P. and Mayhall C. G. Vancomycin-Resistant Enterococci. Clin. Microbiol. Rev., 2000; 13(4): 686–707.
- Chabuck, Z. A., Al-Charrakh, A. H. and Al-Sa’adi, M. A. K. Prevalence of Vancomycin Resistant Enterococci in Hilla City, Iraq. Med. J. Babylon, 2011; 8(3): 326-340.
- Chuang-Smith, O. N., Wells, C.L., Henry, S. M. J. and Dunny, G.M. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. Plose One, 2010; 5: e15798.
- Clark, N. C., Cooksey, R. C., Hill, B. C., Swenson, J. M. and Tenover, F. C. Characterization of glycopeptide-resistant Enterococci fromU.S. Hospitals. Antimicrob. Agents Chemother., 1993; 37: 2311–2317.
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for disk susceptibility tests, 21th ed. Approved standard M100-21. National Committee for Clinical Laboratory Standards, 2011; Wayne, Pa.
- Comerlato, C. B., Carvalho de Resende, M. C., Caierao, J. and d’Azevedo,P. A. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem. Inst. Oswaldo Cruz., 2013; 108(5): 590-595
- Creti, R., Imperi, M., Bertuccini, L., Fabretti, F., Orefici,G., Di Rosa, R. and Baldassarri L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med.Microbiol., 2004; 53: 13–20.
- Dahl, K. H., Simonsen, G. S., Olsvik, Ø. and Sundsfjord, A. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant Enterococci. Antimicrob. Agents Chemother., 1999; 43: 1105–1110.
- Dahle´ n, G., Blomqvist, S., Almsta hl, A. and Carle´ n A. Virulence factors and antibiotic susceptibility in Enterococci isolated from oral mucosal and deep infections. J. Oral Microbiol., 2012; 4: 10855-10862.
- Dale, J. W. and Park, S. F. John Wiley &Sons, Ltd. Molecular Genetics of Bacteria. 4th ed, West Sussex, England.
- Depardieu, F., Perichon, B. and Courvalin, P. Detection of the van alphabet and identification of Enterococci and Staphy- lococci at the species level by multiplex PCR. J. Clin. Microbiol., 2004; 42: 5857 – 5860.
- Di Rosa, R., Creti, R., Venditti, M., D’Amelio, R., Arciola, C. R., Montanaro; L. and Baldassarri, L. Relationship between biofilm formation, the Enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett., 2006; 256: 145–150.
- Diekema, D. J., Coffman, S. L., Marshall, S. A., Beach, M. L., Rolston K. V. I. and Jones, R. N.. Comparison of activities of broad-spectrum ²-lactam compounds against 1,128 gram-positive cocci recently isolated in cancer treatment centers. Antimicrob. Agents Chemother. 1999; 43(4): 940–943.
- Dutka-Malen, S., Evers, S. and Courvalin, P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant Enterococci by PCR. J. clin. Microbiol., 1995; 33(1): 24–27.
- Eaton, T. J. and Gasson, M. J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Env. Microbiol., 2001; 67(4): 1628–1635.
- Fatholahzadeh, B., Hashemi, F. B., Emaneini, M., Aligholi, M., Nakhjavani. F. A. and Kazem, B. Detection of vancomycin resistant Enterococci (VRE) isolated from urinary tract infections (UTI) in Tehran, Iran. DARU., 2006; 14(3): 141-145.
- Fernandes, S.C. and Dhanashree, B. Drug Resistance & virulence determinants in clinical isolates of Enterococcus Species. Indian J. Med. Res., 2013; 137: 981-985.
- French, G. L. Enterococci and vancomycin resistance. Clin. Infect. Dis., 1998; 27(1): 75–83.
- Furumura, M. T., Figueiredo, P. M.S., Carbonell, G. V., Darini, A. L. da C., Yano, T. Virulence-associated characteristics of Entreococcus faecalis strains isolated from clinical sources. Brazilian J. Microbiol., 2006; 37:230-236.
- Garth, A. J., Swogger, E., Wolcott, R., deLancey Pulcini, E., Secor, P., Sestrich, J., Costerton, J. W. and Stewart, O. S. Biofilms in chronic wounds. Wound Rep. Regen, 2008; 16: 37-44.
- Giacometti, A. O., Cirioni, A. M., Schimizzi, M. S., Del Prete, F. Barchiesi, M. M., D’Errico, E., Petrelli and Scalise, G. Epidemiology and microbiology of surgical wound infections. J. Clin. Microbiol., 2000; 38(2): 918.
- Harley, J. P. and Prescott, H. Laboratory exercises in microbiology. 5th ed, The Mcgraw-Hill.French, 2002.
- Howden, B.P. et al. Genomic insights to control the emergence of vancomycin-resistant Enterococci. Microbiol., 2013; 4: 412–413.
- Jett, B. D. and Gilmore, M. S. The growth-inhibitory effect of the Enterococcus faecalis bacteriocin encoded by pAD1 extends to the oral Streptococci. J. Dent. Res., 1990; 69: 1640-1645.
- Jones, M. E., Draghi, D. C., Thornsberry, C., Karlowsky, J. A., Sahm, D. F. and Wenzel, R. P. Emerging resistance among bacterial pathogens in the intensive care unit – a European and North American Surveillance study (2000–2002). Ann. Clin. Microbiol. Antimicrob., 2004; 3: 1-14.
- Kariyama, R., Mitsuhata, R., Chow, J. W., Clewell, D. B. and Kumon, H. Simple and Reliable Multiplex PCR Assay for Surveillance Isolates of Vancomycin-Resistant Enterococci. J. Clin. Microbiol., 2000; 38(8): 3092-3095.
- Lim, S.K., Tanimoto, K., Tomita, H. and Ike, Y. Pheromone-responsive conjugative vancomycin resistance plasmids in Enterococcus faecalis isolates from humans and chicken feces. Appl. Env. Microbial., 2006; 72(10): 6544–6553.
- Mahmoudpour, A., Rahimi, S., Sina, M., Soroush, M. H., Shashisa, S. and Aminabadi, N. A. Isolation and identification of Enterococcus faecalis from necrotic root canals using multiplex PCR. J. Oral Sci., 2007; 49(3): 221-227.
- Mira, M. U., Deana, M., Zora, J., Vera, G., Biljana, M. and Biljana, R. Prevalence of different enterococcal species isolated from blood and their susceptibility to antimicrobial drugs in Vojvodina, Serbia, 2011-2013. African J. Microbiol. Res., 2014; 8(8): 819-824.
- Mousa, A. H. Detection the virulence factors of Enterococcus faecalis isolated from the root canals and study the effect of the purified enterocin in some of these isolates. Al-Mustansiriyah J. Sci., 2012; 23(1): 25-32.
- Mundy, L. M., Sahm, D. F. and Gilmore, M. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev., 2000; 13(4): 513–522.
- Mylonakis, E., and Calderwood, S. B. Infective endocarditis in adults. N. Engl. J. Med., 2001; 345: 1318–1330.
- Nicas, T. I., Wu, C. Y. E., Hobbs Jr, J.N., Preston, D. A. and Allen, N. E. Characterization of vancomycin resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother., 1989; 33(7): 1121-1124.
- Necidova, L., Janstova, B., Karpiskova, S., Cupakova, S., Duskova, M., and Karpiskova, R. Importance of Enterococcus spp. For forming a biofilm. Czech. J. Food Sci., 2009; 27: 354-356.
- Oli, A. K., Raju S., Rajeshwari N. S. and Kelmani C. R. Biofilm formation by multidrug resistant Enterococcus faecalis (MDEF) originated from clinical samples. J. Microbiol. Biotech. Res., 2012; 2(2): 284-288.
- Palmer, K. L. and Gilmore, M. S. Multidrug-resistant enterococci lack CRISPR-cas. Microbiol., 2010; 1(4): 1-10.
- Patel, R., Uhl, J. R., Kohner, P., Hopkins, M. K. and Cockerill, F. R. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 1997; 35(3):703-707.
- Patel, R. Clinical impact of vancomycin-resistant Enterococci. J. Antimicrob. Chemother., 2003; 51(3): 13-21.
- Papp-Wallace, K. M., Endimiani, A., Taracila, M. A. and Bonomo, R. A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother., 2011; 55(11): 4943–4960.
- Paoletti, C., Foglia, G., Princivalli, M.S., Magi, G., Guaglianone, E., Donelli, G., Pruzzo, C., Biovasco, F. and Facinelli, B. Co-transfer of vanA and aggregation substance genes from Enterococcus faecalis isolates in intra and inter- species matings. J. Antimicrob. Chemother., 2007; 59: 1005–1009.
- Paulsen, IT., Banerjei, L., Myers, G.S., Nelson, K.E., Seshadri, R., Read, T.D., Fouts, D.E., Eisen, J.A., Gill, S.R., Heidelberg, J.F., Tettelin, H., Dodson, R.J., Umayam, L., Brinkac, L., Beanan, M., Daugherty, S., DeBoy, R.T., Durkin, S., Kolonay, J., Madupu, R., Nelson, W., Vamathevan, J., Tran, B., Upton, J., Hansen, T., Shetty, J., Khouri, H., Utterback, T., Radune, D., Ketchum, K.A., Dougherty, B. A. and Fraser, C.M. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science, 2003; 299(5615): 2071-4.
- Perichon, B., Reynolds, P. and Courvalin, P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother., 1997; 41: 2016–2018.
- Périchon, B. and Courvalin, P. Glycopeptide resistance in Enterococci. P.229-240 In: Antimicrobial Drug Resistance. Mayers, D.L. (ed.), Humana Press, NewYork, 2009.
- Preethee, T., Kandaswamy, D. and Hannah, R. Molecular identification of an Enterococcus faecalis endocarditis antigen efaA in root canals of therapy-resistant endodontic infections. J. Conserv. Dent., 2012; 15(4): 319–322.
- Raad, I., Hachem, R. and Hanna, H. et al. Prospective, randomized study comparing quinupristin-dalfopristin with linezolid in the treatment of vancomycin-resistant Enterococcus faecium infections. J. Antimicrob. Chemother., 2004; 53: 646–649.
- Reynolds, P. E., Snaith, H. A., Maguire, A. J., Dutka-Malen, S. and Courvalin, P. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J., 1994; 301: 5-8.
- Rice, L. B. Emergence of vancomycin-resistant enterococci. Emerg.Infect. Dis., 2001; 7(2): 183-187.
- Rosvoll, T. C. S. Plasmids, resistance and hospital adaptation in enterococci an epidemiological approach. Ph.D Thesis Faculty of Health Sciences. University of Tromsø, 2012.
- Saifi, M., Pourshafie, M. R., Eshraghian, M. R. and Soltan Dallal M. M. Anti-microbial resistance of enterococci isolated from urinary tract infections in Iran. Iranian Biomedical. J., 2008; 12(3): 185-190.
- Sambrook, J., Fritsch, E.F. and Maniatis,T. Molecular cloning: a laboratory manual, vol. 3.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 1989.
- Satake, S., Clark, N., Rimiland, D., Nolte, F. S. and Tenover, F. C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J. Clin. Microbiol., 1997; 35(9): 2325–2330.
- Scagnelli, M., Pellizer, G. and de Lalla, F. et al. Epidemiological analysis of vancomycin-resistant Enterococci in a large tertiary-care hospital in Northern Italy. Eur. J. Clin. Microbiol. Infect. Dis., 2001; 20: 609–616.
- Schleifer, K. H. and Kilpper-Balz, R. Transfer of Streptococcus faecalis and Streptococcus faecium to the Genus Enterococcus norn. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol., 1984; 34(1): 31-34.
- Sedgley, C., Nage,l A., Dahlén, G., Reit, C. and Molander, A Real-time quantitative polymerase chain reaction and culture analyses of Enterococcus faecalis in root canals. J. Endod., 2006; 32: 173-177.
- Sharifi, Y., Hasani, A., Ghotaslou, R., Varshochi, M., Hasani, A., Aghazadeh, M. and Milani, M. Survey of Virulence Determinants among vancomycin resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens of hospitalized patients of north west of Iran. Open Microbiol. J., 2012; 6: 34-39.
- Sherman, J. M. The Streptococci. Bacteriol. Rev., 1937; 1: 3-97.
- Shlaes, D.M. and Gutmann, E. L. Synergistic killing of vancomycin resistant Enterococci of classes A, B and C by combinations of vancomycin, penicillin and gentamicin. Antimicrob. Agents Chemother., 1991; 35: 776-9.
- Shales, D. M., Etter, L. and Gutmann, L. Synergistic killing of vancomycin-resistant enterococci of classes A, B, and C by combinations of vancomycin, penicillin, and gentamicin. Antimicrob. Agents Chemother., 1991; 35(4): 776-779.
- Shankar, N., Baghdayan, A. S., Huycke, M. M. Lindahl, G. and Gilmore M. S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun., 1999; 67(1):193-200.
- Shankar, N., Lockatell, C. V., Baghdayan, A. S., Drachenberg, C., Gilmore, M. S. and Johnson, D. E. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun., 2001; 69(7):4366-4372.
- Sievert, D. M., Boulton, M. L., Stolman, G., Johnson, D., Stobierski, M. G., Downes, F. P., Somsel, P. A., Rudrik, J. T., Brown, W. J., Hafeez, W., Lund strom, T., Flanagan, E., Johnson, R., Mitchell, J. and Chang, S. Staphylococcus aureus resistant to vancomycin. – United States. Morb. Mortal. Wkly. Rep., 2002; 51: 565–567.
- Sifri, C. D., Mylonakis, E., Singh, K. V., Qin, X., Garsin, D. A., Murray, B. E., Ausubel, F. M. and Calderwood, S. B. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in caenorhabditis elegans and mice. Infect. Immun., 2002; 70: 5647–5650.
- Sillanpaa, J., Nallapareddy, S. R., Singh, K.V., Prakash, V.P., Fothergill, T., Ton-That, H. and Murray, B. E. Characterization of the ebpfm pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence., 2010; 1: 236-246.
- Slaughter, S., Hayden, M.K., Nathan, C., Hu, T.C., Rice, T.J., van-Voortis, J., Matusheka, M., Franklin, C. and Weinstein, R.A. A comparison of the universal use of gloves and gowns with that of glove use alone on the acquisition of vancomycin-resistant Enterococci in a medical intensive-care unit. Ann. Int. Med., 1996; 125: 448-456.
- Song, X., Sun, J., Mikalsen, T., Roberts, A. P. and Sundsfjord, A. Characterisation of the Plasmidome within Enterococcus faecalis Isolated from Marginal Periodontitis Patients in Norway. PLOS ONE, 2013; 8(4): 1-7.
- Stevens, M. P. and Edmond, M. B. Endocarditis Due to Vancomycin-Resistant Enterococci: Case Report and Review of the Literature. Clin. Infect. Dis., 2005; 41: 1134–1142.
- Sunde, P.T., Olsen, I., Debelian, G.J. and Tronstad, L. Microbiota of periapical lesions refractory to endodontic therapy. J. Endod., 2002; 28: 304-310.
- Teixeira, L. M. and Merquior, V. L. C. Enterococcus. P. 18-26. In: de Filippis, I.; McKee, M. L. Molecular typing in bacterial infections. Humana Press Inc, New York, 2013.
- Telkar, A. et al. Change in the prevalence and the antibiotic resistance of the enterococcal species isolated from blood culture. J. Clin. Diag. Res., 2012; 6: 405-407.
- Tellis, R. and Muralidharan, S. A prospective study of antibiotic resistance and virulence factors in Enterococci isolated from patients with end stage renal disease. Int. J. Biomed. Res., 2012; 3(03) 174 180.
- Tendolkar, P. M., Baghdayan, A. S., Gilmore, M. S. and Shankar, N. Enterococcal surface protein Esp enhances biofilm formation by Enterococcus faecalis. Infect. Immun., 2004; 72(10): 6032-6039.
- Tendolkar, P. M., Baghdayan, A. S. and Shankar, N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 2006; 188: 2063–2072.
- Tomita, H. and Ike, Y. Genetic analysis of transfer-related regions of the vancomycin resistance Enterococcus conjugative plasmid pHT_: identification of oriT and a putative relaxase gene. J. Bacteriol., 2005; 187(22): 7727–7737.
- Toledo-Arana, A., Valle, J., Solano, C., Arrizubieta, M. J., Cucarella , C., Lamata, M., Amorena, B., Leiva, J., Penades, J. R. and Lasa, I. The Enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol., 2001; 67: 4538–4545.
- Tomita, H. and Ike, Y. Genetic Analysis of the Enterococcus Vancomycin Resistance Conjugative Plasmid pHT_: Identification of the Region Involved in Cell Aggregation and traB, a Key Regulator Gene for Plasmid Transfer and Cell Aggregation. J. Bacteriol., 2008; 190(23): 7739–7753.
- Tsvetkova, K., Marvaud, J. C. and Lambert, T. Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J. Bacteriol., 2010; 192(3):702-713.
- Tyne, D. V., Martin, M. J. and Gilmore, M. S. Structure, function, and biology of the Enterococcus faecalis cytolysin toxins. 2013; 5: 895-911.
- Upadhyaya, P.M., Ravikumar, G. KL. and Umapathy, B.L. Review of virulence factors of Enterococcus: An emerging nosocomial pathogen. Indian J. Med. Microbiol., 2009; 27(4): 301-305.
- Uysal, H., Ciftci, G. and Ciftci, A. Determination of antigenic glycoproteins of Enterococcus faecalis strains distinguished with aggregation substance, gelatinase and cytolysine. Rev. Méd. Vét., 2013; 164(7): 374-381.
- Vanek, N. N., Simon, S. I., Jacques-Palaz, K., Mariscalco, M. M., Dunny, G. M., Rakita, R. M. Enterococcus faecalis aggregation substance promotes opsoninin dependent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immun. Med. Microbiol., 1999; 26: 49-60.
- Vergis, E. N., Shanker N., Joseph, W. C., Hayden, M. K. and Syndman D. R. et al. Association between the presence of Enterococcal virulence factors gelatinase, haemolysin and Enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin. Infect. Dis., 2002; 35:570-5.
- Vesic, D. and Kristich, C. J. MurAA is required for intrinsic cephalosporin resistance of Enterococcus faecalis. Antimicrob. Agents Chemother., 2012; 56(5):2443-2451.
- Vandepitte, J., Verhagen, J., Engbaek, K., Rohner, P., Piot, P. and Heuck, C. C. Basic laboratory procedure in clinical bacteriology. 2nd ed. World Health Organization, Geneva, 2003.
- Willey, B. M., Jones, R. N., McGeer, A., Witte, W., French, G., Roberts, R. B., Jenkins, S. G., Nadler, H. and Low, D. E. Practical approach to the identification of clinically relevant Enterococcus species. Diagn. Microbiol. Infect. Dis., 1999; 34:165–171.
- Zoletti, G. O., Siqueira, J. F. and Santos, K. R. N. Identification of Enterococcus faecalis in root-filled teeth with or without periradicular lesions by culture dependent and—independent approaches. J. Oral Endodontic., 2006; 32: 722-726.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


